An Insulin Receptor-Binding Multifunctional Protein from Tamarindus indica L. Presents a Hypoglycemic Effect in a Diet-Induced Type 2 Diabetes—Preclinical Study
Abstract
:1. Introduction
2. Materials and Methods
2.1. Isolation of the Trypsin Inhibitor from Tamarind Seed (TTI)
2.2. Experimental Model
2.3. Molecular Docking and Molecular Dynamics Simulations
2.3.1. Sourc of the Studied Molecules
2.3.2. Molecular Docking between TTIp 56/287 and IR
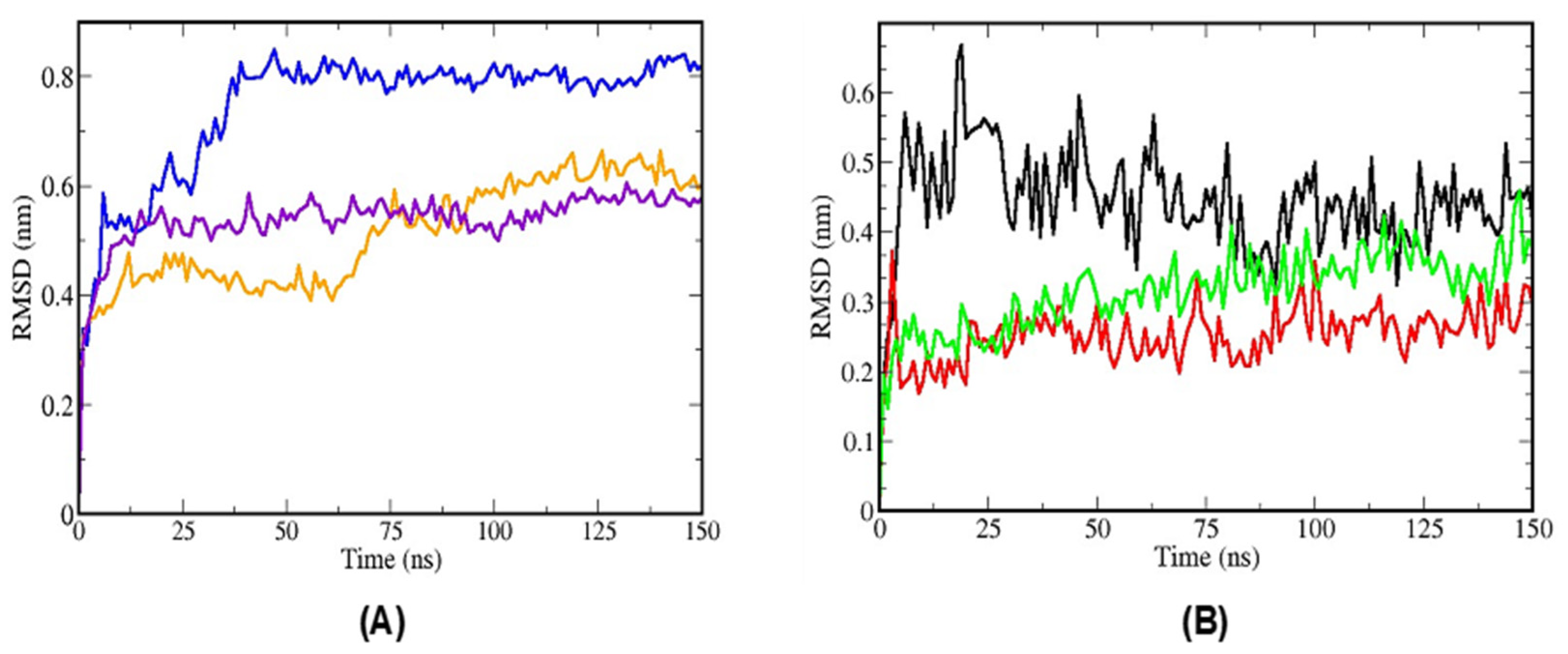
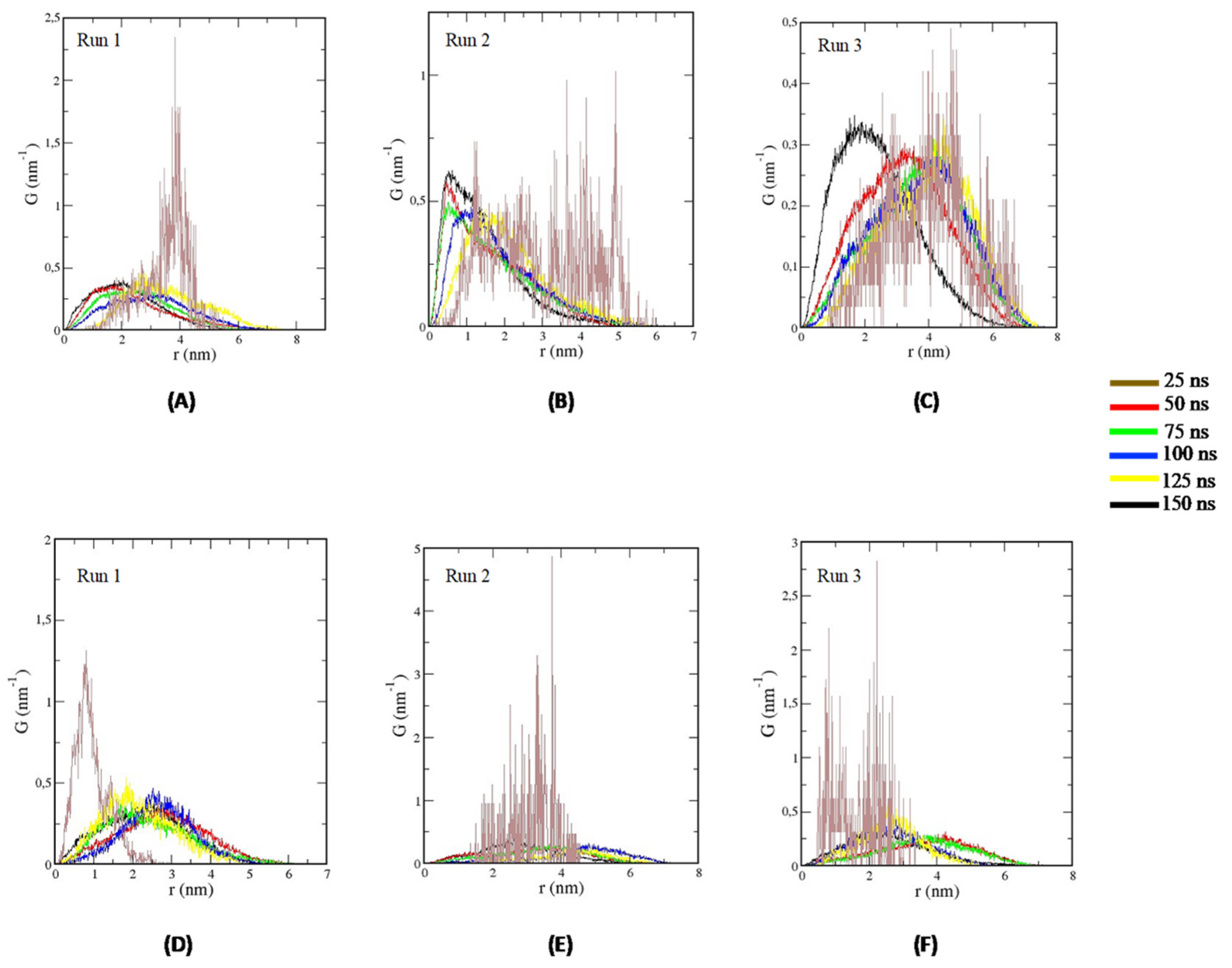
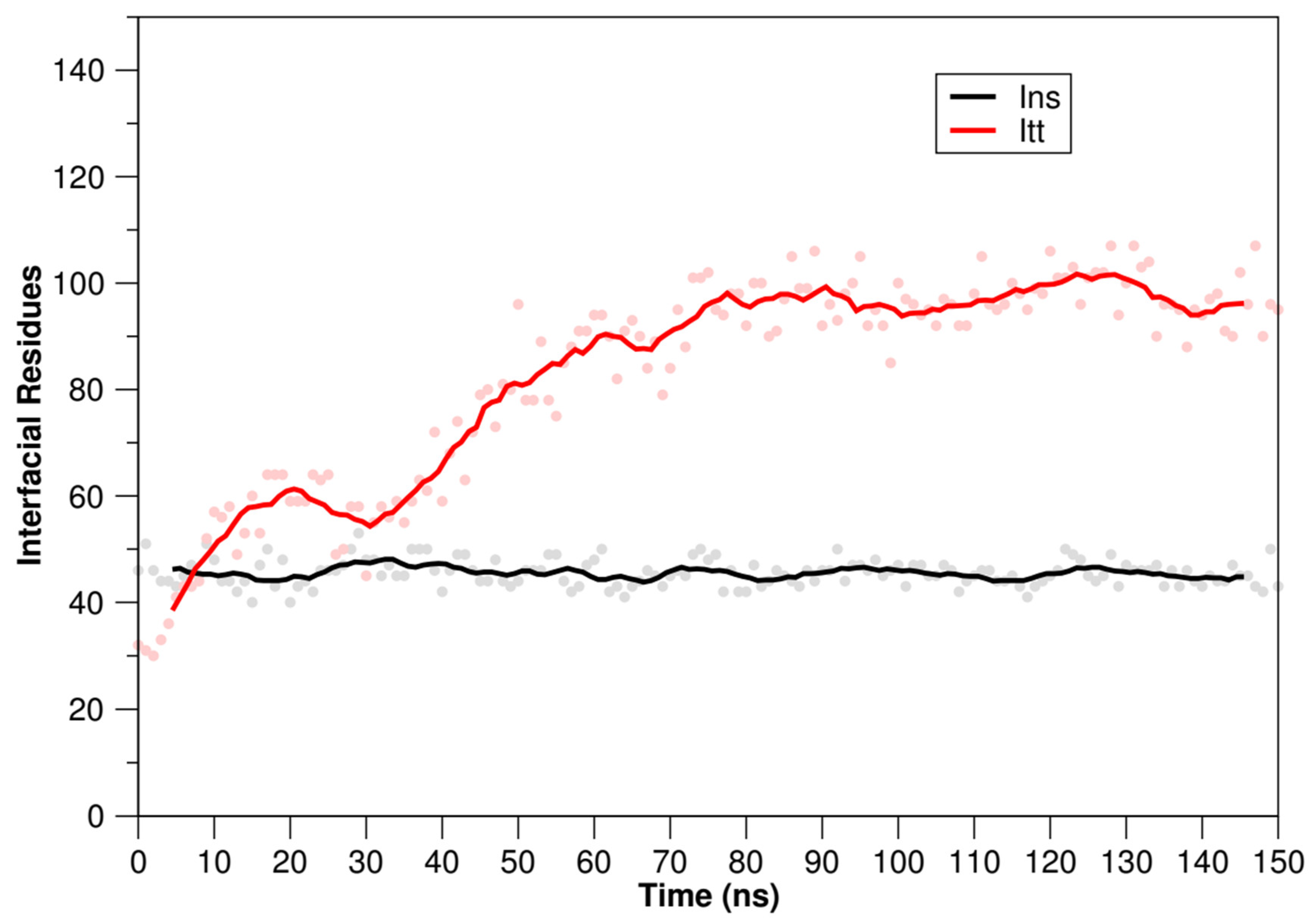
2.3.3. Molecular Dynamics Simulations of TTIp 56/287-IR (PDB ID:4OGA) and Ins-IR
2.4. Statistical Analysis
3. Results
3.1. Obtaining the Trypsin Inhibitor from Tamarind Seeds (TTI)
3.2. Confirmation of T2DM in Wistar Rats after a 17-Week Period Consuming HGLI Diet
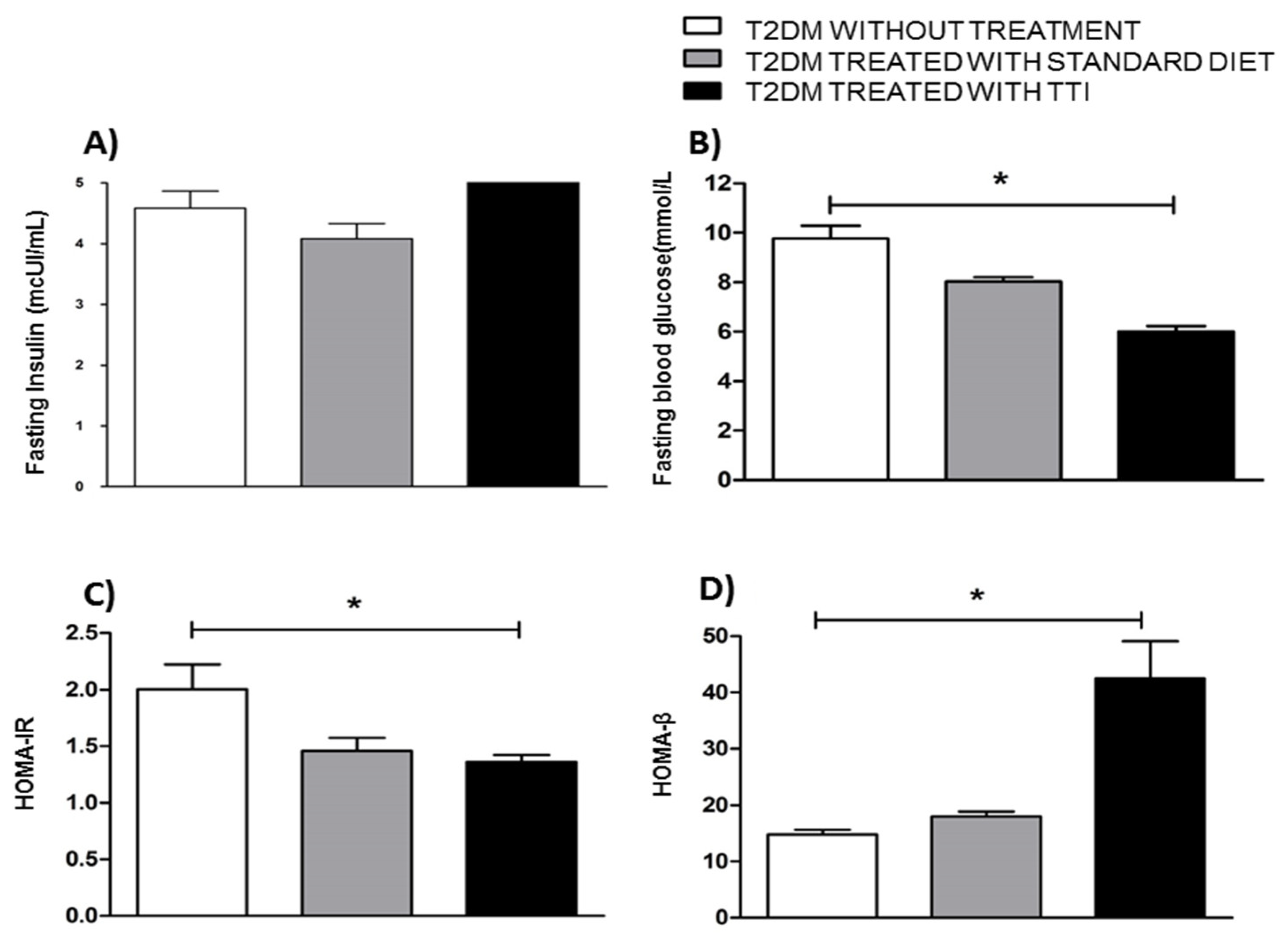
3.3. Evaluation of Fasting Glucose, Fasting Insulinemia and HOMA-IR and HOMA-β Related to the Action and Functionality of Insulin in Wistar Rats with T2DM
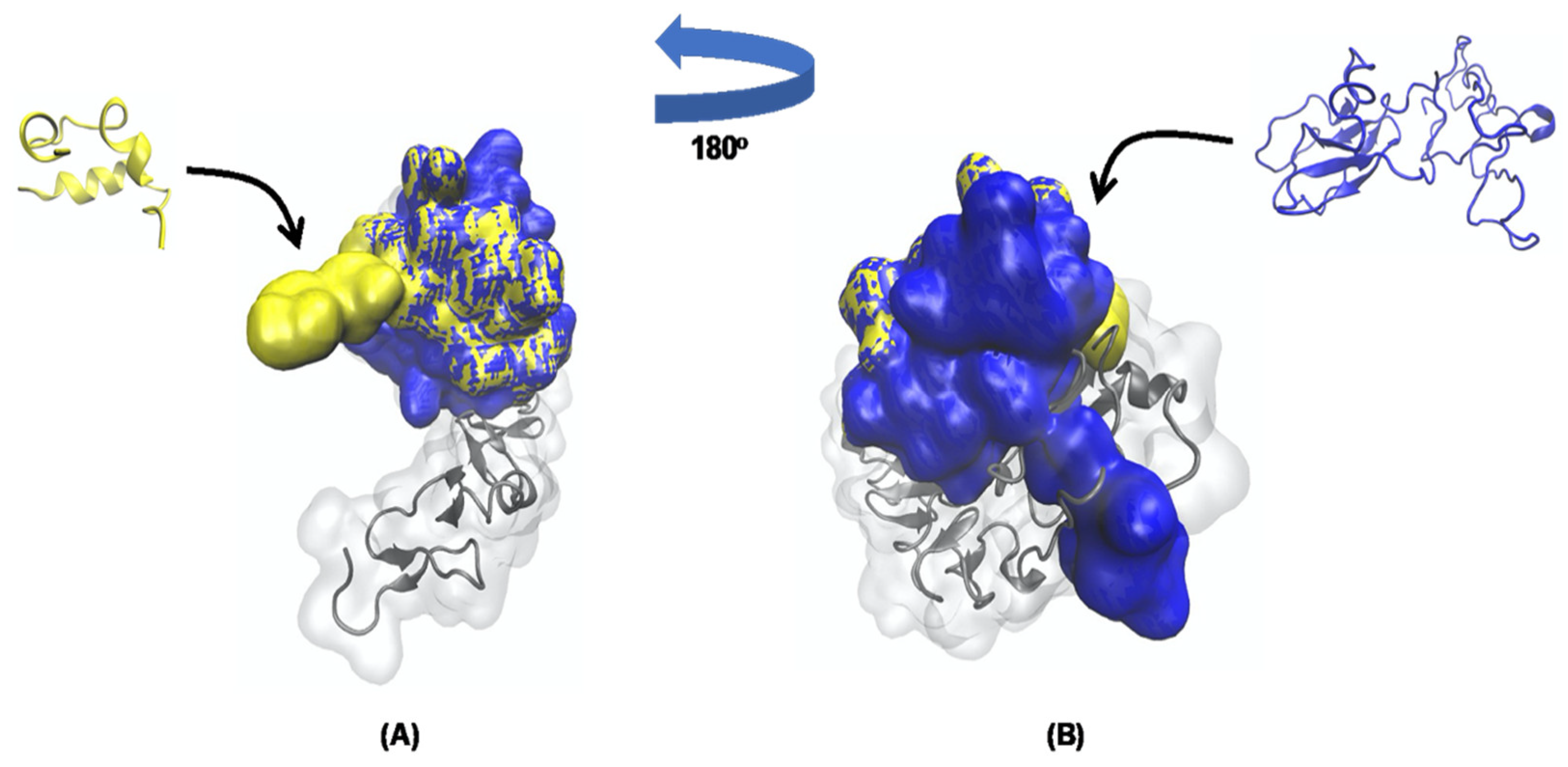
3.4. Molecular Docking Analysis and Molecular Dynamics Simulation between TTIp 56/287 Systems with IR and between Insulin with IR
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- American Diabetes Association. 2. Classification and Diagnosis of Diabetes: Standards of Medical Care in Diabetes—2021. Diabetes Care 2021, 44, S15–S33. [Google Scholar] [CrossRef] [PubMed]
- IDF Diabetes Atlas. 463 People Living with Diabetes Million; International Diabetes Federation (IDF): Brussels, Belgium, 2019. [Google Scholar]
- Chung, W.K.; Erion, K.; Florez, J.C.; Hattersley, A.T.; Hivert, M.; Lee, C.G.; Mccarthy, M.I.; Nolan, J.J.; Norris, J.M.; Pearson, E.R.; et al. Precision Medicine in Diabetes: A Consensus Report from the American Diabetes Association (ADA) and the European Association for the Study of Diabetes (EASD) ADOPT A Diabetes Outcome Progression Trial DPP Diabetes Prevention Program DPP4i Dipeptidyl Pepti. Diabetologia 2020, 63, 1671–1693. [Google Scholar] [CrossRef] [PubMed]
- Saltiel, A.R.; Kahn, C.R. Insulin Signalling and the Regulation of Glucose and Lipid Metabolism. Nature 2001, 414, 799–806. [Google Scholar] [CrossRef]
- Kahn, S.E.; Cooper, M.E.; Del Prato, S. Pathophysiology and Treatment of Type 2 Diabetes: Perspectives on the Past, Present, and Future. Lancet 2014, 383, 1068–1083. [Google Scholar] [CrossRef] [Green Version]
- Costa, I.S.; Medeiros, A.F.; Piuvezam, G.; Medeiros, G.C.B.S.; Maciel, B.L.L.; Morais, A.H.A. Insulin-like Proteins in Plant Sources: A Systematic Review. Diabetes Metab. Syndr. Obes. Targets Ther. 2020, 13, 3421–3431. [Google Scholar] [CrossRef]
