Sensitive Detection of Staphylococcus aureus by a Colorimetric Biosensor Based on Magnetic Separation and Rolling Circle Amplification
Abstract
:1. Introduction
2. Materials and Methods
2.1. Material and Chemicals
2.2. Apparatus
2.3. Bacterial Culture and Counting
2.4. The Procedure for Synthesizing Van-MNPs
2.5. Preparation of DNFs
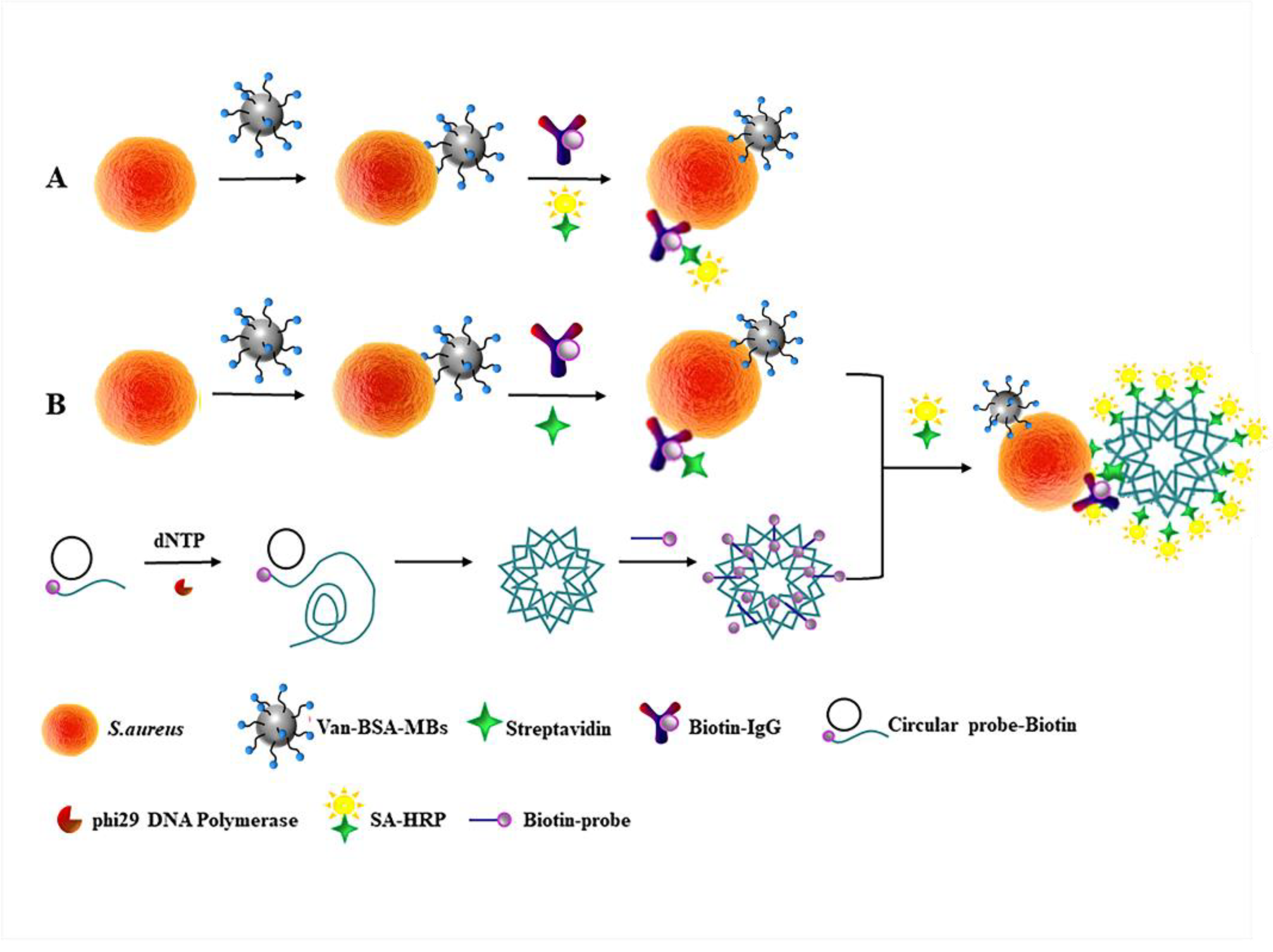
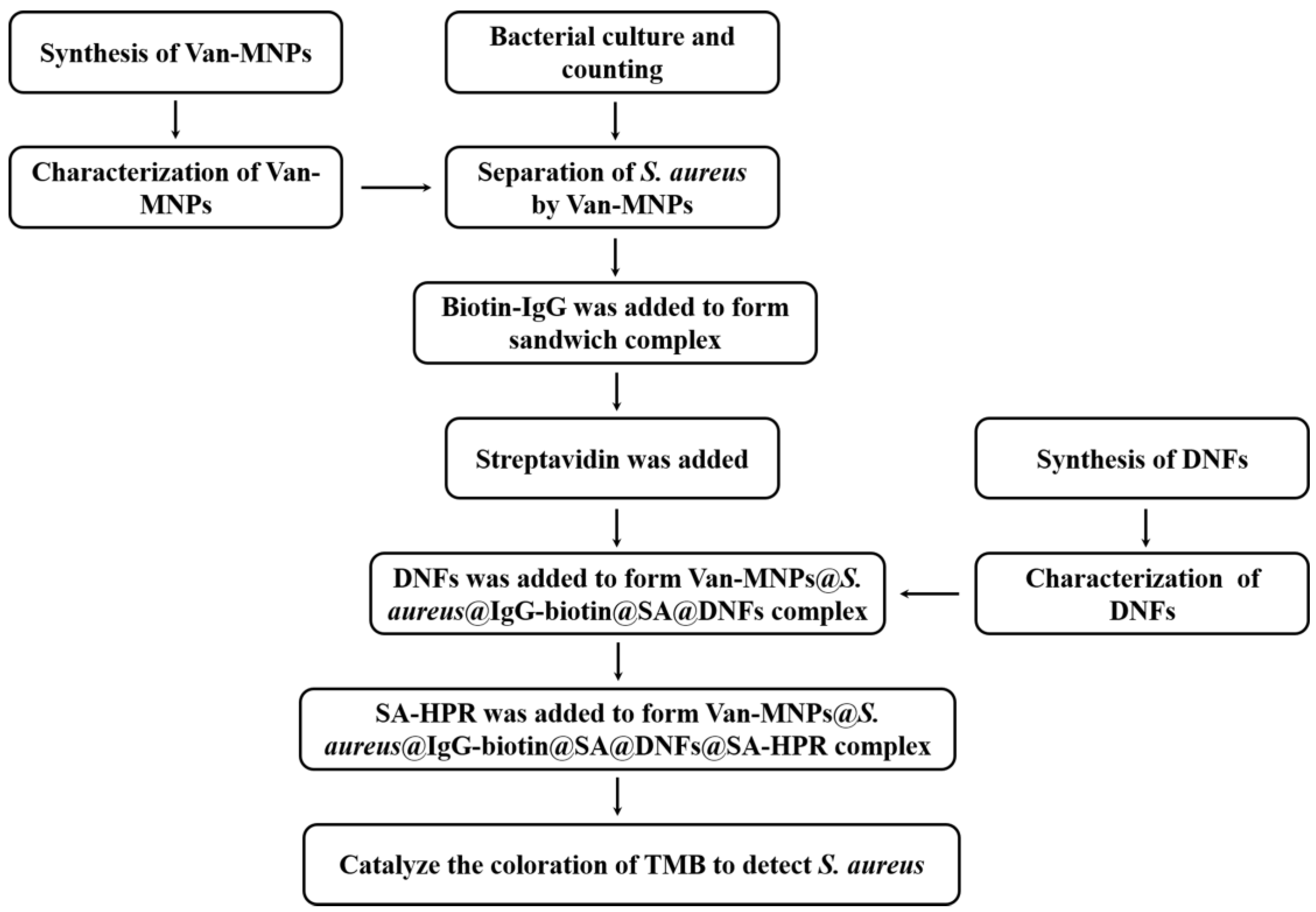
2.6. The Principle of the AMS-DNFs Strategy
2.7. Specificity Analysis
2.8. Detection of S. aureus in Juice Sample
2.9. Statistical Analysis
3. Results and Discussion
3.1. Strategy of AMS-DNFs for the Detection of S. aureus
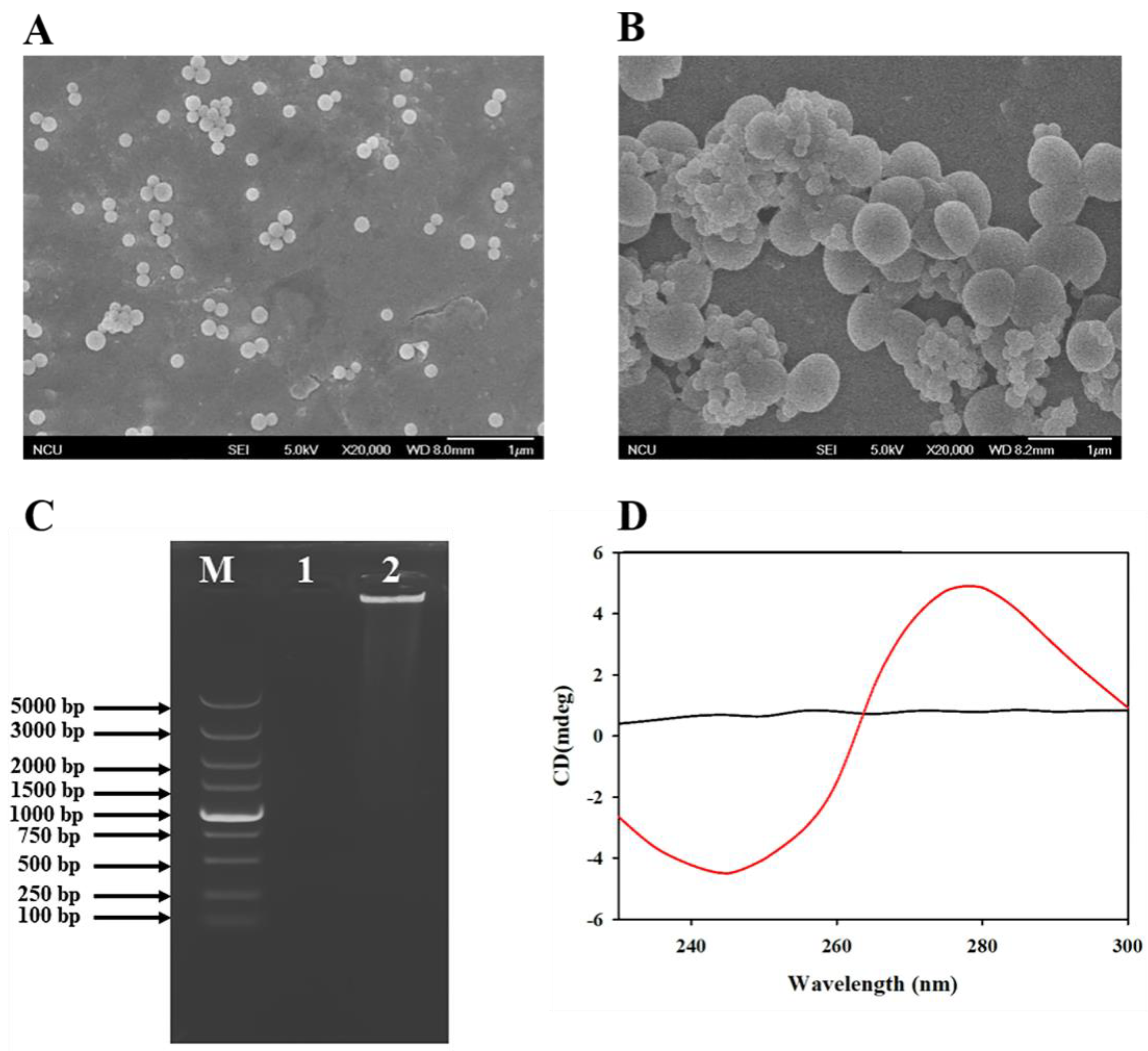
3.2. Characterization of Antibiotic-MBs and DNFs
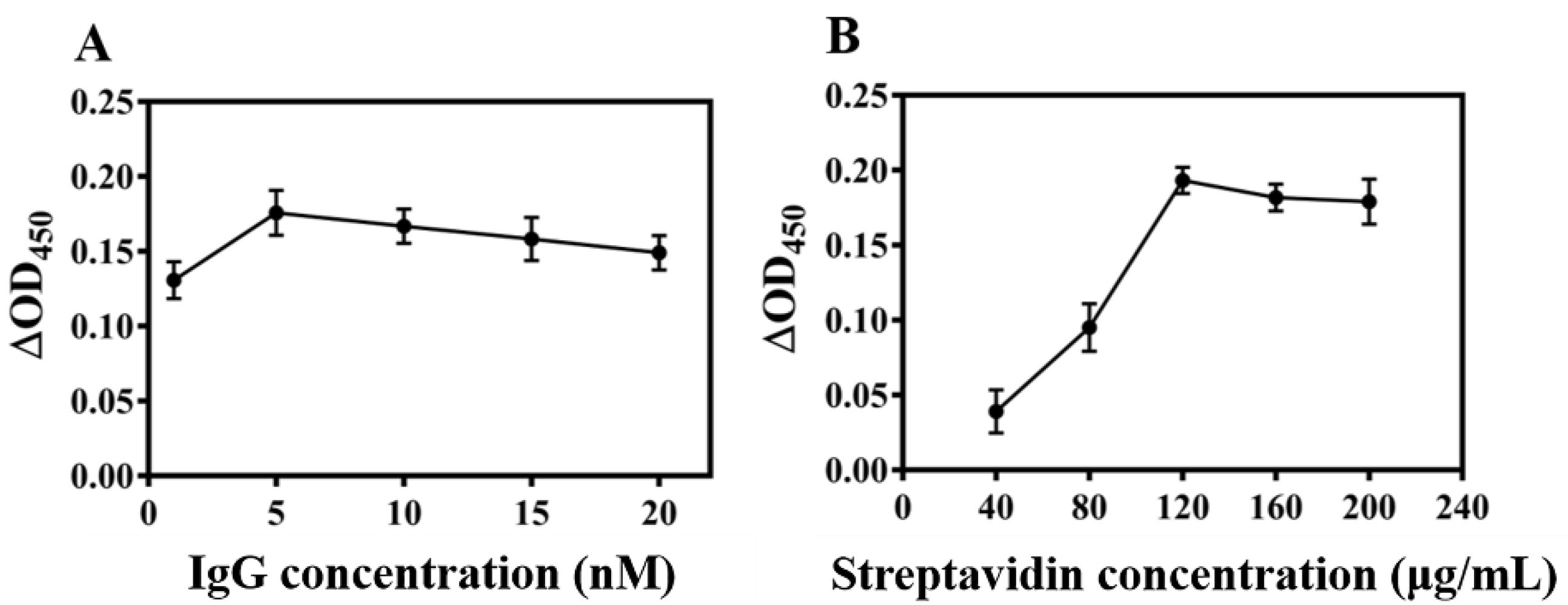
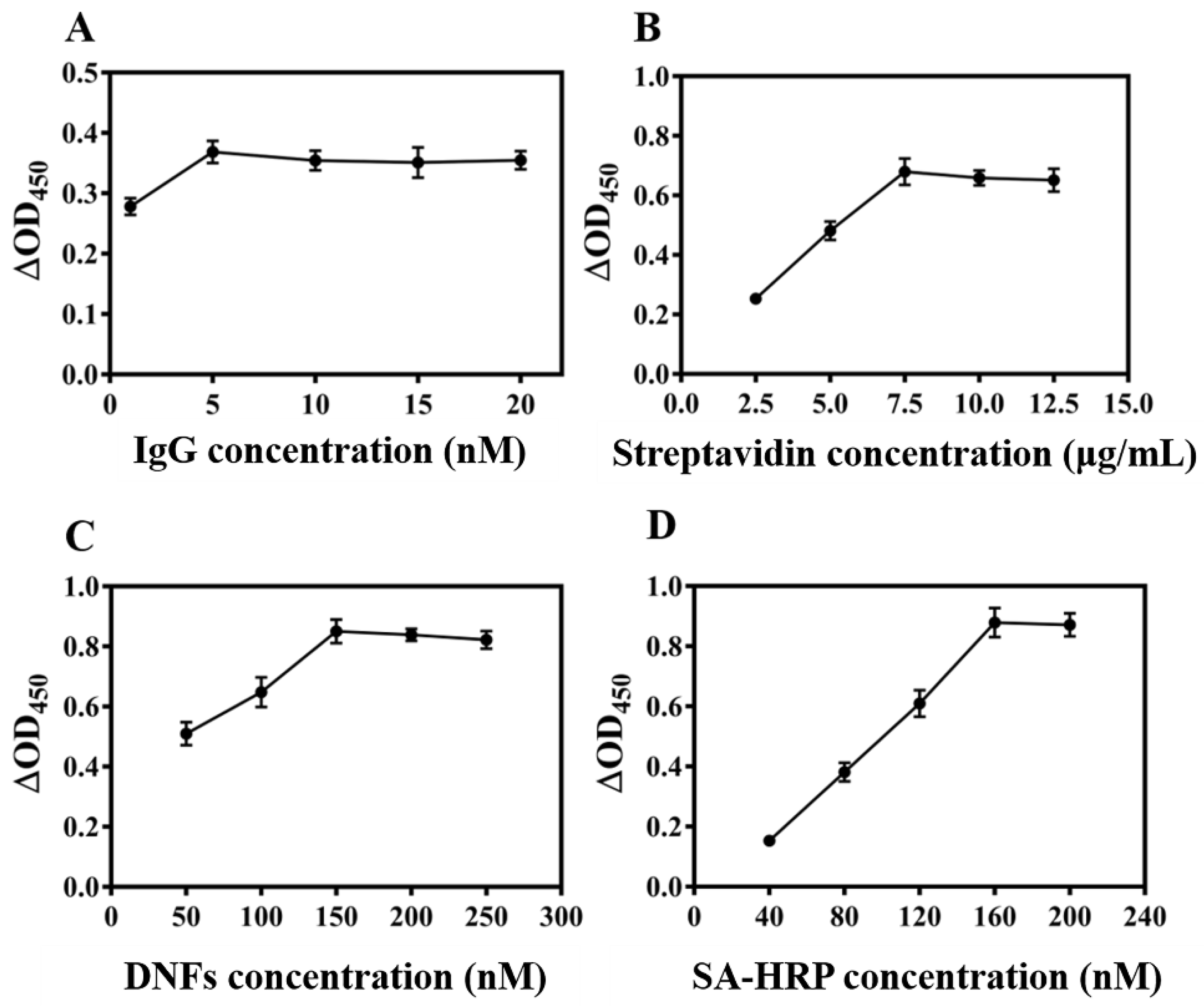
3.3. Optimization of Colorimetry Experimental Conditi
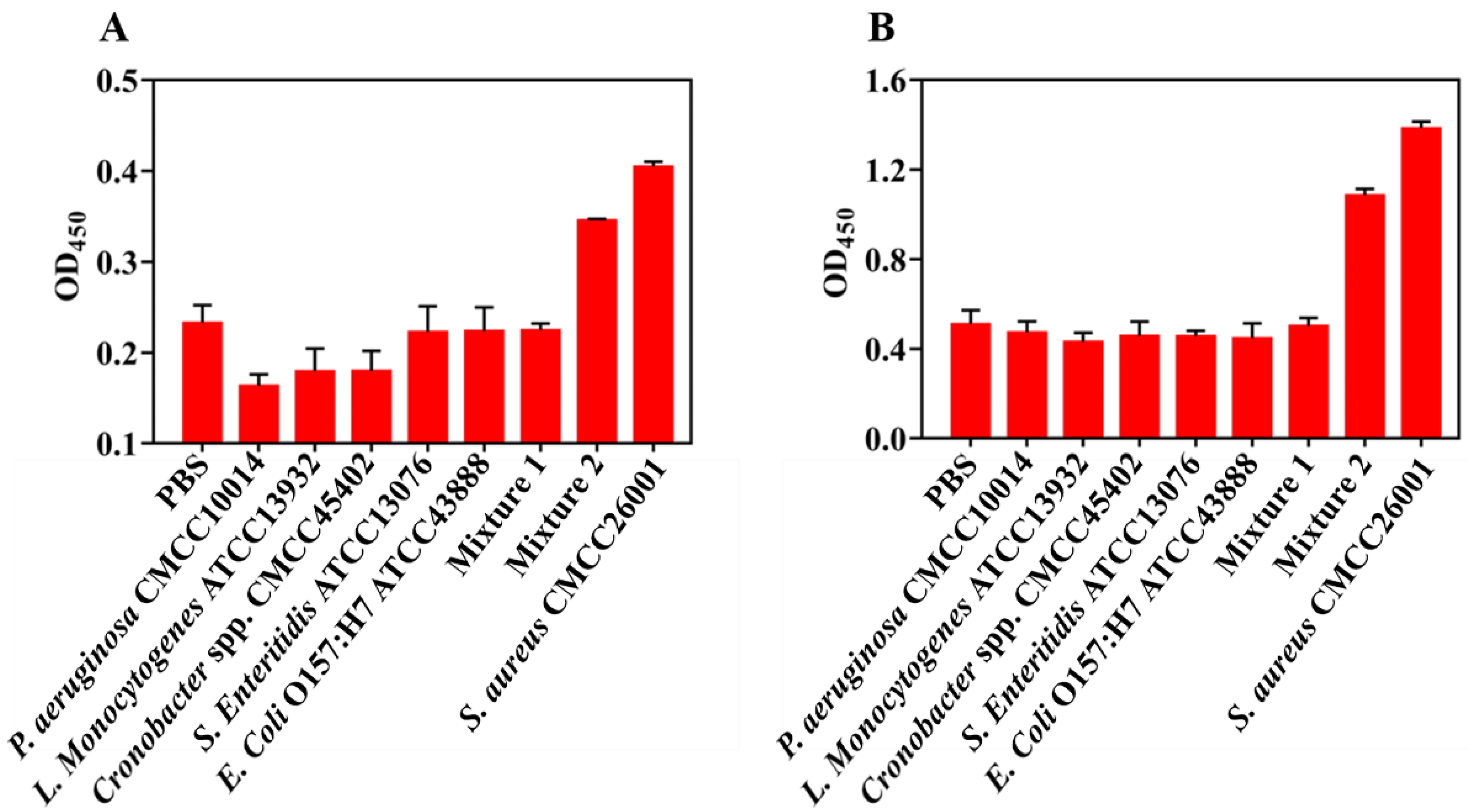
3.4. Specificity of Two Colorimetric Experiment
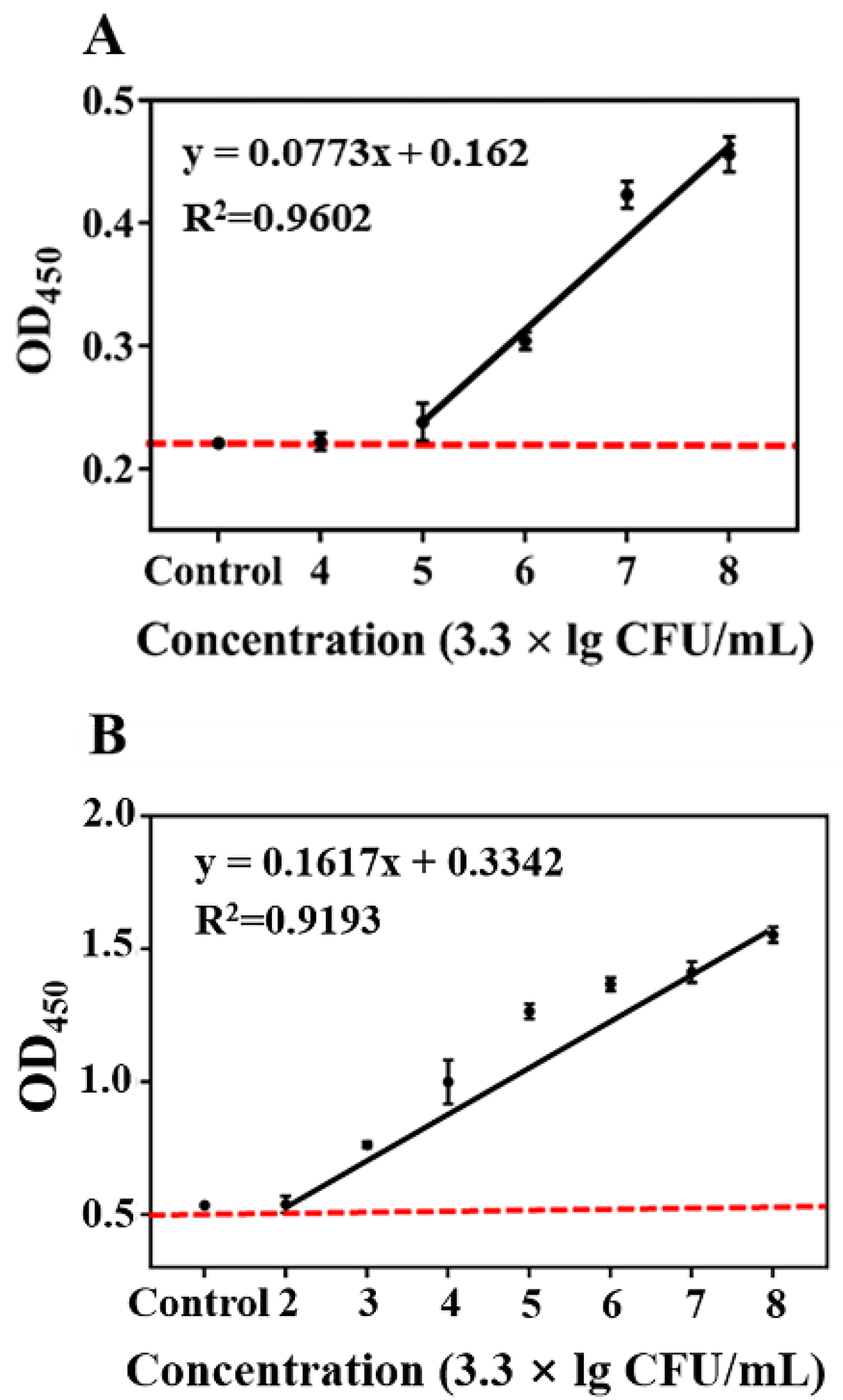
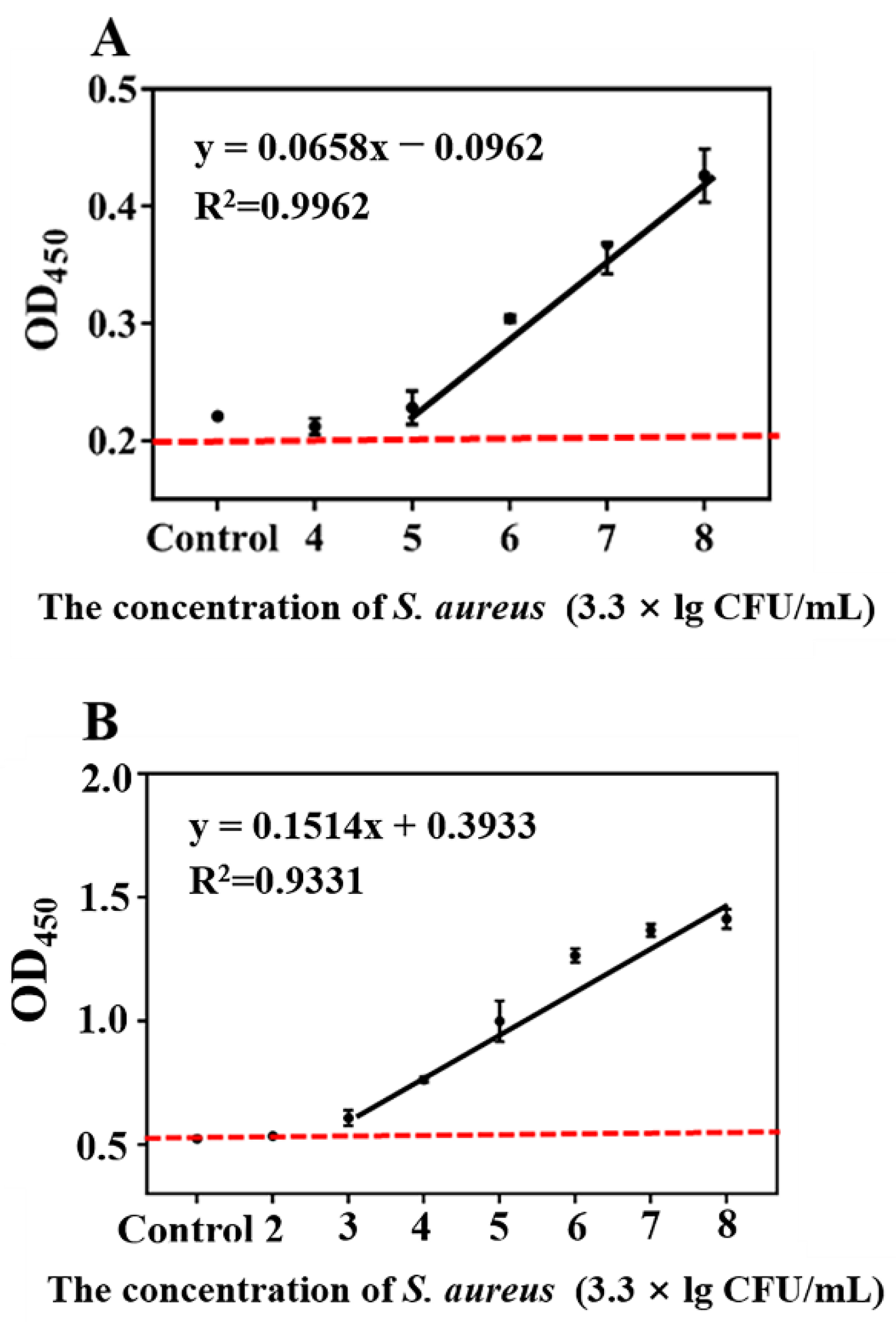
3.5. Evaluation of AMS-DNFs Assay in Juice Samples
4. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Abdalhai, M.H.; Fernandes, A.N.M.; Bashari, M.; Ji, J.; He, Q.; Sun, X. Rapid and sensitive detection of foodborne pathogenic bacteria (Staphylococcus aureus) using an electrochemical DNA genomic biosensor and its application in fresh beef. J. Agric. Food Chem. 2014, 62, 12659–12667. [Google Scholar] [CrossRef] [PubMed]
- Zhang, M.; Wang, X.; Han, L.; Niu, S.; Shi, C.; Ma, C. Rapid detection of foodborne pathogen Listeria monocytogenes by strand exchange amplification. Anal. Biochem. 2018, 545, 38–42. [Google Scholar] [CrossRef] [PubMed]
- Ahmed, A.A.-H.; Maharik, N.M.S.; Valero, A.; Kamal, S.M. Incidence of enterotoxigenic Staphylococcus aureus in milk and Egyptian artisanal dairy products. Food Control 2019, 104, 20–27. [Google Scholar] [CrossRef]
- Valderrama, W.B.; Dudley, E.G.; Doores, S.; Cutter, C.N. Commercially available rapid methods for detection of selected food-borne pathogens. Crit. Rev. Food Sci. Nutr. 2016, 56, 1519–1531. [Google Scholar] [CrossRef]
