Integrated Metabolomic and Transcriptomic Analysis Reveals Differential Flavonoid Accumulation and Its Underlying Mechanism in Fruits of Distinct Canarium album Cultivars
Abstract
:1. Introduction
2. Materials and Methods
2.1. Materials
2.2. Non-Targeted Metabolomics Analysis
2.2.1. Sample Preparation
2.2.2. Chromatographic Conditions
2.2.3. Mass Spectrometry Conditions
2.2.4. Data Analysis
2.3. RNA Sequencing
2.3.1. Total RNA Extraction and Detection
2.3.2. Library Preparation for RNA-Seq
2.3.3. Data Analysis
2.3.4. Identification of MBW (MYB- bHLH-WD40) Complex Genes from Transcriptome Data
2.3.5. qRT-PCR Verification
3. Results
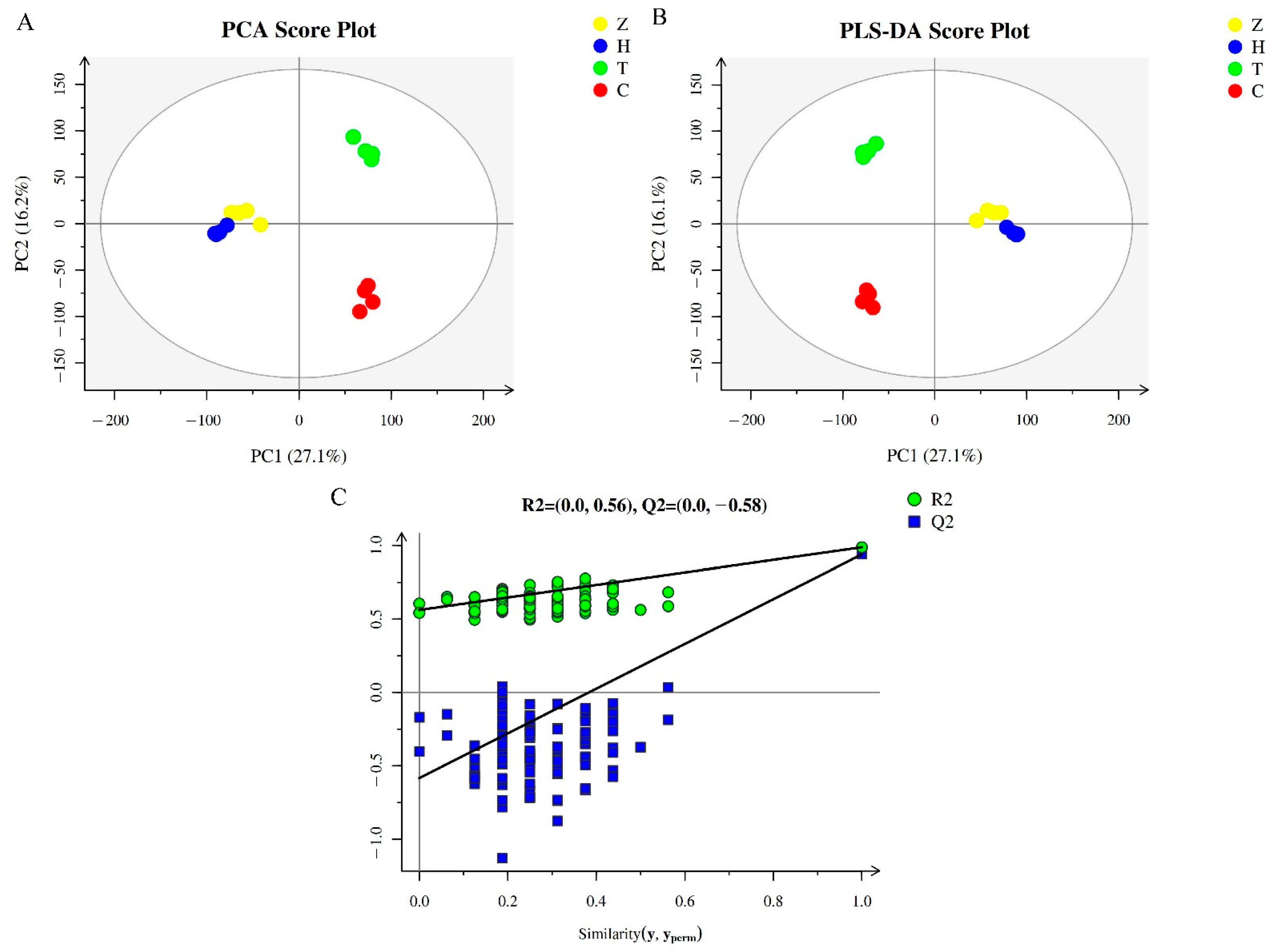
3.1. Quality Control (QC) Analysis of Metabolome Data
3.2. Screening of Differentially Accumulated Metabolites (DAMs) among Fruits from Different C. album Cultivars
3.3. KEGG Pathway Enrichment Analysis of DAMs and Screening of DAFs (Differentially Accumulated Flavonoids)
3.4. Comparative Transcriptomic Analysis
3.4.1. Overview of the RNA-Seq Data
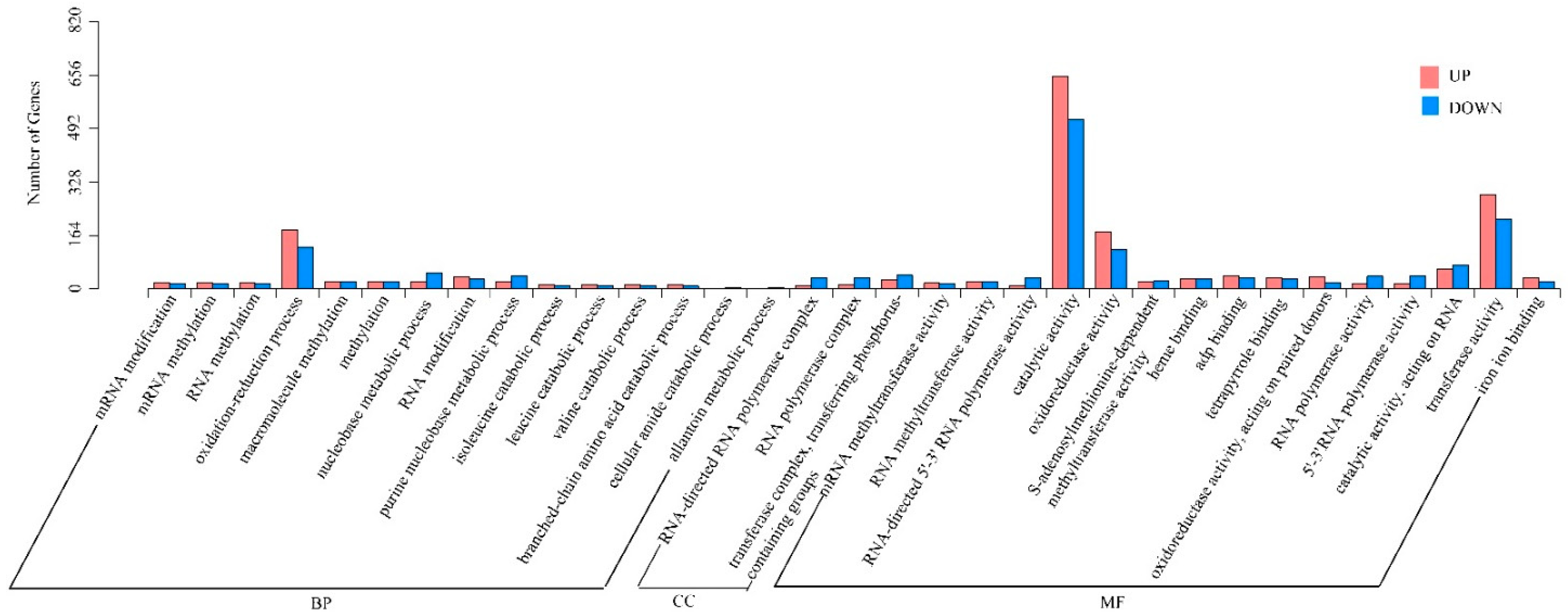
3.4.2. Identification and Enrichment Analysis of DEGs