- McKern, N.M.; Lawrence, M.C.; Streltsov, V.A.; Lou, M.Z.; Adams, T.E.; Lovrecz, G.O.; Elleman, T.C.; Richards, K.M.; Bentley, J.D.; Pilling, P.A.; et al. Structure of the Insulin Receptor Ectodomain Reveals a Folded-over Conformation. Nature 2006, 443, 218–221. [Google Scholar] [CrossRef]
- Boucher, J.; Kleinridders, A.; Kahn, C.R. Insulin Receptor Signaling in Normal. Cold Spring Harb. Perspect. Biol. 2014, 6, a009191. [Google Scholar] [CrossRef] [Green Version]
- De Bem, G.F.; Costa, C.A.; Santos, I.B.; Cristino Cordeiro, V.d.S.; de Carvalho, L.C.R.M.; de Souza, M.A.V.; de Andrade Soares, R.; da Cunha Sousa, P.J.; Ognibene, D.T.; Resende, A.C.; et al. Antidiabetic Effect of Euterpe Oleracea Mart. (Açaí) Extract and Exercise Training on High-Fat Diet and Streptozotocin-Induced Diabetic Rats: A Positive Interaction. PLoS ONE 2018, 13, e0199207. [Google Scholar] [CrossRef]
- Lauritano, C.; Ianora, A. Marine Organisms with Anti-Diabetes Properties. Mar. Drugs 2016, 14, 220. [Google Scholar] [CrossRef]
- Sánchez, A.; Vázquez, A. Bioactive Peptides: A Review. Food Qual. Saf. 2017, 1, 29–46. [Google Scholar] [CrossRef]
- Jirácek, J.; Žáková, L. Structural Perspectives of Insulin Receptor Isoform-Selective Insulin Analogs. Front. Endocrinol. 2017, 8, 167. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Menting, J.G.; Yang, Y.; Chan, S.J.; Phillips, N.B.; Smith, B.J.; Whittaker, J.; Wickramasinghe, N.P.; Whittaker, L.J.; Pandyarajan, V.; Wan, Z.L.; et al. Protective Hinge in Insulin Opens to Enable Its Receptor Engagement. Proc. Natl. Acad. Sci. USA 2014, 111, E3395–E3404. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Carvalho, F.M.C.; Lima, V.C.O.; Costa, I.S.; Medeiros, A.F.; Serquiz, A.C.; Lima, M.C.J.S.; Serquiz, R.P.; Maciel, B.L.L.; Uchôa, A.F.; Santos, E.A.; et al. A Trypsin Inhibitor from Tamarind Reduces Food Intake and Improves Inflammatory Status in Rats with Metabolic Syndrome Regardless of Weight Loss. Nutrients 2016, 8, 544. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Carvalho, F.M.C.; Lima, V.C.O.; Costa, I.S.; Luz, A.B.S.; Ladd, F.V.L.; Serquiz, A.C.; Bortolin, R.H.; Silbiger, V.N.; Maciel, B.L.L.; Santos, E.A.; et al. Anti-TNF-α Agent Tamarind Kunitz Trypsin Inhibitor Improves Lipid Profile of Wistar Rats Presenting Dyslipidemia and Diet-Induced Obesity Regardless of PPAR-γ Induction. Nutrients 2019, 11, 512. [Google Scholar] [CrossRef] [Green Version]