- Reta, N.; Saint, C.P.; Michelmore, A.; Prieto-Simon, B.; Voelcker, N.H. Nanostructured electrochemical biosensors for label-free detection of water-and food-borne pathogens. ACS Appl. Mater. Interf. 2018, 10, 6055–6072. [Google Scholar] [CrossRef]
- Wu, Y. Rapid and Dynamic Detection of Antimicrobial Response of Pathogenic Bacteria using MZO Nanostructure-Modified Quartz Crystal Microbalance. Ph.D. Dissertation, Rutgers The State University of New Jersey, School of Graduate Studies, Piscataway, NJ, USA, 2019. [Google Scholar] [CrossRef]
- Pursley, J.N. Observing the Sedimentation of TiO2 Nanoparticles in Aqueous Solution Using a Quartz Crystal Microbalance with Dissipation Monitoring; University of Missouri: Kansas City, MI, USA, 2016. [Google Scholar] [CrossRef]
- Neethirajan, S.; Jayas, D.S. Nanotechnology for the food and bioprocessing industries. Food Bioproces. Technol. 2011, 4, 39–47. [Google Scholar] [CrossRef]
- Cho, I.-H.; Bhandari, P.; Patel, P.; Irudayaraj, J. Membrane filter-assisted surface enhanced Raman spectroscopy for the rapid detection of E. coli O157: H7 in ground beef. Biosens. Bioelectron. 2015, 64, 171–176. [Google Scholar] [CrossRef]