- 22 upregulated and 22 downregulated DEGs involved in plant hormone signal transduction;
- 9 upregulated and 7 downregulated DEGs involved in carotenoid biosynthesis;
- 14 upregulated and 15 downregulated DEGs involved in phenylpropane biosynthesis;
- 30 upregulated and 3 downregulated DEGs involved in plant pathogen interaction;
- 8 upregulated and 2 downregulated DEGs involved in flavonoid biosynthesis;
- 5 downregulated and 7 upregulated DEGs involved in ABC transport;
- 9 fatty acid elongation related and 14 fatty acid biosynthesis related DEGs, for which 8 and 2 DEGs, respectively, were inhibited;
- 8 genes were found to be inhibited and 19 genes were induced in glycolysis/gluconeogenesis;
- Among the 15 galactose metabolism related DEGs, 5 DEGs were downregulated;
- Stilbenoid, diarylheptanoid, and gingerol biosynthesis contained 1 downregulated gene and 4 upregulated genes;
- Among the 12 and 9 DEGs involved in phenylalanine metabolism and tryptophan metabolism, 7 and 1 DEGs were upregulated, respectively;
- 1 upregulated and 3 downregulated DEGs involved in limonene and pinene degradation;
- 5 out of 6 DEGs involved in sesquiterpenoid and triterpenoid biosynthesis were induced.
