Integrative Physiological and Transcriptome Analysis Reveals the Mechanism for the Repair of Sub-Lethally Injured Escherichia coli O157:H7 Induced by High Hydrostatic Pressure
Abstract
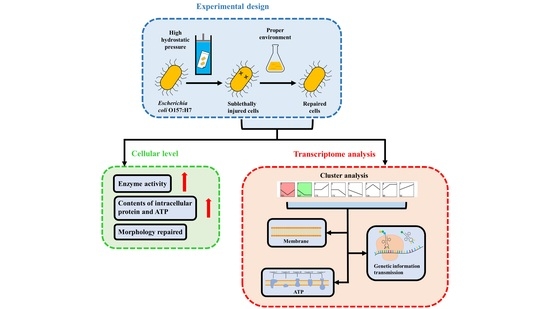
:1. Introduction
2. Materials and Methods
2.1. Strain and Culture Condition
2.2. Determination of Sub-Lethal Treatment Conditions
2.3. Determination of Repair Conditions
2.4. Determination of the Intracellular Protein Content and Adenosine Triphosphate (ATP) Contents
2.5. Observation of the Morphological Changes
2.6. Measurement of Enzyme Activities
2.7. RNA Isolation and Library Construction
2.8. Transcriptomic Analysis
2.9. Real Time Quantitative PCR (qRT-PCR) Validation
2.10. Statistical Analysis
3. Results
3.1. Determination of Sublethal Injury and Repair Conditions
3.2. Physiological Analysis Reveals the Repair Mechanism
3.2.1. Determination of the Intracellular Protein and ATP Contents
3.2.2. Observation of Morphological Changes
3.2.3. Determination of the Enzyme Activities
3.3. Changes in Transcript Levels of E. coli O157:H7 during HHP and Repair
3.3.1. Transcriptional Response to the HHP and Repair
3.3.2. Cluster Analysis, GO, and KEGG Analysis
3.4. Response of DEGs Related to Membrane after HHP and Repair Processes
3.5. Response of DEG Related to Energy Biosynthesis after HHP and Repair
3.6. Response of DEG Related to Genetic Information Transmission after HHP and Repair
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Morata, A.; Loira, I.; Vejarano, R.; Bañuelos, M.A.; Sanz, P.D.; Otero, L.; Suárez-Lepe, J.A. Grape Processing by High Hydrostatic Pressure: Effect on Microbial Populations, Phenol Extraction and Wine Quality. Food Bioprocess Technol. 2015, 8, 277–286. [Google Scholar] [CrossRef] [Green Version]
- Al-Nehlawi, A.; Guri, S.; Guamis, B.; Saldo, J. Synergistic effect of carbon dioxide atmospheres and high hydrostatic pressure to reduce spoilage bacteria on poultry sausages. LWT Food Sci. Technol. 2014, 58, 404–411. [Google Scholar] [CrossRef]
- Huang, H.-W.; Hsu, C.-P.; Wang, C.-Y. Healthy expectations of high hydrostatic pressure treatment in food processing industry. J. Food Drug Anal. 2020, 28, 1–13. [Google Scholar] [CrossRef] [PubMed]
- Huang, H.-W.; Wu, S.-J.; Lu, J.-K.; Shyu, Y.-T.; Wang, C.-Y. Current status and future trends of high-pressure processing in food industry. Food Control 2017, 72, 1–8. [Google Scholar] [CrossRef]
- Sharp, A.; Smout, E.; Byrne, L.; Greenwood, R.; Abdoollah, R.; Hutchinson, C.; Jenkins, C.; Arunachalam, N.; Padfield, S.; Hughes, G.; et al. An outbreak of Shiga toxin-producing Escherichia coli O157:H7 linked to a mud-based obstacle course, England, August 2018. Zoonoses Public Health 2020, 67, 467–473. [Google Scholar] [CrossRef]
- Mulchandani, R.; Brehmer, C.; Butt, S.; Vishram, B.; Harrison, M.; Marchant, E.; Ferris, S.; Jorgensen, F.; Smith, R.; Godbole, G.; et al. Outbreak of Shiga toxin-producing Escherichia coli O157 linked with consumption of a fast-food product containing imported cucumbers, United Kingdom, August 2020. Int. J. Infect. Dis. 2021, 110, S62–S68. [Google Scholar] [CrossRef]
- Heiman, K.E.; Mody, R.K.; Johnson, S.D.; Griffin, P.M.; Gould, L.H. Escherichia coli O157 Outbreaks in the United States, 2003–2012. Emerg. Infect. Dis. 2015, 21, 1293–1301. [Google Scholar] [CrossRef]