- Meng, X.; Li, F.; Li, F.; Xiong, Y.; Xu, H. Vancomycin modified PEGylated-magnetic nanoparticles combined with PCR for efficient enrichment and detection of Listeria monocytogenes. Sens. Actuators B Chem. 2017, 247, 546–555. [Google Scholar] [CrossRef]
- Zhang, F.; Nangreave, J.; Liu, Y.; Yan, H. Structural DNA nanotechnology: State of the art and future perspective. J. Am. Chem. Soc. 2014, 136, 11198–11211. [Google Scholar] [CrossRef] [Green Version]
- Hu, Q.; Li, H.; Wang, L.; Gu, H.; Fan, C. DNA nanotechnology-enabled drug delivery systems. Chem. Rev. 2018, 119, 6459–6506. [Google Scholar] [CrossRef]
- Kim, E.; Agarwal, S.; Kim, N.; Hage, F.S.; Leonardo, V.; Gelmi, A.; Stevens, M.M. Bioinspired Fabrication of DNA–Inorganic Hybrid Composites Using Synthetic DNA. ACS Nano 2019, 13, 2888–2900. [Google Scholar] [CrossRef] [PubMed]
- Ali, M.M.; Li, F.; Zhang, Z.; Zhang, K.; Kang, D.-K.; Ankrum, J.A.; Le, X.C.; Zhao, W. Rolling circle amplification: A versatile tool for chemical biology, materials science and medicine. Chem. Soc. Rev. 2014, 43, 3324–3341. [Google Scholar] [CrossRef] [PubMed]