3.5. Conjoint Analysis of Metabolomics and Transcriptomics Data
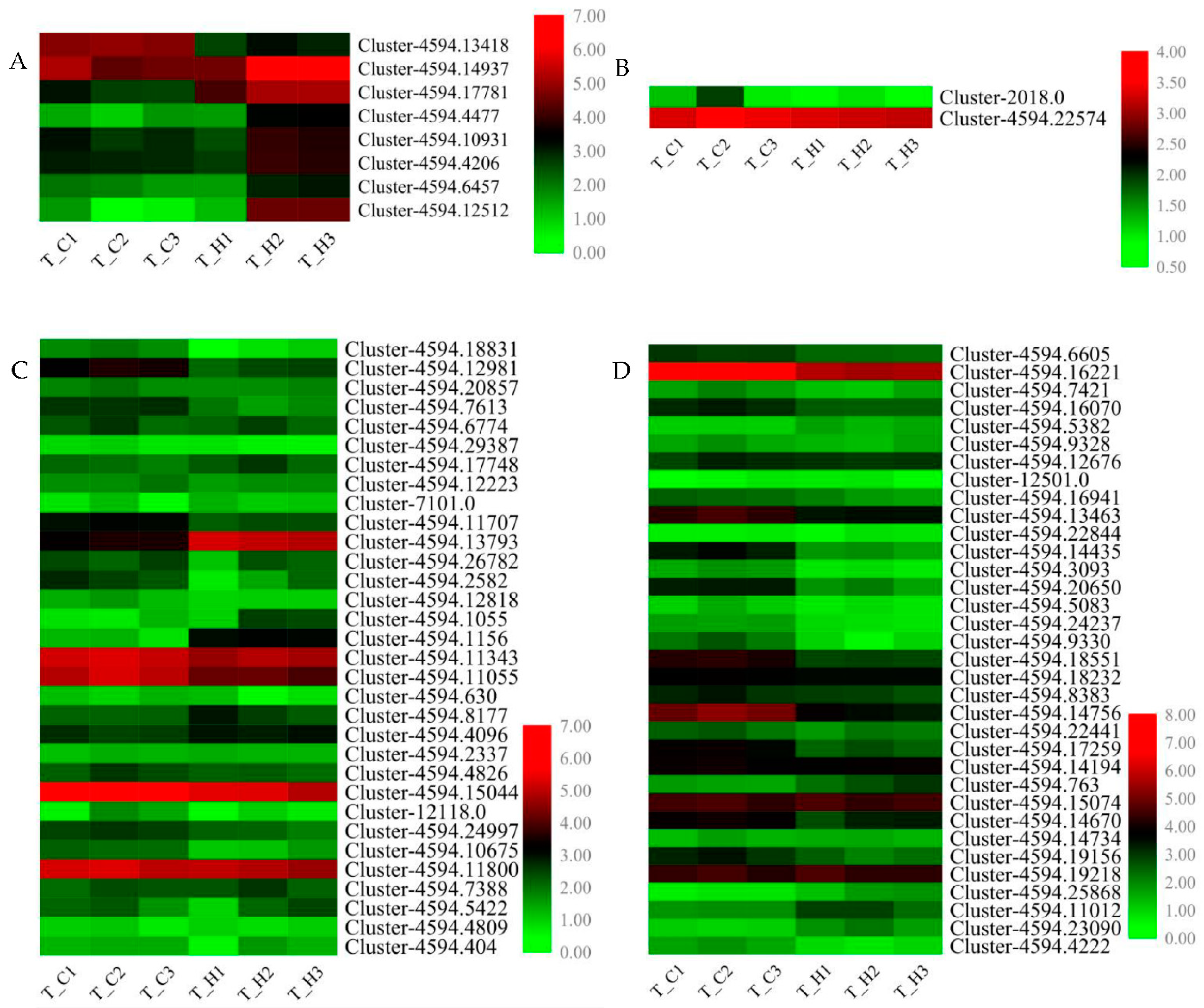
3.6. Identification and Quantitative Real-Time PCR Verification of Flavonoid Biosynthesis Related Structural Genes and Transcription Factor Genes
4. Discussion
4.1. Metabolomics Analysis Distinguished the C. album Cultivars Well and Can Provide a Basis for the Rational Utilization of C. album
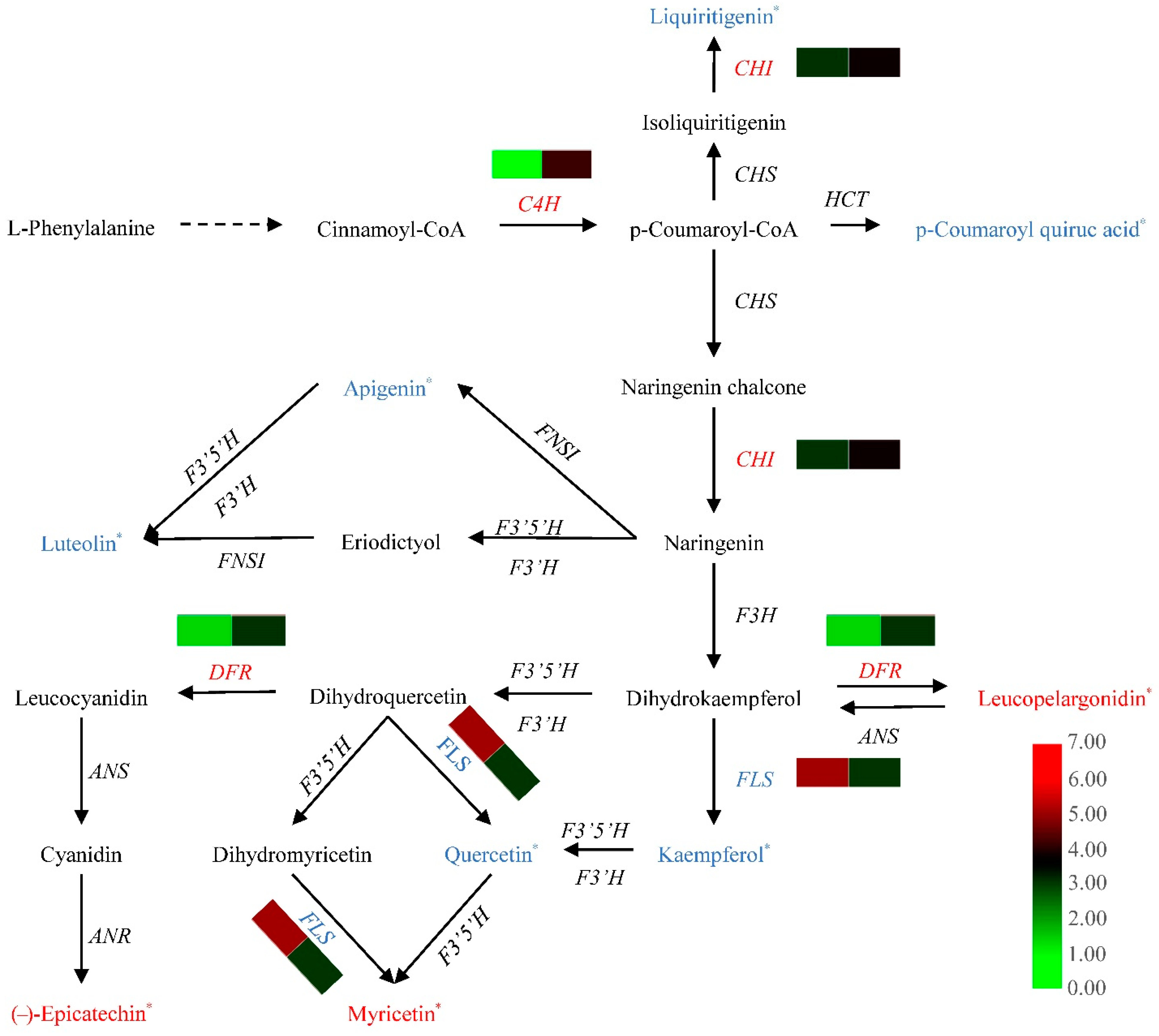
4.2. Flavonoid Accumulation Is a Key Determinant of the Metabolome Differences among C. album Cultivars
4.3. Transcription of Flavonoid Biosynthetic Structural Genes and Transcription Factor Genes Contributed Greatly to the Metabolome Differences among Fruits of Different C. album Cultivars
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- He, C. Canarium album (Lour.) Raeusch. (Qingguo, Chinese olive). In Dietary Chinese Herbs; Springer: Vienna, Austria, 2015; pp. 307–313. [Google Scholar]
- Yeh, Y.T.; Lu, T.J.; Lian, G.T.; Lung, M.C.; Lee, Y.L.; Chiang, A.N.; Hsieh, S.C. Chinese olive (Canarium album L.) fruit regulates glucose utilization by activating AMP-activated protein kinase. FASEB J. 2020, 34, 7866–7884. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yeh, Y.T.; Chiang, A.N.; Hsieh, S.C. Chinese olive (Canarium album L.) fruit extract attenuates metabolic dysfunction in diabetic rats. Nutrients 2017, 9, 1123. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, N.N.; Guo, W.H.; Hu, H.; Zhou, A.R.; Liu, Q.P.; Zheng, B.D.; Zeng, S.X. Effect of a polyphenol-rich Canarium album extract on the composition of the gut microbiota of mice fed a high-fat diet. Molecules 2018, 23, 2188. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yeh, Y.T.; Cho, Y.Y.; Hsieh, S.C.; Chiang, A.N. Chinese olive extract ameliorates hepatic lipid accumulation in vitro and in vivo by regulating lipid metabolism. Sci. Rep. 2018, 8, 1057. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kuo, Y.H.; Yeh, Y.T.; Pan, S.Y.; Hsieh, S.C. Identification and structural elucidation of anti-inflammatory compounds from Chinese olive (Canarium album L.) fruit extracts. Foods 2019, 8, 441. [Google Scholar] [CrossRef] [Green Version]
- Li, J.; Wang, R.; Wang, Y.; Zeng, J.; Xu, Z.; Xu, J.; He, X. Anti-inflammatory benzofuran neolignans from the fruits of Canarium album (Chinese olive). J. Agric. Food Chem. 2022, 70, 1122–1133. [Google Scholar] [CrossRef]
- Zeng, H.; Miao, S.; Zheng, B.; Lin, S.; Jian, Y.; Chen, S.; Zhang, Y. Molecular structural characteristics of polysaccharide fractions from Canarium album (Lour.) Raeusch and their antioxidant activities. J. Food Sci. 2015, 80, H2585–H2596. [Google Scholar] [CrossRef]
- Yan, J.; Peng, C.; Chen, P.; Zhang, W.; Jiang, C.; Sang, S.; Zhu, W.; Yuan, Y.; Hong, Y.; Yao, M. In-vitro anti-Helicobacter pylori activity and preliminary mechanism of action of Canarium album Raeusch. fruit extracts. J. Ethnopharmacol. 2022, 283, 114578. [Google Scholar] [CrossRef]
- Zhang, S.; Huang, Y.; Li, Y.; Wang, Y.; He, X. Anti-neuroinflammatory and antioxidant phenylpropanoids from Chinese olive. Food Chem. 2019, 286, 421–427. [Google Scholar] [CrossRef]
- He, Z.; Xia, W. Microwave-assisted extraction of phenolics from Canarium album L. and identification of the main phenolic compound. Nat. Prod. Res. 2011, 25, 85–92. [Google Scholar] [CrossRef]
