Molecular Cloning and Functional Analysis of DXS and FPS Genes from Zanthoxylum bungeanum Maxim
Abstract
:1. Introduction
2. Materials and Methods
2.1. Plant Materials
2.2. RNA Extraction and cDNA Synthesis
2.3. Gene Cloning and Sequence Analysis
2.4. Recombinant Protein and Enzymatic Activity Assay
2.5. Transient Overexpression in Tobacco Leaves
2.6. Volatile Analysis by HS-SPME–GC-MS
2.7. Real-Time Quantitative PCR
2.8. Bioinformatics Analysis
3. Results
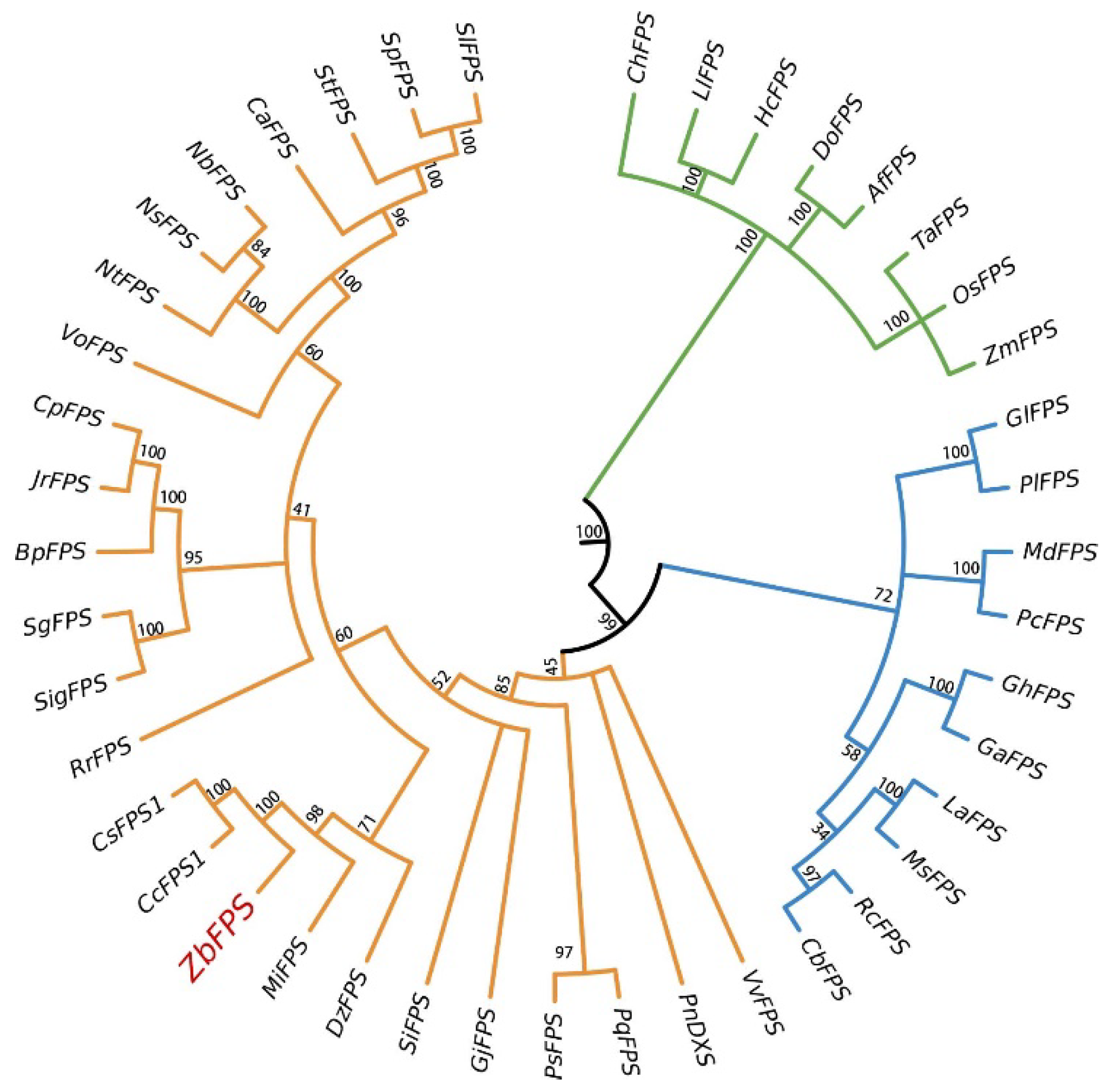
3.1. Molecular Cloning and Sequences Analysis
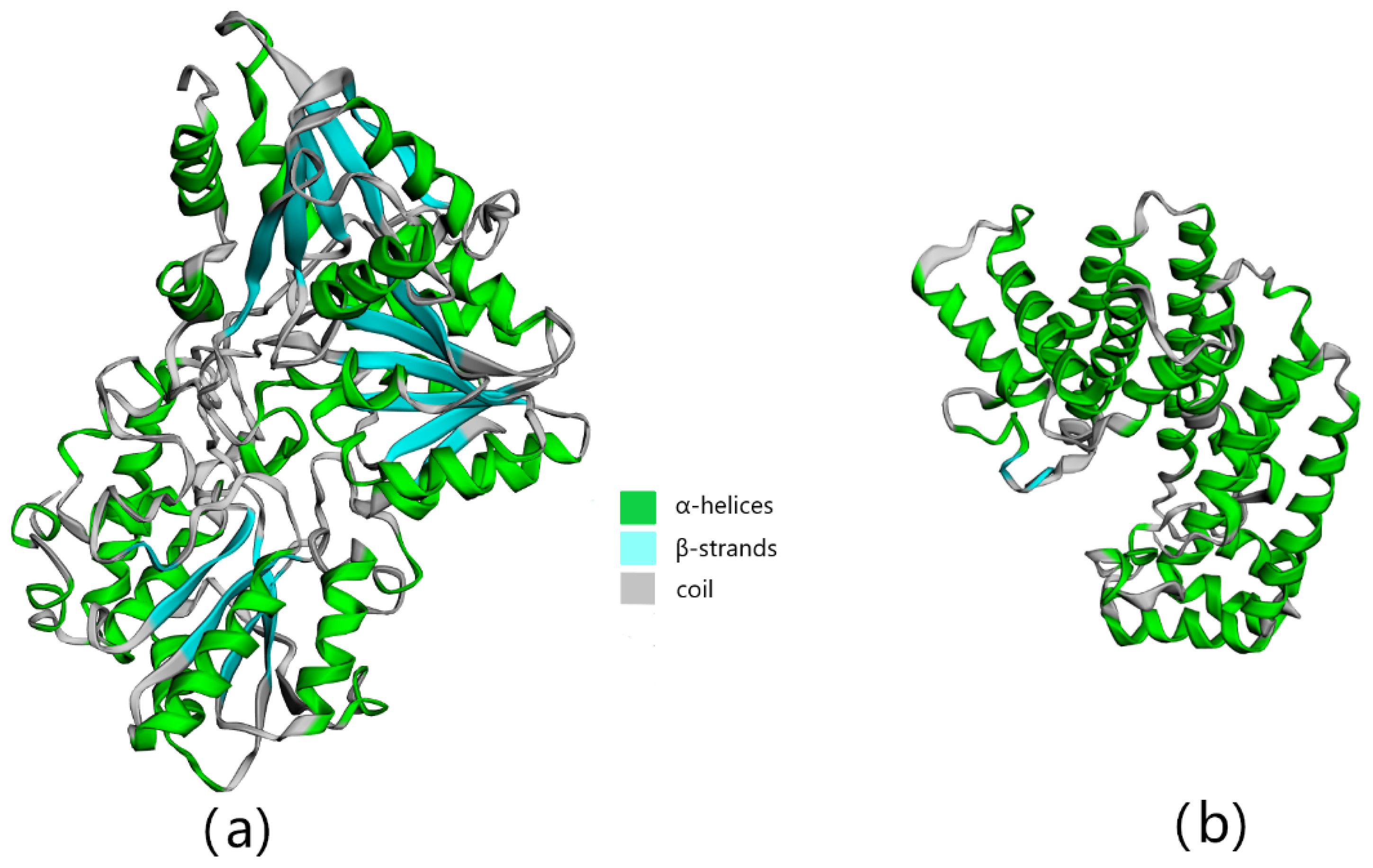
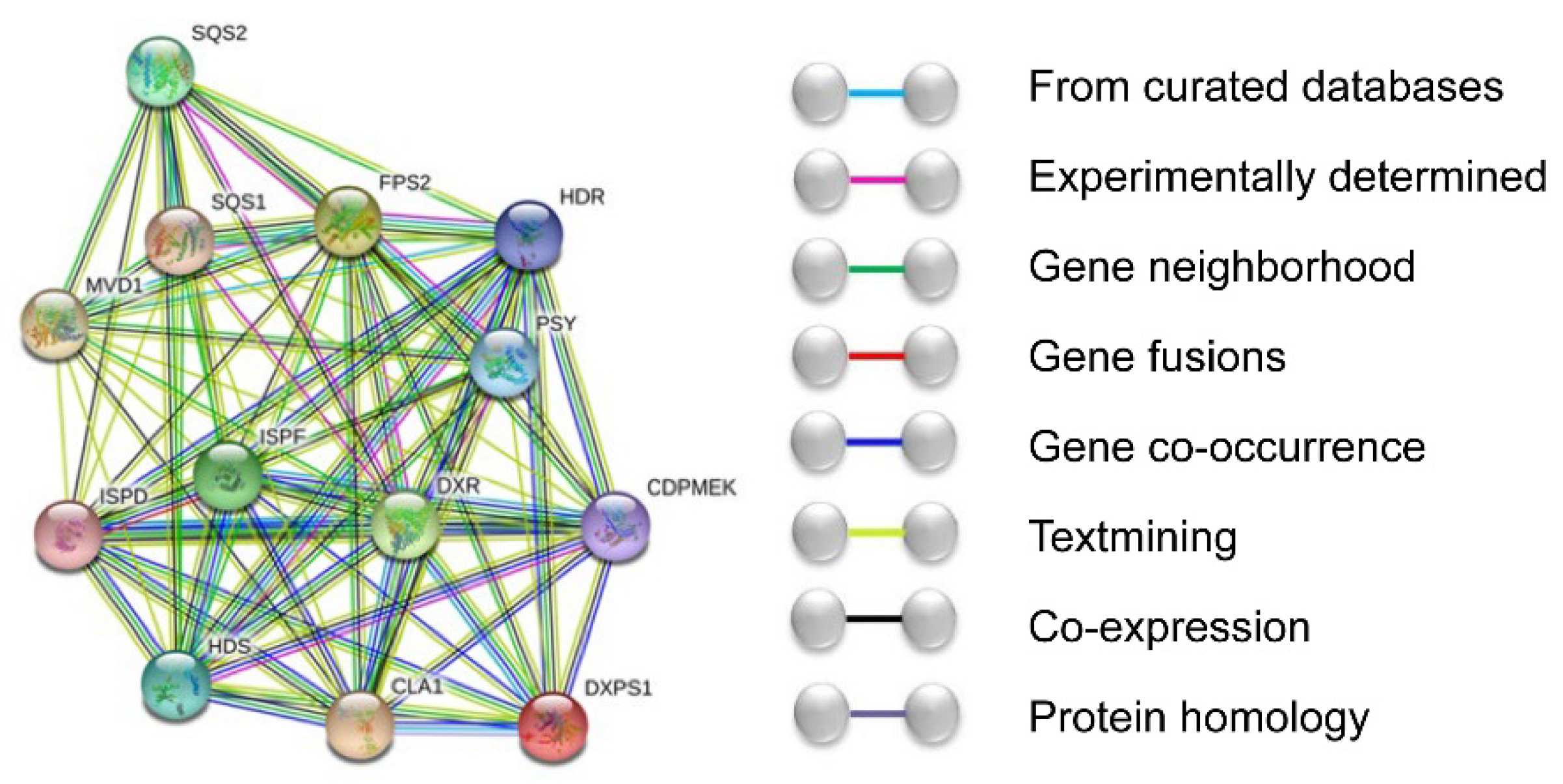
3.2. Bioinformatics Analysis
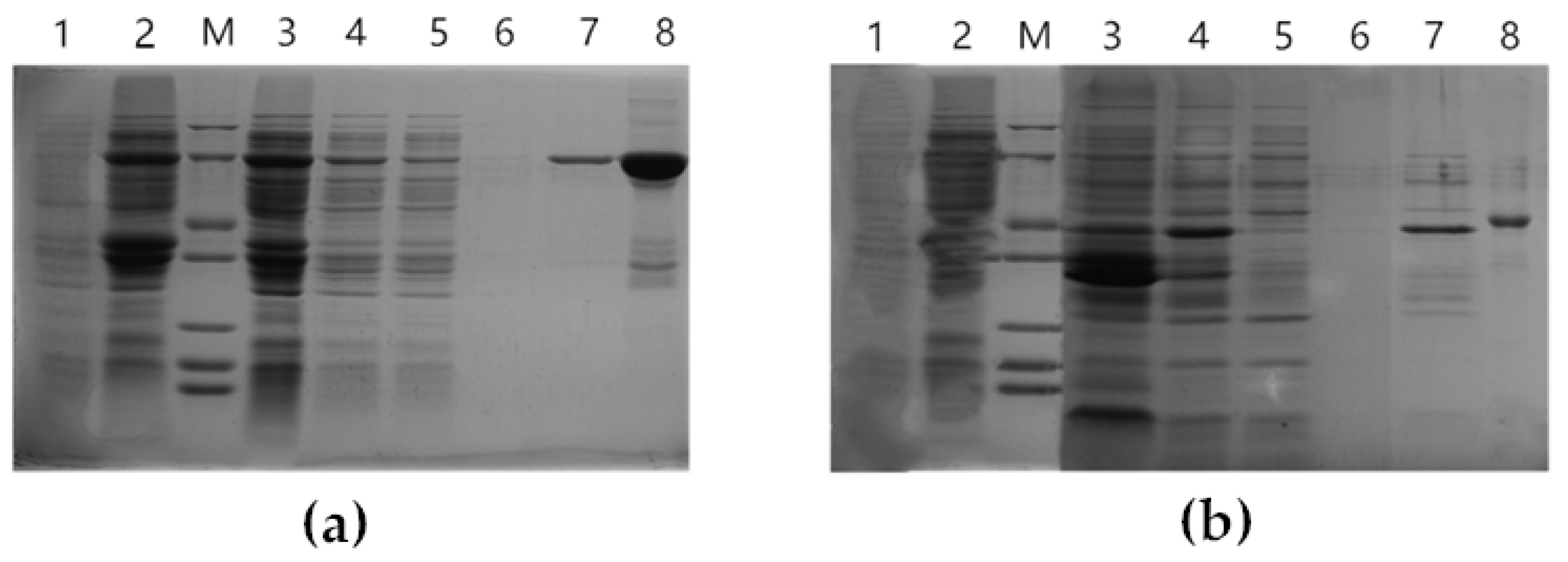
3.3. Protein Purification, Induction, and In Vitro Enzymatic Activity Testing
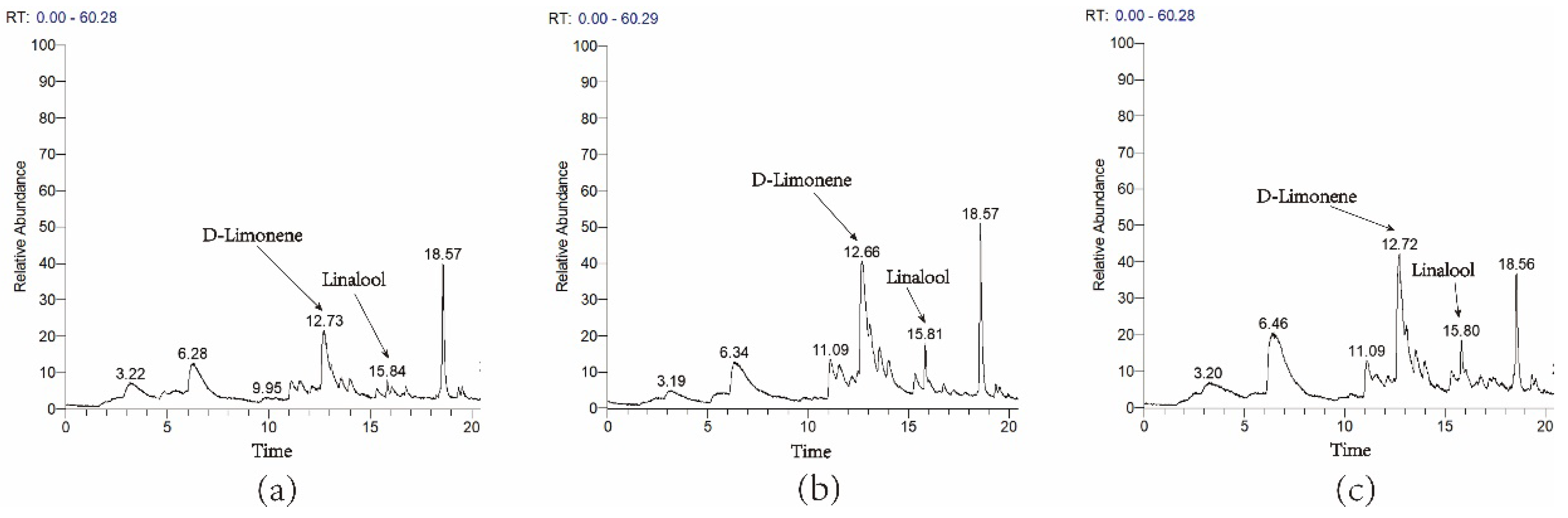
3.4. Transient Overexpression of ZbDXS and ZbFPS Increased Production of Terpene
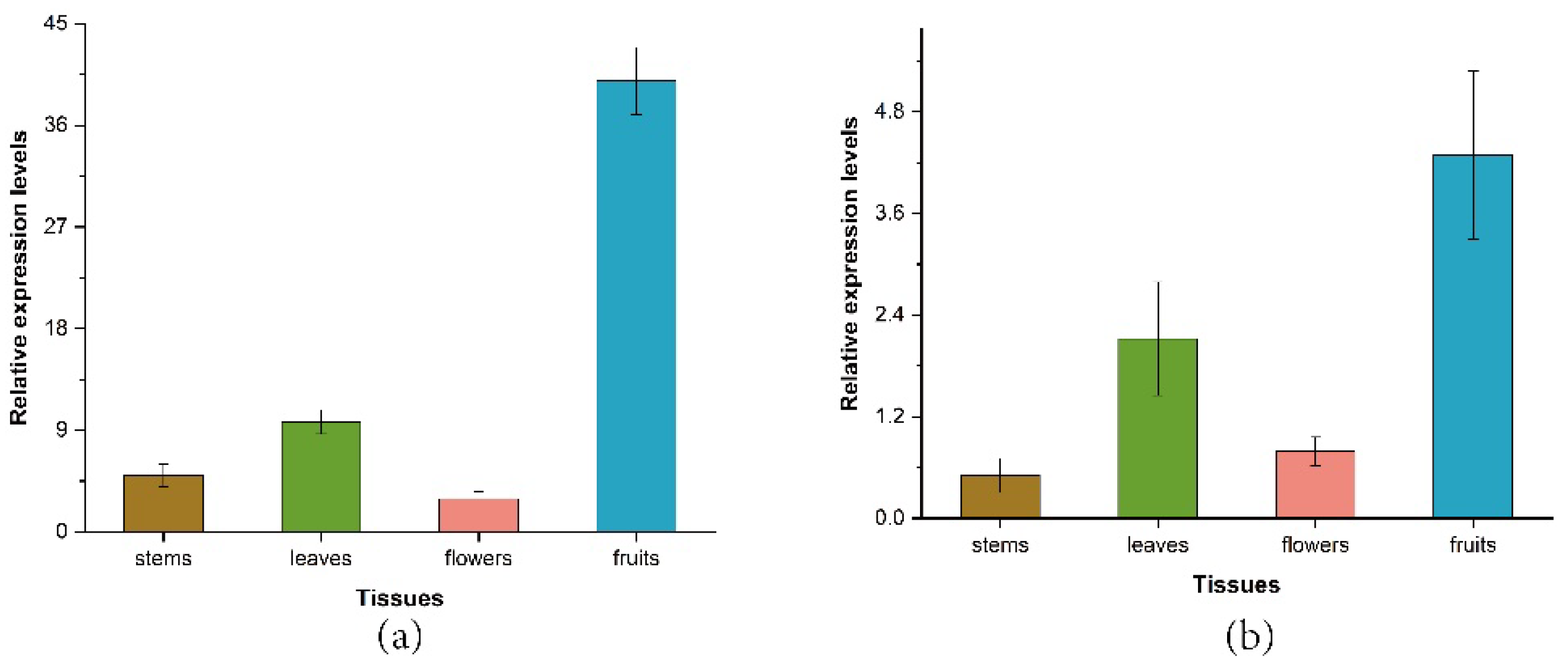
3.5. Expression Profile of ZbDXS and ZbFPS in Different Tissues
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Kutchan, T.M. Ecological arsenal and developmental dispatcher. The paradigm of secondary metabolism. Plant Physiol. 2001, 125, 58–60. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- McFrederick, Q.S.; Fuentes, J.D.; Roulston, T.a.; Kathilankal, J.C.; Lerdau, M. Effects of air pollution on biogenic volatiles and ecological interactions. Oecologia 2009, 160, 411–420. [Google Scholar] [CrossRef] [PubMed]
- Holopainen, J.K.; Gershenzon, J. Multiple stress factors and the emission of plant VOCs. Trends Plant Sci. 2010, 15, 176–184. [Google Scholar] [CrossRef] [PubMed]