- Matias, L.L.R.; Costa, R.O.A.; Passos, T.S.; Queiroz, J.L.C.; Serquiz, A.C.; Maciel, B.L.L.; Santos, P.P.A.; Camillo, C.S.; Gonçalves, C.; Amado, I.R.; et al. Tamarind Trypsin Inhibitor in Chitosan-Whey Protein Nanoparticles Reduces Fasting Blood Glucose Levels without Compromising Insulinemia: A Preclinical Study. Nutrients 2019, 11, 2770. [Google Scholar] [CrossRef] [Green Version]
- Aguiar, A.J.F.C.; de Queiroz, J.L.C.; Santos, P.P.A.; Camillo, C.S.; Serquiz, A.C.; Costa, I.S.; Oliveira, G.S.; Gomes, A.F.T.; Matias, L.L.R.; Costa, R.O.A.; et al. Beneficial Effects of Tamarind Trypsin Inhibitor in Chitosan–Whey Protein Nanoparticles on Hepatic Injury Induced High Glycemic Index Diet: A Preclinical Study. Int. J. Mol. Sci. 2021, 22, 9968. [Google Scholar] [CrossRef]
- De Medeiros, A.F.; de Sousa Costa, I.; de Carvalho, F.M.C.; Kiyota, S.; de Souza, B.B.P.; Sifuentes, D.N.; Serquiz, R.P.; Maciel, B.L.L.; Uchôa, A.F.; dos Santos, E.A.; et al. Biochemical Characterisation of a Kunitz-Type Inhibitor from Tamarindus indica L. Seeds and Its Efficacy in Reducing Plasma Leptin in an Experimental Model of Obesity. J. Enzyme Inhib. Med. Chem. 2018, 33, 334–348. [Google Scholar] [CrossRef]
- Medeiros, A.F.; de Souza, B.B.P.; Coutinho, L.P.; Murad, A.M.; dos Santos, P.I.M.; de Monteiro, N.K.V.; dos Santos, E.A.; Maciel, B.L.L.; de Araújo Morais, A.H. Structural Insights and Molecular Dynamics into the Inhibitory Mechanism of a Kunitz-Type Trypsin Inhibitor from Tamarindus indica L. J. Enzyme Inhib. Med. Chem. 2021, 36, 480–490. [Google Scholar] [CrossRef]
- Lima, V.C.O.; Luz, A.B.S.; do Amarante, M.S.M.; Lima, M.C.J.S.; Carvalho, F.M.C.; Figueredo, J.B.S.; Santos, P.P.A.; Camillo, C.S.; Ladd, F.V.L.; Maciel, B.L.L.; et al. Tamarind Multifunctional Protein: Safety and Anti-Inflammatory Potential in Intestinal Mucosa and Adipose Tissue in a Preclinical Model of Diet-Induced Obesity. Obes. Facts 2021, 14, 357–369. [Google Scholar] [CrossRef]
- Vieth, M.; Hirst, J.D.; Kolinski, A.; Brooks, C.L. Assessing Energy Functions for Flexible Docking. J. Comput. Chem. 1998, 19, 1612–1622. [Google Scholar] [CrossRef]
- Sharma, S.; Singh, R.; Rana, S. Bioactive Peptides: A Review. Int. J. Bioautom. 2011, 15, 223–250. [Google Scholar]
- Bechaux, J.; Gatellier, P.; Le Page, J.-F.; Drillet, Y.; Sante-Lhoutellier, V. A comprehensive review of bioactive peptides obtained from animal byproducts and their applications. Food Funct. 2019, 10, 6244–6266. [Google Scholar] [CrossRef] [PubMed]
- Cabral, H. Peptides: Molecular and Biotechnological Aspects. Biomolecules 2021, 11, 52. [Google Scholar] [CrossRef] [PubMed]