- Lee, C.-H.; Kang, J.-H.; Woo, H.-J.; Song, K.B. Inactivation of Listeria monocytogenes and Escherichia coli O157:H7 inoculated on fresh-cut romaine lettuce by peanut skin extract/benzethonium chloride emulsion washing. Food Control 2021, 119, 107479. [Google Scholar] [CrossRef]
- Espina, L.; García-Gonzalo, D.; Laglaoui, A.; Mackey, B.M.; Pagán, R. Synergistic combinations of high hydrostatic pressure and essential oils or their constituents and their use in preservation of fruit juices. Int. J. Food Microbiol. 2013, 161, 23–30. [Google Scholar] [CrossRef]
- Yang, B.; Shi, Y.; Xia, X.; Xi, M.; Wang, X.; Ji, B.; Meng, J. Inactivation of foodborne pathogens in raw milk using high hydrostatic pressure. Food Control 2012, 28, 273–278. [Google Scholar] [CrossRef]
- Toledo, J.; Pérez Pulido, R.; Abriouel, H.; Grande, M.J.; Gálvez, A. Inactivation of Salmonella enterica cells in Spanish potato omelette by high hydrostatic pressure treatments. Innov. Food Sci. Emerg. Technol. 2012, 14, 25–30. [Google Scholar] [CrossRef]
- Shi, H.; Chen, Z.; Chen, D.; Kan, J. Sublethal injury and recovery of Escherichia coli O157:H7 and K-12 after exposure to lactic acid. Food Control 2017, 82, 190–195. [Google Scholar] [CrossRef]
- Nasilowska, J.; Sokolowska, B.; Fonberg-Broczek, M. Behavior of Listeria innocua Strains Under Pressure Treatment—Inactivation and Sublethal Injury. Polish J. Food Nutr. Sci. 2019, 69, 45–52. [Google Scholar] [CrossRef]
- Sokolowska, B.; Skapska, S.; Niezgoda, J.; Rutkowska, M.; Dekowska, A.; Rzoska, S. Inactivation and Sublethal Injury of Escherichia coli and Listeria Innocua by High Hydrostatic Pressure. In Proceedings of the 51 EHPRG International Meeting, London, UK, 1–6 September 2013. [Google Scholar]
- Shao, L.; Tian, X.; Yu, Q.; Xu, L.; Li, X.; Dai, R. Inactivation and recovery kinetics of Escherichia coli O157:H7 treated with ohmic heating in broth. LWT 2019, 110, 1–7. [Google Scholar] [CrossRef]
- Kobayashi, H.; Miyamoto, T.; Hashimoto, Y.; Kiriki, M.; Motomatsu, A.I.; Honjoh, K.-I.; Iio, M. Identification of Factors Involved in Recovery of Heat-Injured Salmonella Enteritidis. J. Food Prot. 2005, 68, 932–941. [Google Scholar] [CrossRef] [PubMed]
- Hui, S.; Chen, Z.; Pang, Y.; Kan, J. Technology. Study on Lactic Acid-Induced Sublethal Injury and Resuscitation of Foodborne Pathogen Escherichia coli O157:H7. J. Chin. Inst. Food Sci. Technol. 2018, 18, 9–15. [Google Scholar]
- Suo, B.; Wang, Y. Evaluation of a multiplex selective enrichment broth SEL for simultaneous detection of injured Salmonella, Escherichia coli O157:H7 and Listeria monocytogenes. Braz. J. Microbiol. 2014, 44, 737–742. [Google Scholar] [CrossRef] [Green Version]
- Sokołowska, B.; Skąpska, S.; Niezgoda, J.; Rutkowska, M.; Dekowska, A.; Rzoska, S.J. Inactivation and sublethal injury of Escherichia coli and Listeria innocua by high hydrostatic pressure in model suspensions and beetroot juice. High Press. Res. 2014, 34, 147–155. [Google Scholar] [CrossRef]
- Abe, A.; Furukawa, S.; Migita, Y.; Tanaka, M.; Ogihara, H.; Morinaga, Y. Sublethal High Hydrostatic Pressure Treatment Reveals the Importance of Genes Coding Cytoskeletal Protein in Escherichia coli Morphogenesis. Curr. Microbiol. 2013, 67, 515–521. [Google Scholar] [CrossRef]
- Yamamoto, K.; Zhang, X.; Inaoka, T.; Morimatsu, K.; Kimura, K.; Nakaura, Y. Bacterial Injury Induced by High Hydrostatic Pressure. Food Eng. Rev. 2021, 13, 442–453. [Google Scholar] [CrossRef]