- Yang, L.P.; Gu, X.L.; Chen, J.X.; Yang, J.; Tan, S.Y.; Duan, W.J. Chemical constituents from Canarium album Raeusch and their anti-influenza A virus activities. J. Nat. Med. 2018, 72, 808–815. [Google Scholar] [CrossRef] [PubMed]
- Chen, F.; Yang, L.; Huang, Y.; Chen, Y.; Sang, H.; Duan, W.; Yang, J. Isocorilagin, isolated from Canarium album (Lour.) Raeusch, as a potent neuraminidase inhibitor against influenza A virus. Biochem. Biophys. Res. Commun. 2020, 523, 183–189. [Google Scholar] [CrossRef] [PubMed]
- Chen, F.; Yang, L.; Zhai, L.; Huang, Y.; Chen, F.; Duan, W.; Yang, J. Methyl brevifolincarboxylate, a novel influenza virus PB2 inhibitor from Canarium album (Lour.) Raeusch. Chem. Biol. Drug. Des. 2020, 96, 1280–1291. [Google Scholar] [CrossRef]
- Chang, Q.; Su, M.H.; Chen, Q.X.; Zeng, B.Y.; Li, H.H.; Wang, W. Physicochemical properties and antioxidant capacity of Chinese olive (Canarium album L.) cultivars. J. Food Sci. 2017, 82, 1369–1377. [Google Scholar] [CrossRef] [PubMed]
- Alawiye, T.T.; Babalola, O.O. Metabolomics: Current application and prospects in crop production. Biologia 2020, 76, 227–239. [Google Scholar] [CrossRef]
- Ku, K.M.; Becker, T.M.; Juvik, J.A. Transcriptome and metabolome analyses of glucosinolates in two broccoli cultivars following jasmonate treatment for the induction of glucosinolate defense to Trichoplusia ni (hubner). Int. J. Mol. Sci. 2016, 17, 1135. [Google Scholar] [CrossRef] [Green Version]
- Li, Y.; Fang, J.; Qi, X.; Lin, M.; Zhong, Y.; Sun, L.; Cui, W. Combined analysis of the fruit metabolome and transcriptome reveals candidate genes involved in flavonoid biosynthesis in Actinidia arguta. Int. J. Mol. Sci. 2018, 19, 1471. [Google Scholar] [CrossRef] [Green Version]
- Min, K.; Yi, G.; Lee, J.G.; Kim, H.S.; Hong, Y.; Choi, J.H.; Lim, S.; Lee, E.J. Comparative transcriptome and metabolome analyses of two strawberry cultivars with different storability. PLoS ONE 2020, 15, e0242556. [Google Scholar]
- Li, J.; Yan, G.; Duan, X.; Zhang, K.; Zhang, X.; Zhou, Y.; Wu, C.; Zhang, X.; Tan, S.; Hua, X.; et al. Research progress and trends in metabolomics of fruit trees. Front. Plant Sci. 2022, 13, 881856. [Google Scholar] [CrossRef]
- Zhao, J.; Li, H.; Yin, Y.; An, W.; Qin, X.; Wang, Y.; Li, Y.; Fan, Y.; Cao, Y. Transcriptomic and metabolomic analyses of Lycium ruthenicum and Lycium barbarum fruits during ripening. Sci. Rep. 2020, 10, 4354. [Google Scholar] [CrossRef] [Green Version]
- Jia, L.; Wang, J.; Wang, R.; Duan, M.; Qiao, C.; Chen, X.; Ma, G.; Zhou, X.; Zhu, M.; Jing, F.; et al. Comparative transcriptomic and metabolomic analyses of carotenoid biosynthesis reveal the basis of white petal color in Brassica napus. Planta 2021, 253, 8. [Google Scholar] [CrossRef] [PubMed]
- Bai, Q.; He, B.; Cai, Y.; Lian, H.; Zhang, Q. Transcriptomic and metabolomic analyses reveal several critical metabolic pathways and candidate genes involved in resin biosynthesis in Pinus massoniana. Mol. Genet. Genom. 2020, 295, 327–341. [Google Scholar] [CrossRef] [PubMed]
- Zhu, T.; Zhang, M.; Su, H.; Li, M.; Wang, Y.; Jin, L.; Li, M. Integrated metabolomic and transcriptomic analysis reveals differential mechanism of flavonoid biosynthesis in two cultivars of Angelica sinensis. Molecules 2022, 27, 306. [Google Scholar] [CrossRef] [PubMed]