- Mohsen, M.G.; Kool, E.T. The discovery of rolling circle amplification and rolling circle transcription. Acc. Chem. Res. 2016, 49, 2540–2550. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yan, Y.; Li, J.; Li, W.; Wang, Y.; Song, W.; Bi, S. DNA flower-encapsulated horseradish peroxidase with enhanced biocatalytic activity synthesized by an isothermal one-pot method based on rolling circle amplification. Nanoscale 2018, 10, 22456–22465. [Google Scholar] [CrossRef]
- Shim, J.W.; Tan, Q.; Gu, L.-Q. Single-molecule detection of folding and unfolding of the G-quadruplex aptamer in a nanopore nanocavity. Nucleic Acids Res. 2009, 37, 972–982. [Google Scholar] [CrossRef] [Green Version]
- Krufczik, M.; Sievers, A.; Hausmann, A.; Lee, J.-H.; Hildenbrand, G.; Schaufler, W.; Hausmann, M. Combining low temperature fluorescence DNA-hybridization, immunostaining, and super-resolution localization microscopy for nano-structure analysis of ALU elements and their influence on chromatin structure. Int. J. Mol. Sci. 2017, 18, 1005. [Google Scholar] [CrossRef] [Green Version]
- Lu, W.; Wang, Y.; Song, S.; Chen, C.; Yao, B.; Wang, M. A fishhook probe-based rolling circle amplification (FP-RCA) assay for efficient isolation and detection of microRNA without total RNA extraction. Analyst 2018, 143, 5046–5053. [Google Scholar] [CrossRef]
- Zhu, Y.; Wang, H.; Wang, L.; Zhu, J.; Jiang, W. Cascade signal amplification based on copper nanoparticle-reported rolling circle amplification for ultrasensitive electrochemical detection of the prostate cancer biomarker. ACS Appl. Mater. Interfaces 2016, 8, 2573–2581. [Google Scholar] [CrossRef]
- Bi, S.; Ji, B.; Zhang, Z.; Zhang, S. A chemiluminescence imaging array for the detection of cancer cells by dual-aptamer recognition and bio-bar-code nanoprobe-based rolling circle amplification. Chem. Commun. 2013, 49, 3452–3454. [Google Scholar] [CrossRef] [Green Version]
- Sjödahl, J. Structural studies on the four repetitive Fc-binding regions in protein A from Staphylococcus aureus. Eur. J. Biochem. 1977, 78, 471–490. [Google Scholar] [CrossRef]