- Bi, X.; Wang, Y.; Zhao, F.; Sun, Z.; Hu, X.; Liao, X. Sublethal injury and recovery of Escherichia coli O157:H7 by high pressure carbon dioxide. Food Control 2015, 50, 705–713. [Google Scholar] [CrossRef]
- Noriega, E.; Velliou, E.; Van Derlinden, E.; Mertens, L.; Van Impe, J.F.M. Effect of cell immobilization on heat-induced sublethal injury of Escherichia coli, Salmonella Typhimurium and Listeria innocua. Food Microbiol. 2013, 36, 355–364. [Google Scholar] [CrossRef] [PubMed]
- Lan, L.; Shi, H. New insights into the formation and recovery of sublethally injured Escherichia coli O157:H7 induced by lactic acid. Food Microbiol. 2022, 102, 103918. [Google Scholar] [CrossRef]
- Somolinos, M.; García, D.; Pagán, R.; Mackey, B. Relationship between sublethal injury and microbial inactivation by the combination of high hydrostatic pressure and citral or tert-butyl hydroquinone. Appl. Environ. Microbiol. 2008, 74, 7570–7577. [Google Scholar] [CrossRef] [Green Version]
- Phuvasate, S.; Su, Y.-C. Alteration of structure and cellular protein profiles of Vibrio parahaemolyticus cells by high pressure treatment. Food Control 2015, 50, 831–837. [Google Scholar] [CrossRef]
- Ma, J.; Wang, H.; Yu, L.; Yuan, W.; Fu, W.; Gao, F.; Jiang, Y. Dynamic self-recovery of injured Escherichia coli O157:H7 induced by high pressure processing. LWT 2019, 113, 108308. [Google Scholar] [CrossRef]
- Inaoka, T.; Kimura, K.; Morimatsu, K.; Yamamoto, K. Characterization of high hydrostatic pressure-injured Bacillus subtilis cells. Biosci. Biotechnol. Biochem. 2017, 81, 1235–1240. [Google Scholar] [CrossRef] [Green Version]
- Aertsen, A.; De Spiegeleer, P.; Vanoirbeek, K.; Lavilla, M.; Michiels, C.W. Induction of oxidative stress by high hydrostatic pressure in Escherichia coli. Appl. Environ. Microbiol. 2005, 71, 2226–2231. [Google Scholar] [CrossRef] [Green Version]
- Bravim, F.; Mota, M.; Fernandes, A.; Fernandes, P. High hydrostatic pressure leads to free radicals accumulation in yeast cells triggering oxidative stress. FEMS Yeast Res. 2016, 16, fow052. [Google Scholar] [CrossRef] [Green Version]
- Pinske, C.; Sargent, F. Exploring the directionality of Escherichia coli formate hydrogenlyase: A membrane-bound enzyme capable of fixing carbon dioxide to organic acid. MicrobiologyOpen 2016, 5, 721–737. [Google Scholar] [CrossRef] [PubMed]
- Sokol, K.P.; Robinson, W.E.; Oliveira, A.R.; Zacarias, S.; Lee, C.-Y.; Madden, C.; Bassegoda, A.; Hirst, J.; Pereira, I.A.C.; Reisner, E. Reversible and Selective Interconversion of Hydrogen and Carbon Dioxide into Formate by a Semiartificial Formate Hydrogenlyase Mimic. J. Am. Chem. Soc. 2019, 141, 17498–17502. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Martín, M.G.; Sender, P.D.; Peirú, S.; de Mendoza, D.; Magni, C. Acid-inducible transcription of the operon encoding the citrate lyase complex of Lactococcus lactis Biovar diacetylactis CRL264. J. Bacteriol. 2004, 186, 5649–5660. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Figueira, R.; Brown, D.R.; Ferreira, D.; Eldridge, M.J.G.; Burchell, L.; Pan, Z.; Helaine, S.; Wigneshweraraj, S. Adaptation to sustained nitrogen starvation by Escherichia coli requires the eukaryote-like serine/threonine kinase YeaG. Sci. Rep. 2015, 5, 17524. [Google Scholar] [CrossRef]