- Lai, R.; Feng, X.; Chen, J.; Zhang, Y.; Wei, X.; Chen, Y.; Cheng, C.; Wu, R. De novo transcriptome assembly and comparative transcriptomic analysis provide molecular insights into low temperature stress response of Canarium album. Sci. Rep. 2021, 11, 10561. [Google Scholar] [CrossRef] [PubMed]
- Lai, R.; Guan, Q.; Shen, C.; Feng, X.; Zhang, Y.; Chen, Y.; Cheng, C.; Wu, R. Integrated sRNA-seq and RNA-seq analysis reveals the regulatory roles of miRNAs in the low-temperature responses of Canarium album. Horticulturae 2022, 8, 667. [Google Scholar] [CrossRef]
- Chen, X.; Wu, Q.; Chen, Z.; Li, T.; Zhang, Z.; Gao, H.; Yun, Z.; Jiang, Y. Changes in pericarp metabolite profiling of four litchi cultivars during browning. Food Res. Int. 2019, 120, 339–351. [Google Scholar] [CrossRef]
- Eriksson, L.; Byrne, T.; Johansson, E.; Trygg, J.; Vikström, C. Multi- and Megavariate Data Analysis Basic Principles and Applications; MKS Umetrics AB: Malmö, Sweden, 2013. [Google Scholar]
- Wang, X.C.; Wu, J.; Guan, M.L.; Zhao, C.H.; Geng, P.; Zhao, Q. Arabidopsis MYB4 plays dual roles in flavonoid biosynthesis. Plant J. 2020, 101, 637–652. [Google Scholar] [CrossRef]
- Wei, Z.; Cheng, Y.; Zhou, C.; Li, D.; Gao, X.; Zhang, S.; Chen, M. Genome-wide identification of direct targets of the TTG1-bHLH-MYB complex in regulating trichome formation and flavonoid accumulation in Arabidopsis Thaliana. Int. J. Mol. Sci. 2019, 20, 5014. [Google Scholar] [CrossRef] [Green Version]
- Premathilake, A.T.; Ni, J.; Bai, S.; Tao, R.; Ahmad, M.; Teng, Y. R2R3-MYB transcription factor PpMYB17 positively regulates flavonoid biosynthesis in pear fruit. Planta 2020, 252, 59. [Google Scholar] [CrossRef]
- Wu, M.; Xu, X.; Hu, X.; Liu, Y.; Cao, H.; Chan, H.; Gong, Z.; Yuan, Y.; Luo, Y.; Feng, B.; et al. SlMYB72 regulates the metabolism of chlorophylls, carotenoids, and flavonoids in tomato fruit. Plant Physiol. 2020, 183, 854–868. [Google Scholar] [CrossRef]
- Li, Z.; Peng, R.; Yao, Q. SlMYB14 promotes flavonoids accumulation and confers higher tolerance to 2,4,6-trichlorophenol in tomato. Plant Sci. 2021, 303, 110796. [Google Scholar] [CrossRef]
- Anwar, M.; Yu, W.; Yao, H.; Zhou, P.; Allan, A.C.; Zeng, L. NtMYB3, an R2R3-MYB from narcissus, regulates flavonoid biosynthesis. Int. J. Mol. Sci. 2019, 20, 5456. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhu, L.; Guan, Y.; Zhang, Z.; Song, A.; Chen, S.; Jiang, J.; Chen, F. CmMYB8 encodes an R2R3 MYB transcription factor which represses lignin and flavonoid synthesis in chrysanthemum. Plant Physiol. Biochem. 2020, 149, 217–224. [Google Scholar] [CrossRef] [PubMed]
- Zhai, R.; Wang, Z.; Zhang, S.; Meng, G.; Song, L.; Wang, Z.; Li, P.; Ma, F.; Xu, L. Two MYB transcription factors regulate flavonoid biosynthesis in pear fruit (Pyrus bretschneideri Rehd.). J. Exp. Bot. 2016, 67, 1275–1284. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ma, D.; Reichelt, M.; Yoshida, K.; Gershenzon, J.; Constabel, C.P. Two R2R3-MYB proteins are broad repressors of flavonoid and phenylpropanoid metabolism in poplar. Plant J. 2018, 96, 949–965. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, W.; Xu, F.; Cheng, S.; Liao, Y. Characterization and functional analysis of a MYB gene (GbMYBFL) related to flavonoid accumulation in Ginkgo biloba. Genes Genom. 2018, 40, 49–61. [Google Scholar] [CrossRef]
- Xu, F.; Ning, Y.; Zhang, W.; Liao, Y.; Li, L.; Cheng, H.; Cheng, S. An R2R3-MYB transcription factor as a negative regulator of the flavonoid biosynthesis pathway in Ginkgo biloba. Funct. Integr. Genom. 2014, 14, 177–189. [Google Scholar] [CrossRef]