- Kakade, M.; Simons, N.; Liener, I. An Evaluation of Natural vs. Synthetic Substrates for Measuring the Antitryptic Activity of Soybean Samples. Cereal Chem. 1969, 46, 518–526. [Google Scholar]
- Bradford, M.M. A Rapid and Sensitive Method for the Quantitation of Microgram Quantities of Protein Utilizing the Principle of Protein-Dye Binding. Anal. Biochem. 1976, 72, 248–254. [Google Scholar] [CrossRef]
- Laemmli, U.K. Cleavage of Structural Proteins during the Assembly of the Head of Bacteriophage T4. Nature 1970, 227, 680–685. [Google Scholar] [CrossRef]
- Oakley, B.R.; Kirsch, D.R.; Morris, N.R. A Simplified Ultrasensitive Silver Stain for Detecting Proteins in Polyacrylamide Gels. Anal. Biochem. 1980, 105, 361–363. [Google Scholar] [CrossRef]
- Roy, A.; Kucukural, A.; Zhang, Y. I-TASSER: A Unified Platform for Automated Protein Structure and Function Prediction. Nat. Protoc. 2010, 5, 725–738. [Google Scholar] [CrossRef] [Green Version]
- Kilkenny, C.; Browne, W.J.; Cuthill, I.C.; Emerson, M.; Altman, D.G. Improving Bioscience Research Reporting: The ARRIVE Guidelines for Reporting Animal Research. PLoS Biol. 2010, 8, e1000412. [Google Scholar] [CrossRef]
- Matthews, D.R.; Hosker, J.P.; Rudenski, A.S.; Naylor, B.A.; Treacher, D.F.; Turner, R.C. Homeostasis Model Assessment: Insulin Resistance and β-Cell Function from Fasting Plasma Glucose and Insulin Concentrations in Man. Diabetologia 1985, 28, 412–419. [Google Scholar] [CrossRef] [Green Version]
- Berman, H.M.; Bluhm, W.F.; Philip, E.; Marvin, J.; Weissig, H.; John, D. The Protein Data Bank. Acta Crystallogr. D Biol. Crystallogr. 2002, D58, 899–907. [Google Scholar] [CrossRef] [PubMed]
- Vienberg, S.G.; Bouman, S.D.; Sørensen, H.; Stidsen, C.E.; Kjeldsen, T.; Glendorf, T.; Sørensen, A.R.; Olsen, G.S.; Andersen, B.; Nishimura, E. Receptor-Isoform-Selective Insulin Analogues Give Tissue-Preferential Effects. Biochem. J. 2011, 440, 301–308. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Schneidman-Duhovny, D.; Inbar, Y.; Nussinov, R.; Wolfson, H.J. PatchDock and SymmDock: Servers for Rigid and Symmetric Docking. Nucleic Acids Res. 2005, 33 (Suppl. 2), 363–367. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Andrusier, N.; Nussinov, R.; Wolfson, H.J. FireDock: Fast Interaction Refinement in Molecular Docking. Proteins Struct. Funct. Bioinform. 2007, 69, 139–159. [Google Scholar] [CrossRef] [PubMed]
- Berendsen, H.J.C.; van der Spoel, D.; van Drunen, R. GROMACS: A Message-Passing Parallel Molecular Dynamics Implementation. Comput. Phys. Commun. 1995, 91, 43–56. [Google Scholar] [CrossRef]