- Dudareva, N.; Klempien, A.; Muhlemann, J.K.; Kaplan, I. Biosynthesis, function, and metabolic engineering of plant volatile organic compounds. New Phytol. 2013, 198, 16–32. [Google Scholar] [CrossRef]
- Chen, F.; Tholl, D.; Bohlmann, J.; Pichersky, E. The family of terpene synthases in plants: A mid-size family of genes for specialized metabolism that is highly diversified throughout the kingdom. Plant J. 2011, 66, 212–229. [Google Scholar] [CrossRef]
- Rodriguez-Concepcion, M.; Boronat, A. Elucidation of the methylerythritol phosphate pathway for isoprenoid biosynthesis in bacteria and plastids. A metabolic milestone achieved through genomics. Plant Physiol. 2002, 130, 1079–1089. [Google Scholar] [CrossRef] [Green Version]
- Suire, C.; Bouvier, F.; Backhaus, R.A.; Begu, D.; Bonneau, M.; Camara, B. Cellular localization of isoprenoid biosynthetic enzymes in Marchantia polymorpha. Uncovering a new role of oil bodies. Plant Physiol. 2000, 124, 971–978. [Google Scholar] [CrossRef] [Green Version]
- Chandler, S.; Tanaka, Y. Genetic modification in floriculture. Crit. Rev. Plant Sci. 2007, 26, 169–197. [Google Scholar] [CrossRef]
- Fujihashi, M.; Zhang, Y.W.; Higuchi, Y.; Li, X.Y.; Koyama, T.; Miki, K. Crystal structure of cis-prenyl chain elongating enzyme, undecaprenyl diphosphate synthase. Proc. Natl. Acad. Sci. USA 2001, 98, 4337–4342. [Google Scholar] [CrossRef] [Green Version]
- Lichtenthaler, H.K. The plants’ 1-deoxy-D-xylulose-5-phosphate pathway for biosynthesis of isoprenoids. Fett-Lipid 1998, 100, 128–138. [Google Scholar] [CrossRef]