- Zhao, Y.; Zhang, Y.Y.; Liu, H.; Zhang, X.S.; Ni, R.; Wang, P.Y.; Gao, S.; Lou, H.X.; Cheng, A.X. Functional characterization of a liverworts bHLH transcription factor involved in the regulation of bisbibenzyls and flavonoids biosynthesis. BMC Plant Biol. 2019, 19, 497. [Google Scholar] [CrossRef]
- Arlotta, C.; Puglia, G.D.; Genovese, C.; Toscano, V.; Karlova, R.; Beekwilder, J.; De Vos, R.C.H.; Raccuia, S.A. MYB5-like and bHLH influence flavonoid composition in pomegranate. Plant Sci. 2020, 298, 110563. [Google Scholar] [CrossRef]
- Zhao, L.; Gao, L.; Wang, H.; Chen, X.; Wang, Y.; Yang, H.; Wei, C.; Wan, X.; Xia, T. The R2R3-MYB, bHLH, WD40, and related transcription factors in flavonoid biosynthesis. Funct. Integr. Genom. 2013, 13, 75–98. [Google Scholar] [CrossRef]
- Xu, W.; Dubos, C.; Lepiniec, L. Transcriptional control of flavonoid biosynthesis by MYB-bHLH-WDR complexes. Trends Plant Sci. 2015, 20, 176–185. [Google Scholar] [CrossRef] [PubMed]
- Ghisoni, S.; Lucini, L.; Rocchetti, G.; Chiodelli, G.; Farinelli, D.; Tombesi, S.; Trevisan, M. Untargeted metabolomics with multivariate analysis to discriminate hazelnut (Corylus avellana L.) cultivars and their geographical origin. J. Sci. Food Agric. 2020, 100, 500–508. [Google Scholar] [CrossRef] [PubMed]
- Mi, S.; Yu, W.; Li, J.; Liu, M.; Wang, X. Characterization and discrimination of chilli peppers based on multi-element and non-targeted metabolomics analysis. LWT-Food Sci. Technol. 2020, 131, 109742. [Google Scholar] [CrossRef]
- Guijas, C.; Montenegro-Burke, J.R.; Warth, B.; Spilker, M.E.; Siuzdak, G. Metabolomics activity screening for identifying metabolites that modulate phenotype. Nat. Biotechnol. 2018, 36, 316–320. [Google Scholar] [CrossRef]
- Xi, W.; Feng, J.; Liu, Y.; Zhang, S.; Zhao, G. The R2R3-MYB transcription factor PaMYB10 is involved in anthocyanin biosynthesis in apricots and determines red blushed skin. BMC Plant Biol. 2019, 19, 287. [Google Scholar] [CrossRef]
- Wang, J.G.; Gao, X.M.; Ma, Z.L.; Chen, J.; Liu, Y.N.; Shi, W.Q. Metabolomic and transcriptomic profiling of three types of litchi pericarps reveals that changes in the hormone balance constitute the molecular basis of the fruit cracking susceptibility of Litchi chinensis cv. Baitangying. Mol. Biol. Rep. 2019, 46, 5295–5308. [Google Scholar] [CrossRef]
- Qiu, W.; Su, W.; Cai, Z.; Dong, L.; Li, C.; Xin, M.; Fang, W.; Liu, Y.; Wang, X.; Huang, Z.; et al. Combined analysis of transcriptome and metabolome reveals the potential mechanism of coloration and fruit quality in yellow and purple Passiflora edulis Sims. J. Agric. Food Chem. 2020, 68, 12096–12106. [Google Scholar] [CrossRef]
- Ghisoni, S.; Lucini, L.; Angilletta, F.; Rocchetti, G.; Farinelli, D.; Tombesi, S.; Trevisan, M. Discrimination of extra-virgin-olive oils from different cultivars and geographical origins by untargeted metabolomics. Food Res. Int. 2019, 121, 746–753. [Google Scholar] [CrossRef]
- Monasterio, R.P.; Olmo-Garcia, L.; Bajoub, A.; Fernandez-Gutierrez, A.; Carrasco-Pancorbo, A. Phenolic compounds profiling of virgin olive oils from different varieties cultivated in mendoza, argentina, by using liquid chromatography-mass spectrometry. J. Agric. Food Chem. 2017, 65, 8184–8195. [Google Scholar] [CrossRef]