- Zhang, W.; Yang, R.; Cieplak, P.; Luo, R.; Lee, T.; Caldwell, J.; Wang, J.; Kollman, P. A Point-Charge Force Field for Molecular Mechanics Simulations of Proteins Based on Condensed-Phase. J. Comput. Chem. 2003, 24, 1999. [Google Scholar]
- Jorgensen, W.L.; Chandrasekhar, J.; Madura, J.D.; Impey, R.W.; Klein, M.L. Comparison of Simple Potential Functions for Simulating Liquid Water. J. Chem. Phys. 1983, 79, 926–935. [Google Scholar] [CrossRef]
- Van Gunsteren, W.F.; Berendsen, H.J.C. A Leap-Frog Algorithm for Stochastic Dynamics. Mol. Simul. 1988, 1, 173–185. [Google Scholar] [CrossRef]
- Darden, T.; York, D.; Pedersen, L. Particle Mesh Ewald: An N·log(N) Method for Ewald Sums in Large Systems. J. Chem. Phys. 1993, 98, 10089–10092. [Google Scholar] [CrossRef] [Green Version]
- Hess, B.; Bekker, H.; Berendsen, H.J.C.; Fraaije, J.G.E.M. LINCS: A Linear Constraint Solver for Molecular Simulations. J. Comput. Chem. 1997, 18, 1463–1472. [Google Scholar] [CrossRef]
- Hoover, W.G. Canonical Dynamics: Equilibrium Phase-Space Distributions. Phys. Rev. A 1985, 31, 1695–1697. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nosé, S.; Klein, M.L. Constant Pressure Molecular Dynamics for Molecular Systems. Mol. Phys. 1983, 50, 1055–1076. [Google Scholar] [CrossRef]
- Hestenes, M.R.; Stiefel, E. Methods of Conjugate Gradients for Solving Linear Systems. J. Res. Natl. Bur. Stand. 1952, 49, 409. [Google Scholar] [CrossRef]
- Petrou, S.; Kwon, J.; Madan, J. A Practical Guide to Conducting a Systematic Review and Meta-Analysis of Health State Utility Values. Pharmacoeconomics 2018, 36, 1043–1061. [Google Scholar] [CrossRef] [Green Version]
- Amorim-Carmo, B.; Daniele-Silva, A.; Parente, A.M.S.; Furtado, A.A.; Carvalho, E.; Oliveira, J.W.F.; Santos, E.C.G.; Silva, M.S.; Silva, S.R.B.; Silva-Júnior, A.A.; et al. Potent and Broad-Spectrum Antimicrobial Activity of Analogs from the Scorpion Peptide Stigmurin. Int. J. Mol. Sci. 2019, 20, 623. [Google Scholar] [CrossRef] [Green Version]
- Humphrey, W.; Dalke, A.; Schulten, K. VMD: Visual Molecular Dynamics. J. Mol. Graph. 1996, 14, 33–38. [Google Scholar] [CrossRef]
- Tschöp, M.; Heiman, M.L. Rodent Obesity Models: An Overview. Exp. Clin. Endocrinol. Diabetes 2001, 109, 307–319. [Google Scholar] [CrossRef]
- Cochran, W.G. Sampling Techniques, 3rd ed.; Wiley: Hoboken, NJ, USA, 1977. [Google Scholar]
- Luz, A.B.S.; dos Santos Figueredo, J.B.; Salviano, B.D.P.D.; Aguiar, A.J.F.C.; Pinheiro, L.G.S.D.; Krause, M.F.D.; Da Silva Camillo, C.; Ladd, F.V.L.; Bortolin, R.H.; Silbiger, V.N.; et al. Adipocytes and Intestinal Epithelium Dysfunctions Linking Obesity to Inflammation Induced by High Glycemic Index Pellet-Diet in Wistar Rats. Biosci. Rep. 2018, 38, BSR20180304. [Google Scholar] [CrossRef] [Green Version]