- Carretero-Paulet, L.; Cairo, A.; Talavera, D.; Saura, A.; Imperial, S.; Rodriguez-Concepcion, M.; Campos, N.; Boronat, A. Functional and evolutionary analysis of DXL1, a non-essential gene encoding a 1-deoxy-D-xylulose 5-phosphate synthase-like protein in Arabidopsis thaliana. Gene 2013, 524, 40–53. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Walter, M.H.; Hans, J.; Strack, D. Two distantly related genes encoding 1-deoxy-D-xylulose 5-phosphate synthases: Differential regulation in shoots and apocarotenoid-accumulating mycorrhizal roots. Plant J. 2002, 31, 243–254. [Google Scholar] [CrossRef] [PubMed]
- Cordoba, E.; Porta, H.; Arroyo, A.; San Roman, C.; Medina, L.; Rodriguez-Concepcion, M.; Leon, P. Functional characterization of the three genes encoding 1-deoxy-D-xylulose 5-phosphate synthase in maize. J. Exp. Bot. 2011, 62, 2023–2038. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Xu, C.; Wei, H.; Movahedi, A.; Sun, W.; Ma, X.; Li, D.; Yin, T.; Zhuge, Q. Evaluation, characterization, expression profiling, and functional analysis of DXS and DXR genes of Populus trichocarpa. Plant Physiol. Bioch. 2019, 142, 94–105. Available online: https://www.mdpi.com/2227-9717/7/1/45 (accessed on 15 May 2022). [CrossRef] [PubMed]
- Jadaun, J.S.; Sangwan, N.S.; Narnoliya, L.K.; Singh, N.; Bansal, S.; Mishra, B.; Sangwan, R.S. Over-expression of DXS gene enhances terpenoidal secondary metabolite accumulation in rose-scented geranium and Withania somnifera: Active involvement of plastid isoprenogenic pathway in their biosynthesis. Physiol. Plant. 2017, 159, 381–400. [Google Scholar] [CrossRef] [PubMed]