- Woo, J.S.; Choo, G.S.; Yoo, E.S.; Kim, S.H.; Lee, J.H.; Han, S.H.; Kim, H.J.; Jung, S.H.; Park, Y.S.; Kim, B.S.; et al. Apigenin induces apoptosis by regulating akt and mapk pathways in human melanoma cell a375sm. Mol. Med. Rep. 2020, 22, 4877–4889. [Google Scholar] [CrossRef]
- Patel, D.K.; Prasad, S.K.; Kumar, R.; Hemalatha, S. An overview on antidiabetic medicinal plants having insulin mimetic property. Asian Pac. J. Trop. Biomed. 2012, 2, 320–330. [Google Scholar] [CrossRef] [Green Version]
- Nani, A.; Murtaza, B.; Sayed Khan, A.; Khan, N.A.; Hichami, A. Antioxidant and anti-inflammatory potential of polyphenols contained in mediterranean diet in obesity: Molecular mechanisms. Molecules 2021, 26, 985. [Google Scholar] [CrossRef] [PubMed]
- Xie, Q.; Cai, X.; Dong, X.; Wang, Y.; Sun, M.; Tai, L.; Xu, Y. Effects of epigallocatechin-3-gallate combined with ascorbic acid and glycerol on the stability and uric acid-lowering activity of epigallocatechin-3-gallate. Pharm. Biol. 2021, 59, 157–166. [Google Scholar] [CrossRef]
- Lee, H.S.; Jeong, G.S. Aromadendrin inhibits t cell activation via regulation of calcium influx and nfat activity. Molecules 2020, 25, 4590. [Google Scholar] [CrossRef] [PubMed]
- Aminzadeh, A.; Salarinejad, A. Effects of myricetin against cadmium-induced neurotoxicity in pc12 cells. Toxicol. Res. 2021, 10, 84–90. [Google Scholar] [CrossRef]
- Liang, F.; Zhang, H.; Gao, H.; Cheng, D.; Zhang, N.; Du, J.; Yue, J.; Du, P.; Zhao, B.; Yin, L. Liquiritigenin decreases tumorigenesis by inhibiting dnmt activity and increasing BRCA1 transcriptional activity in triple-negative breast cancer. Exp. Biol. Med. 2021, 246, 459–466. [Google Scholar] [CrossRef] [PubMed]
- Liu, W.; Feng, Y.; Yu, S.; Fan, Z.; Li, X.; Li, J.; Yin, H. The flavonoid biosynthesis network in plants. Int. J. Mol. Sci. 2021, 22, 12824. [Google Scholar] [CrossRef]
- Tian, J.; Zhang, J.; Han, Z.Y.; Song, T.T.; Li, J.Y.; Wang, Y.R.; Yao, Y.C. McMYB12 transcription factors co-regulate proanthocyanidin and anthocyanin biosynthesis in Malus crabapple. Sci. Rep. 2017, 7, 43715. [Google Scholar] [CrossRef] [Green Version]
- Vimolmangkang, S.; Han, Y.; Wei, G.; Korban, S.S. An apple MYB transcription factor, MdMYB3, is involved in regulation of anthocyanin biosynthesis and flower development. BMC Plant Biol. 2013, 13, 176. [Google Scholar] [CrossRef] [Green Version]
- Li, H.; Tian, J.; Yao, Y.Y.; Zhang, J.; Song, T.T.; Li, K.T.; Yao, Y.C. Identification of leucoanthocyanidin reductase and anthocyanidin reductase genes involved in proanthocyanidin biosynthesis in Malus crabapple plants. Plant Physiol. Biochem. 2019, 139, 141–151. [Google Scholar] [CrossRef]
- Park, J.S.; Kim, J.B.; Cho, K.J.; Cheon, C.I.; Sung, M.K.; Choung, M.G.; Roh, K.H. Arabidopsis R2R3-MYB transcription factor AtMYB60 functions as a transcriptional repressor of anthocyanin biosynthesis in lettuce (Lactuca sativa). Plant Cell Rep. 2008, 27, 985–994. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Deng, M.; Zhang, X.; Luo, J.; Liu, H.; Wen, W.; Luo, H.; Yan, J.; Xiao, Y. Metabolomics analysis reveals differences in evolution between maize and rice. Plant J. 2020, 103, 1710–1722. [Google Scholar] [CrossRef] [PubMed]