- Genser, L.; Rossario, J.; Mariolo, C.; Castagneto-Gissey, L.; Panagiotopoulos, S.; Rubino, F. Obesity, Type 2 Diabetes, and the Metabolic Syndrome: Pathophysiologic Relationships and Guidelines for Surgical Intervention. Surg. Clin. N. Am. 2016, 96, 681–701. [Google Scholar] [CrossRef]
- Mahmoud, F.; Al-Ozairi, E. Inflammatory Cytokines and the Risk of Cardiovascular Complications in Type 2 Diabetes. Dis. Mark. 2013, 35, 235–241. [Google Scholar] [CrossRef] [Green Version]
- Winer, D.A.; Luck, H.; Tsai, S.; Winer, S. The Intestinal Immune System. Cell Metab. 2016, 23, 413–426. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ribeiro, J.; Serquiz, A.; Silva, P.; Barbosa, P.; Sampaio, T.; Araujo Junior, R.; Oliveira, A.; Machado, R.; Maciel, B.; Uchoa, A.; et al. Trypsin Inhibitor from Tamarindus indica L. Seeds Reduces Weight Gain and Food Consumption and Increases Plasmatic Cholecystokinin Levels. Clinics 2015, 70, 136–143. [Google Scholar] [CrossRef]
- Batista, T.M.; Haider, N.; Kahn, C.R. Defining the Underlying Defect in Insulin Action in Type 2 Diabetes. Diabetologia 2021, 64, 994–1006. [Google Scholar] [CrossRef]
- Paula, P.C.; Oliveira, J.T.A.; Sousa, D.O.B.; Alves, B.G.T.; Carvalho, A.F.U.; Franco, O.L.; Vasconcelos, I.M. Insulin-like Plant Proteins as Potential Innovative Drugs to Treat Diabetes—The Moringa Oleifera Case Study. New Biotechnol. 2017, 39, 99–109. [Google Scholar] [CrossRef] [PubMed]
- de Araújo Costa, R.O.; Matias, L.L.R.; Passos, T.S.; de Queiroz, J.L.C.; de Carvalho, F.M.C.; Maciel, B.L.L.; Uchôa, A.F.; Amado, I.R.; Gonçalves, C.; Pastrana, L.; et al. Safety and Potential Functionality of Nanoparticles Loaded with a Trypsin Inhibitor Isolated from Tamarind Seeds. Futur. Foods 2020, 1–2, 100001. [Google Scholar] [CrossRef]
- Xu, M.; Sun, B.; Li, D.; Mao, R.; Li, H.; Li, Y.; Wang, J. Beneficial Effects of Small Molecule Oligopeptides Isolated from Panax Ginseng Meyer on Pancreatic Beta-Cell Dysfunction and Death in Diabetic Rats. Nutrients 2017, 9, 1061. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Acquah, C.; Dzuvor, C.K.O.; Tosh, S.; Agyei, D. Anti-Diabetic Effects of Bioactive Peptides: Recent Advances and Clinical Implications. Crit. Rev. Food Sci. Nutr. 2020, 62, 2158–2171. [Google Scholar] [CrossRef]