- Tholl, D. Biosynthesis and Biological Functions of Terpenoids in Plants. Adv. Biochem. Eng. Biotechnol. 2015, 148, 63–106. [Google Scholar]
- Manzano, D.; Andrade, P.; Caudepon, D.; Altabella, T.; Arro, M.; Ferrer, A. Suppressing Farnesyl Diphosphate Synthase Alters Chloroplast Development and Triggers Sterol-Dependent Induction of Jasmonate-and Fe-Related Responses. Plant Physiol. 2016, 172, 93–117. [Google Scholar] [CrossRef]
- Peng, G.; Wang, C.; Song, S.; Fu, X.; Azam, M.; Grierson, D.; Xu, C. The role of 1-deoxy-D-xylulose-5-phosphate synthase and phytoene synthase gene family in citrus carotenoid accumulation. Plant Physiol. Biochem. 2013, 71, 67–76. [Google Scholar] [CrossRef]
- Adler, L.S.; Irwin, R.E. What you smell is more important than what you see? Natural selection on floral scent. New Phytol. 2012, 195, 510–511. [Google Scholar] [CrossRef]
- Fei, X.; Hou, L.; Shi, J.; Yang, T.; Liu, Y.; Wei, A. Patterns of Drought Response of 38 WRKY Transcription Factors of Zanthoxylum bungeanum Maxim. Int. J. Mol. Sci. 2019, 20, 68. [Google Scholar] [CrossRef] [Green Version]
- Lu, D.; Lo, V. Scent, and synaesthesia: The medical use of spice bags in early China. J. Ethnopharmacol. 2015, 167, 38–46. [Google Scholar] [CrossRef] [PubMed]
- Shi, J.; Fei, X.; Hu, Y.; Liu, Y.; Wei, A. Identification of Key Genes in the Synthesis Pathway of Volatile Terpenoids in Fruit of Zanthoxylum bungeanum Maxim. Forests 2019, 10, 328. [Google Scholar] [CrossRef] [Green Version]