- Dong, X.-X.; Yuan, L.-P.; Liu, Y.-X.; Wu, M.-F.; Liu, B.; Sun, Y.-M.; Shen, Y.-D.; Xu, Z.-L. Development of a progesterone immunosensor based on thionine-graphene oxide composites platforms: Improvement by biotin-streptavidin-amplified system. Talanta 2017, 170, 502–508. [Google Scholar] [CrossRef] [PubMed]
- Guo, P.; Xiong, J.; Zheng, D.; Zhang, W.; Liu, L.; Wang, S.; Gu, H. A biosensor based on a film bulk acoustic resonator and biotin–avidin system for the detection of the epithelial tumor marker mucin 1. RSC Adv. 2015, 5, 66355–66359. [Google Scholar] [CrossRef]
- Zhan, Z.; Li, H.; Liu, J.; Xie, G.; Xiao, F.; Wu, X.; Aguilar, Z.P.; Xu, H. A competitive enzyme linked aptasensor with rolling circle amplification (ELARCA) assay for colorimetric detection of Listeria monocytogenes. Food Control 2020, 107, 106806. [Google Scholar] [CrossRef]
- Zhu, G.; Hu, R.; Zhao, Z.; Chen, Z.; Zhang, X.; Tan, W. Noncanonical self-assembly of multifunctional DNA nanoflowers for biomedical applications. J. Am. Chem. Soc. 2013, 135, 16438–16445. [Google Scholar] [CrossRef] [PubMed] [Green