- Wang, F.J.; Song, H.L.; Wang, X.M.; Zhang, W.J.; Wang, B.L.; Zhao, J.; Hu, Z.B. Tandem Multimer Expression and Preparation of Hypoglycemic Peptide MC6 from Momordica Charantia in Escherichia Coli. Appl. Biochem. Biotechnol. 2012, 166, 612–619. [Google Scholar] [CrossRef]
- Tian, M.; Zeng, X.Q.; Song, H.L.; Hu, S.X.; Wang, F.J.; Zhao, J.; Hu, Z.B. Molecular Diversity and Hypoglycemic Polypeptide-P Content of Momordica Charantia in Different Accessions and Different Seasons. J. Sci. Food Agric. 2015, 95, 1328–1335. [Google Scholar] [CrossRef]
- Lo, H.Y.; Ho, T.Y.; Li, C.C.; Chen, J.C.; Liu, J.J.; Hsiang, C.Y. A Novel Insulin Receptor-Binding Protein from Momordica Charantia Enhances Glucose Uptake and Glucose Clearance in Vitro and in Vivo through Triggering Insulin Receptor Signaling Pathway. J. Agric. Food Chem. 2014, 62, 8952–8961. [Google Scholar] [CrossRef]
- Lo, H.Y.; Ho, T.Y.; Lin, C.; Li, C.C.; Hsiang, C.Y. Momordica Charantia and Its Novel Polypeptide Regulate Glucose Homeostasis in Mice via Binding to Insulin Receptor. J. Agric. Food Chem. 2013, 61, 2461–2468. [Google Scholar] [CrossRef]
- Costa, I.S.; Medeiros, A.F.; Carvalho, F.M.C.; Lima, V.C.O.; Serquiz, R.P.; Serquiz, A.C.; Silbiger, V.N.; Bortolin, R.H.; Maciel, B.L.L.; Santos, E.A.; et al. Satietogenic Protein from Tamarind Seeds Decreases Food Intake, Leptin Plasma and CCK-1r Gene Expression in Obese Wistar Rats. Obes. Facts 2018, 11, 440–453. [Google Scholar] [CrossRef] [PubMed]
- Gutmann, T.; Kim, K.H.; Grzybek, M.; Walz, T.; Coskun, Ü. Visualization of Ligand-Induced Transmembrane Signaling in the Full-Length Human Insulin Receptor. J. Cell Biol. 2018, 217, 1643–1649. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Scapin, G.; Dandey, V.P.; Zhang, Z.; Prosise, W.; Hruza, A.; Kelly, T.; Mayhood, T.; Strickland, C.; Potter, C.S.; Carragher, B. Structure of the Insulin Receptor-Insulin Complex by Single-Particle Cryo-EM Analysis. Nature 2018, 556, 122–125. [Google Scholar] [CrossRef] [PubMed]
- Weis, F.; Menting, J.G.; Margetts, M.B.; Chan, S.J.; Xu, Y.; Tennagels, N.; Wohlfart, P.; Langer, T.; Müller, C.W.; Dreyer, M.K.; et al. The Signalling Conformation of the Insulin Receptor Ectodomain. Nat. Commun. 2018, 9, 4420. [Google Scholar] [CrossRef]
- Kong, K.; Smith, B.J.; Watson, C.J.; Žáková, L.; Kletvíková, E.; Chan, S.J.; Steiner, D.F.; Dodson, G.G.; Andrzej, M.; Hall, E.; et al. How Insulin Engages Its Primary Binding Site on the Insulin Receptor. Nature 2014, 493, 241–245. [Google Scholar] [CrossRef] [Green Version]
- Wallis, M. Growth Hormone & IGF Research Coevolution of Insulin-like Growth Factors, Insulin and Their Receptors and Binding Proteins in New World Monkeys. Growth Horm. IGF Res. 2015, 25, 158–167. [Google Scholar] [CrossRef] [Green Version]