- Tian, J.; Feng, S.; Liu, Y.; Zhao, L.; Tian, L.; Hu, Y.; Yang, T.; Wei, A. Single-Molecule Long-Read Sequencing of Zanthoxylum bungeanum Maxim. Transcriptome: Identification of Aroma-Related Genes. Forests 2018, 9, 765. [Google Scholar] [CrossRef] [Green Version]
- Salvatierra, A.; Pimentel, P.; Alejandra Moya-Leon, M.; Herrera, R. Increased accumulation of anthocyanins in Fragaria chiloensis fruits by transient suppression of FcMYB1 gene. Phytochemistry 2013, 90, 25–36. [Google Scholar] [CrossRef] [PubMed]
- Ma, Y.; Wang, Y.; Li, X.; Hou, L.X.; Wei, A.Z. Sensory Characteristics and Antioxidant Activity of Zanthoxylum bungeanum Maxim. Pericarps. Chem. Biodivers. 2019, 16, e1800238. [Google Scholar] [CrossRef] [PubMed]
- Fei, X.; Shi, Q.; Yang, T.; Fei, Z.; Wei, A. Expression Stabilities of Ten Candidate Reference Genes for RT-qPCR in Zanthoxylum bungeanum Maxim. Molecules 2018, 23, 802. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tamura, K.; Peterson, D.; Peterson, N.; Stecher, G.; Nei, M.; Kumar, S. MEGA5: Molecular Evolutionary Genetics Analysis Using Maximum Likelihood, Evolutionary Distance, and Maximum Parsimony Methods. Mol. Biol. Evol. 2011, 1851, 2731–2739. [Google Scholar] [CrossRef] [Green Version]
- Studer, G.; Tauriello, G.; Bienert, S.; Waterhouse, A.; Bertoni, M.; Bordoli, L.; Schwede, T.; Lepore, R. Modeling of Protein Tertiary and Quaternary Structures Based on Evolutionary Information. In Computational Methods in Protein Evolution; Sikosek, T., Ed.; Springer Science+Business Media, LLC: Berlin/Heidelberg, Germany, 2019; pp. 301–316. [Google Scholar] [CrossRef]