- Schneider, B.L.; Kiupakis, A.K.; Reitzer, L.J. Arginine catabolism and the arginine succinyltransferase pathway in Escherichia coli. J. Bacteriol. 1998, 180, 4278–4286. [Google Scholar] [CrossRef] [Green Version]
- Barbosa, M.D.F.S.; Yang, G.; Fang, J.; Kurilla, M.G.; Pompliano, D.L. Development of a whole-cell assay for peptidoglycan biosynthesis inhibitors. Antimicrob. Agents Chemother. 2002, 46, 943–946. [Google Scholar] [CrossRef] [Green Version]
- Shi, T.; Ma, Q.; Liu, X.; Hao, Y.; Li, Y.; Xu, Q.; Xie, X.; Chen, N. Double deletion of murA and murB induced temperature sensitivity in Corynebacterium glutamicum. Bioengineered 2019, 10, 561–573. [Google Scholar] [CrossRef] [Green Version]
- Gao, C.; Dai, Y.; Chang, W.; Fang, C.; Wang, Z.; Ma, X. VraSR has an important role in immune evasion of Staphylococcus aureus with low level vancomycin resistance. Microbes Infect. 2019, 21, 361–367. [Google Scholar] [CrossRef]
- Campbell, J.W.; Cronan, J.E., Jr. Escherichia coli FadR positively regulates transcription of the fabB fatty acid biosynthetic gene. J. Bacteriol. 2001, 183, 5982–5990. [Google Scholar] [CrossRef] [Green Version]
- Zhang, H.; Wang, P.; Qi, Q. Molecular effect of FadD on the regulation and metabolism of fatty acid in Escherichia coli. FEMS Microbiol. Lett. 2006, 259, 249–253. [Google Scholar] [CrossRef] [Green Version]
- Tran, Q.H.; Bongaerts, J.; Vlad, D.; Unden, G. Requirement for the Proton-Pumping NADH Dehydrogenase I of Escherichia coli in Respiration of NADH to Fumarate and Its Bioenergetic Implications. Eur. J. Biochem. 1997, 244, 155–160. [Google Scholar] [CrossRef] [PubMed]
- Brudecki, L.E.; Grindstaff, J.J.; Ahmad, Z. Role of αPhe-291 residue in the phosphate-binding subdomain of catalytic sites of Escherichia coli ATP synthase. Arch. Biochem. Biophys. 2008, 471, 168–175. [Google Scholar] [CrossRef] [PubMed]
- Cordero, T.; Aragonés, V.; Daròs, J.-A. Large-scale Production of Recombinant RNAs on a Circular Scaffold Using a Viroid-derived System in Escherichia coli. JoVE 2018, 141, e58472. [Google Scholar] [CrossRef] [PubMed]
- Johnson, D.B.F.; Xu, J.; Shen, Z.; Takimoto, J.K.; Schultz, M.D.; Schmitz, R.J.; Xiang, Z.; Ecker, J.R.; Briggs, S.P.; Wang, L. RF1 knockout allows ribosomal incorporation of unnatural amino acids at multiple sites. Nat. Chem. Biol. 2011, 7, 779–786. [Google Scholar] [CrossRef] [Green Version]
- Pan, Y.; Cheng, J.-H.; Lv, X.; Sun, D.-W. Assessing the inactivation efficiency of Ar/O2 plasma treatment against Listeria monocytogenes cells: Sublethal injury and inactivation kinetics. LWT 2019, 111, 318–327. [Google Scholar] [CrossRef]
- Nguyen, H.T.M.; Akanuma, G.; Hoa, T.T.M.; Nakai, Y.; Kimura, K.; Yamamoto, K.; Inaoka, T. Ribosome Reconstruction during Recovery from High-Hydrostatic-Pressure-Induced Injury in Bacillus subtilis. Appl. Environ. Microbiol. 2019, 86, e01640-19. [Google Scholar] [CrossRef] [PubMed]
- Guo, M.; Liu, J.; Xu, Z.; Wang, J.; Li, T.; Lei, H.; Duan, X.; Sun, Y.; Zhang, X.; Huang, R. 2-Methoxy-1,4-naphthoquinone Induces Metabolic Shifts in Penicillium Digitatum Revealed by High-Dimensional Biological Data. J. Agric. Food Chem. 2020, 68, 9697–9706. [Google Scholar] [CrossRef]
- Sarker, M.R.; Akimoto, S.; Ono, T.; Kinouchi, T.; Ohnishi, Y. Molecular Cloning of the leuB Gene from Bacteroides fragilis by Functional Complementation in Escherichia coli. Microbiol. Immunol. 1995, 39, 19–25. [Google Scholar] [CrossRef]