Version]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wang, Y.; Wang, Z.; Zhan, Z.; Yan, L.; Wang, L.; Xu, H. Sensitive Detection of Staphylococcus aureus by a Colorimetric Biosensor Based on Magnetic Separation and Rolling Circle Amplification. Foods 2022, 11, 1852. https://doi.org/10.3390/foods11131852
Wang Y, Wang Z, Zhan Z, Yan L, Wang L, Xu H. Sensitive Detection of Staphylococcus aureus by a Colorimetric Biosensor Based on Magnetic Separation and Rolling Circle Amplification. Foods. 2022; 11(13):1852. https://doi.org/10.3390/foods11131852
Chicago/Turabian StyleWang, Yutong, Zhengzheng Wang, Zhongxu Zhan, Leina Yan, Lijun Wang, and Hengyi Xu. 2022. "Sensitive Detection of Staphylococcus aureus by a Colorimetric Biosensor Based on Magnetic Separation and Rolling Circle Amplification" Foods 11, no. 13: 1852. https://doi.org/10.3390/foods11131852
APA StyleWang, Y., Wang, Z., Zhan, Z., Yan, L., Wang, L., & Xu, H. (2022). Sensitive Detection of Staphylococcus aureus by a Colorimetric Biosensor Based on Magnetic Separation and Rolling Circle Amplification. Foods, 11(13), 1852. https://doi.org/10.3390/foods11131852