- Silva, L.B. A Protein with Amino Acid Sequence Homology to Bovine Insulin Is Present in the Legume Vigna Unguiculata (Cowpea). Braz. J. Med. Biol. Res. 2003, 36, 1167–1173. [Google Scholar]
- Joshi, B.N.; Munot, H.; Hardikar, M.; Kulkarni, A.A. Orally Active Hypoglycemic Protein from Costus igneus N. E. Br.: An in Vitro and in Vivo Study. Biochem. Biophys. Res. Commun. 2013, 436, 278–282. [Google Scholar] [CrossRef]
- Rosa Lovati, M.; Manzoni, C.; Castiglioni, S.; Parolari, A.; Magni, C.; Duranti, M. Lupin Seed γ-Conglutin Lowers Blood Glucose in Hyperglycaemic Rats and Increases Glucose Consumption of HepG2 Cells. Br. J. Nutr. 2012, 107, 67–73. [Google Scholar] [CrossRef] [Green Version]
- Bhaskar, V.; Goldfine, I.D.; Bedinger, D.H.; Lau, A.; Kuan, H.F.; Gross, L.M.; Handa, M.; Maddux, B.A.; Watson, S.R.; Zhu, S.; et al. A Fully Human, Allosteric Monoclonal Antibody That Activates the Insulin Receptor and Improves Glycemic Control. Diabetes 2012, 61, 1263–1271. [Google Scholar] [CrossRef] [PubMed] [Green Version]

| Rats | Fasting Glucose (mmol/L) |
|---|---|
| Rat 1 | 9.30 |
| Rat 2 | 7.85 |
| Rat 3 | 8.92 |
| Rat 4 | 7.73 |
| Rat 5 | 7.85 |
| Rat 6 | 8.51 |
| Rat 7 | 9.37 |
| Rat 8 | 9.73 |
| Rat 9 | 7.54 |
| Rat 10 | 10.23 |
| Rat 11 | 12.00 |
| Rat 12 | 9.44 |
| Rat 13 | 9.41 |
| Rat 14 | 8.38 |
| Rat 15 | 11.14 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Costa, I.; Lima, M.; Medeiros, A.; Bezerra, L.; Santos, P.; Serquiz, A.; Lima, M.; Oliveira, G.; Santos, E.; Maciel, B.; et al. An Insulin Receptor-Binding Multifunctional Protein from Tamarindus indica L. Presents a Hypoglycemic Effect in a Diet-Induced Type 2 Diabetes—Preclinical Study. Foods 2022, 11, 2207. https://doi.org/10.3390/foods11152207
Costa I, Lima M, Medeiros A, Bezerra L, Santos P, Serquiz A, Lima M, Oliveira G, Santos E, Maciel B, et al. An Insulin Receptor-Binding Multifunctional Protein from Tamarindus indica L. Presents a Hypoglycemic Effect in a Diet-Induced Type 2 Diabetes—Preclinical Study. Foods. 2022; 11(15):2207. https://doi.org/10.3390/foods11152207
Chicago/Turabian StyleCosta, Izael, Mayara Lima, Amanda Medeiros, Lucas Bezerra, Paula Santos, Alexandre Serquiz, Maíra Lima, Gerciane Oliveira, Elizeu Santos, Bruna Maciel, and et al. 2022. "An Insulin Receptor-Binding Multifunctional Protein from Tamarindus indica L. Presents a Hypoglycemic Effect in a Diet-Induced Type 2 Diabetes—Preclinical Study" Foods 11, no. 15: 2207. https://doi.org/10.3390/foods11152207