- Mewalal, R.; Rai, D.K.; Kainer, D.; Chen, F.; Kulheim, C.; Peter, G.F.; Tuskan, G.A. Plant-Derived Terpenes: A Feedstock for Specialty Biofuels. Trends Biotechnol. 2017, 35, 227–240. [Google Scholar] [CrossRef] [Green Version]
- Kirby, J.; Keasling, J.D. Biosynthesis of Plant Isoprenoids: Perspectives for Microbial Engineering. Annu. Rev. Plant Biol. 2009, 60, 335–355. [Google Scholar] [CrossRef]
- Wang, X.; Ort, D.R.; Yuan, J.S. Photosynthetic terpene hydrocarbon production for fuels and chemicals. Plant Biotechnol. J. 2015, 13, 137–146. [Google Scholar] [CrossRef] [Green Version]
- Dhar, M.K.; Koul, A.; Kaul, S. Farnesyl pyrophosphate synthase: A key enzyme in isoprenoid biosynthetic pathway and potential molecular target for drug development. New Biotechnol. 2013, 30, 114–123. [Google Scholar] [CrossRef] [PubMed]
- Qian, J.; Liu, Y.; Chao, N.; Ma, C.; Chen, Q.; Sun, J.; Wu, Y. Positive selection and functional divergence of farnesyl pyrophosphate synthase genes in plants. BMC Mol. Bio. 2017, 18, 3. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Grunler, J.; Ericsson, J.; Dallner, G. Branch-Point Reactions in the Biosynthesis of Cholesterol, Dolichol, Ubiquinone, and Prenylated Proteins. Biochim. Biophys. Acta 1994, 1212, 259–277. [Google Scholar] [CrossRef]
- Runquist, M.; Ericsson, J.; Thelin, A.; Chojnacki, T.; Dallner, G. Isoprenoid Biosynthesis in Rat-Liver Mitochondr in-Studies on Farnesyl Pyrophosphate Synthase and Trans-Prenyltransferase. J. Biol. Chem. 1994, 269, 5804–5809. [Google Scholar] [CrossRef]
- Cunillera, N.; Boronat, A.; Ferrer, A. The Arabidopsis thaliana FPS1 gene generates a novel mRNA that encodes a mitochondrial farnesyl-diphosphate synthase isoform. J. Biol. Chem. 1997, 272, 15381–15388. [Google Scholar] [CrossRef] [Green Version]