- Ricca, E.; Calvo, J.M. The nucleotide sequence of leuA from Salmonella typhimurium. Nucleic Acids Res. 1990, 18, 1290. [Google Scholar] [CrossRef] [Green Version]
- Bajerski, F.; Stock, J.; Hanf, B.; Darienko, T.; Heine-Dobbernack, E.; Lorenz, M.; Naujox, L.; Keller, E.R.J.; Schumacher, H.M.; Friedl, T.; et al. ATP Content and Cell Viability as Indicators for Cryostress Across the Diversity of Life. Front. Physiol. 2018, 9, 921. [Google Scholar] [CrossRef]
- Gao, X.; Yang, Y.; Wang, J.; Zhang, L.; Sun, C.; Wang, Y.; Zhang, J.; Dong, H.; Zhang, H.; Gao, C.; et al. Inhibition of mitochondria NADH–Ubiquinone oxidoreductase (complex I) sensitizes the radioresistant glioma U87MG cells to radiation. Biomed. Pharmacother. 2020, 129, 110460. [Google Scholar] [CrossRef] [PubMed]
- Gürgan, M.; Erkal, N.A.; Özgür, E.; Gündüz, U.; Eroglu, I.; Yücel, M. Transcriptional Profiling of Hydrogen Production Metabolism of Rhodobacter capsulatus under Temperature Stress by Microarray Analysis. Int. J. Mol. Sci. 2015, 16, 13781–13797. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- García, D.; Mañas, P.; Gómez, N.; Raso, J.; Pagán, R. Biosynthetic requirements for the repair of sublethal membrane damage in Escherichia coli cells after pulsed electric fields. J. Appl. Microbiol. 2006, 100, 428–435. [Google Scholar] [CrossRef]
- Chilton, P.; Isaacs, N.S.; Mañas, P.; Mackey, B.M. Biosynthetic requirements for the repair of membrane damage in pressure-treated Escherichia coli. Int. J. Food Microbiol. 2001, 71, 101–104. [Google Scholar] [CrossRef]
- Nikparvar, B.; Subires, A.; Capellas, M.; Hernandez, M.; Bar, N. A Dynamic Model of Membrane Recovery Mechanisms in Bacteria following High Pressure Processing. IFAC-PapersOnLine 2019, 52, 243–250. [Google Scholar] [CrossRef]
- Arroyo, C.; Somolinos, M.; Cebrián, G.; Condón, S.; Pagán, R. Pulsed electric fields cause sublethal injuries in the outer membrane of Enterobacter sakazakii facilitating the antimicrobial activity of citral. Lett. Appl. Microbiol. 2010, 51, 525–531. [Google Scholar] [CrossRef]
- Yu, L.; Zhang, S.; Xu, Y.; Mi, X.; Xing, T.; Li, J.; Zhang, L.; Gao, F.; Jiang, Y. Acid resistance of E. coli O157:H7 and O26:H11 exposure to lactic acid revealed by transcriptomic analysis. LWT 2021, 136, 110352. [Google Scholar] [CrossRef]






Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Hao, J.-Y.; Lei, Y.-Q.; Shi, J.-Y.; Zhao, W.-B.; Gan, Z.-L.; Hu, X.; Sun, A.-D. Integrative Physiological and Transcriptome Analysis Reveals the Mechanism for the Repair of Sub-Lethally Injured Escherichia coli O157:H7 Induced by High Hydrostatic Pressure. Foods 2022, 11, 2377. https://doi.org/10.3390/foods11152377
Hao J-Y, Lei Y-Q, Shi J-Y, Zhao W-B, Gan Z-L, Hu X, Sun A-D. Integrative Physiological and Transcriptome Analysis Reveals the Mechanism for the Repair of Sub-Lethally Injured Escherichia coli O157:H7 Induced by High Hydrostatic Pressure. Foods. 2022; 11(15):2377. https://doi.org/10.3390/foods11152377
Chicago/Turabian StyleHao, Jing-Yi, Yu-Qing Lei, Jun-Yan Shi, Wan-Bin Zhao, Zhi-Lin Gan, Xin Hu, and Ai-Dong Sun. 2022. "Integrative Physiological and Transcriptome Analysis Reveals the Mechanism for the Repair of Sub-Lethally Injured Escherichia coli O157:H7 Induced by High Hydrostatic Pressure" Foods 11, no. 15: 2377. https://doi.org/10.3390/foods11152377