- Martin, D.; Piulachs, M.D.; Cunillera, N.; Ferrer, A.; Belles, X. Mitochondrial targeting of farnesyl diphosphate synthase is a widespread phenomenon in eukaryotes. Biochim. Biophys. Acta 2007, 1773, 419–426. [Google Scholar] [CrossRef] [Green Version]
- Zhang, Y.L.; Li, Z.X. Two different farnesyl diphosphate synthase genes exist in the genome of the green peach aphid, Myzus persicae. Genome 2008, 51, 501–510. [Google Scholar] [CrossRef]
- Gupta, S.D.; Mehan, R.S.; Tansey, T.R.; Chen, H.T.; Goping, G.; Goldberg, I.; Shechter, I. Differential binding of proteins to peroxisomes in rat hepatoma cells: Unique association of enzymes involved in isoprenoid metabolism. J. Lipid Res. 1999, 40, 1572–1584. [Google Scholar] [CrossRef]
- Ferella, M.; Li, Z.H.; Andersson, B.; Docampo, R. Farnesyl diphosphate synthase localizes to the cytoplasm of Trypanosoma cruzi and T-brucei. Exp. Parasitol. 2008, 119, 308–312. [Google Scholar] [CrossRef] [Green Version]
- Taban, A.H.; Tittiger, C.; Blomquist, G.J.; Welch, W.H. Isolation And Characterization of Farnesyl Diphosphate Synthase From the Cotton Boll Weevil, Anthonomus grandis. Arch. Insect Biochem. 2009, 71, 88–104. [Google Scholar] [CrossRef]
- Blas, A.L.; Ming, R.; Liu, Z.; Veatch, O.J.; Paull, R.E.; Moore, P.H.; Yu, Q. Cloning of the Papaya Chromoplast-Specific Lycopene beta-Cyclase, CpCYC-b, Controlling Fruit Flesh Color Reveals Conserved Microsynteny and a Recombination Hot Spot. Plant Physiol. 2010, 152, 2013–2022. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Su, S.S.; Zhang, H.M.; Liu, X.Y.; Pan, G.F.; Ling, S.P.; Zhang, X.S.; Yang, X.M.; Tai, Y.L.; Yuan, Y. Cloning and characterization of a farnesyl pyrophosphate synthase from Matricaria recutita L. and its upregulation by methyl jasmonate. Genet. Mol. Res. 2015, 14, 349–361. [Google Scholar] [CrossRef] [PubMed]
- Wu, C.T.; Lan, L.; Li, Y.; Nie, Z.Y.; Zeng, R.Z. The relationship between latex metabolism gene expression with rubber yield and related traits in Hevea brasiliensis. BMC Genom. 2018, 19, 897. [Google Scholar] [CrossRef] [PubMed] [Green Version]
| Gene Name | Substrates | Content(U/L) ± SD |
|---|---|---|
| ZbDXS | TMB | 0.29 ± 0.01 |
| ZbFPS | TMB | 0.73 ± 0.03 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Tian, L.; Shi, J.; Yang, L.; Wei, A. Molecular Cloning and Functional Analysis of DXS and FPS Genes from Zanthoxylum bungeanum Maxim. Foods 2022, 11, 1746. https://doi.org/10.3390/foods11121746
Tian L, Shi J, Yang L, Wei A. Molecular Cloning and Functional Analysis of DXS and FPS Genes from Zanthoxylum bungeanum Maxim. Foods. 2022; 11(12):1746. https://doi.org/10.3390/foods11121746
Chicago/Turabian StyleTian, Lu, Jingwei Shi, Lin Yang, and Anzhi Wei. 2022. "Molecular Cloning and Functional Analysis of DXS and FPS Genes from Zanthoxylum bungeanum Maxim" Foods 11, no. 12: 1746. https://doi.org/10.3390/foods11121746
APA StyleTian, L., Shi, J., Yang, L., & Wei, A. (2022). Molecular Cloning and Functional Analysis of DXS and FPS Genes from Zanthoxylum bungeanum Maxim. Foods, 11(12), 1746. https://doi.org/10.3390/foods11121746