- Zhao, P.; Li, X.; Jia, J.; Yuan, G.; Chen, S.; Qi, D.; Cheng, L.; Liu, G. BHLH92 from sheepgrass acts as a negative regulator of anthocyanin/proanthocyandin accumulation and influences seed dormancy. J. Exp. Bot. 2019, 70, 269–284. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, F.; Zhu, H.; Chen, D.; Li, Z.; Peng, R.; Yao, Q. A grape bHLH transcription factor gene, VvbHLH1, increases the accumulation of flavonoids and enhances salt and drought tolerance in transgenic Arabidopsis thaliana. Plant Cell Tissue Organ Cult. 2016, 125, 387–398. [Google Scholar] [CrossRef]


| Metabolites | H vs. C | T vs. C | Z vs. C | H vs. T | Z vs. T | Z vs. H | ||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| VIP | p | VIP | p | VIP | p | VIP | p | VIP | p | VIP | p | |
| (−)-Epigallocatechin * | 1.064 | 0.061 | 0.175 | 0.885 | 0.825 | 0.312 | 1.325 | 0.030 | 1.266 | 0.030 | 0.950 | 0.312 |
| (+)-Pinoresinol | 1.064 | 0.061 | 0.922 | 0.112 | 1.267 | 0.030 | 1.371 | 0.030 | 1.491 | 0.030 | 0.763 | 0.194 |
| (1S,2R,4S)-(-)-Bornyl acetate | 0.664 | 0.194 | 0.544 | 0.470 | 1.469 | 0.030 | 0.431 | 0.665 | 1.494 | 0.030 | 1.593 | 0.030 |
| (2S)-Liquiritigenin * | 1.125 | 0.021 | 0.479 | 0.312 | 1.198 | 0.021 | 1.146 | 0.021 | 1.252 | 0.021 | NA | NA |
| (S)-Abscisic acid | 0.864 | 0.030 | 1.105 | 0.112 | 1.351 | 0.030 | 0.364 | 0.665 | 1.403 | 0.030 | 1.576 | 0.030 |
| 10-Hydroxydecanoic acid | 1.188 | 0.030 | 1.627 | 0.030 | 1.345 | 0.030 | 1.397 | 0.030 | 1.271 | 0.030 | 0.070 | 0.665 |
| 1-Dehydro-[6]-gingerdione | 1.295 | 0.030 | 1.251 | 0.030 | 1.220 | 0.030 | 1.083 | 0.112 | 0.915 | 0.194 | 0.859 | 0.112 |
| 1H-Indole-3-acetamide | 1.357 | 0.030 | 1.399 | 0.030 | 1.346 | 0.030 | 1.364 | 0.030 | 1.417 | 0.030 | 0.972 | 0.312 |
| 1-O-Galloyl-beta-D-glucose | 1.051 | 0.030 | 0.154 | 0.885 | 0.963 | 0.061 | 0.753 | 0.030 | 0.734 | 0.030 | 0.950 | 0.112 |
| 2,3,4,4,6-Peptahydroxychalcone 4-O-glucoside | 1.278 | 0.021 | 1.121 | 0.194 | 1.377 | 0.021 | 1.269 | 0.021 | 1.365 | 0.021 | NA | NA |
| 4-Acetamidobutanoic acid | 1.262 | 0.030 | 1.295 | 0.030 | 1.267 | 0.030 | 0.571 | 0.312 | 0.049 | 0.470 | 1.016 | 0.112 |
| 4-Methoxyflavanone | 0.601 | 0.312 | 0.578 | 0.885 | 1.449 | 0.030 | 0.391 | 0.665 | 1.339 | 0.030 | 0.347 | 0.885 |
| 6-Phosphogluconic acid | 1.334 | 0.021 | NA | NA | 0.938 | 0.021 | 1.336 | 0.021 | 1.067 | 0.021 | NA | NA |
| 9(S)-HPOT | 1.260 | 0.030 | 0.255 | 1.000 | 1.358 | 0.030 | 1.046 | 0.030 | 1.126 | 0.030 | 1.415 | 0.030 |
| Acacetin | 1.259 | 0.030 | 0.709 | 0.665 | 1.399 | 0.030 | 0.317 | 1.000 | 1.206 | 0.030 | 0.805 | 0.312 |
| Alanine | 1.353 | 0.030 | 1.650 | 0.030 | 1.210 | 0.030 | 1.393 | 0.030 | 1.449 | 0.030 | 1.574 | 0.030 |
| Alpha-Linolenic acid | 1.176 | 0.030 | 1.340 | 0.061 | 1.398 | 0.030 | 0.404 | 0.312 | 1.194 | 0.030 | 1.104 | 0.030 |
| Apigenin * | 1.296 | 0.030 | 1.477 | 0.030 | 1.414 | 0.030 | 0.981 | 0.112 | 1.259 | 0.030 | 0.950 | 0.194 |
| Apocynin | 1.271 | 0.030 | 1.141 | 0.061 | 1.343 | 0.030 | 1.194 | 0.030 | 1.187 | 0.061 | 0.284 | 0.665 |
| Aromadendrin * | 1.347 | 0.030 | 1.637 | 0.030 | 1.427 | 0.030 | 1.198 | 0.030 | 0.610 | 0.470 | 0.989 | 0.112 |
| Ascorbate | 1.229 | 0.030 | 0.600 | 0.470 | 1.121 | 0.030 | 1.106 | 0.030 | 1.121 | 0.030 | 1.484 | 0.030 |
| Astragalin * | 0.279 | 0.301 | 0.633 | 0.453 | 1.452 | 0.030 | 0.519 | 0.301 | 1.358 | 0.021 | 1.308 | 0.021 |
| Azelaic acid | 1.319 | 0.030 | 0.844 | 0.312 | 1.418 | 0.030 | 0.892 | 0.194 | 0.935 | 0.312 | 0.989 | 0.112 |
| Betonicine | 1.364 | 0.030 | 1.301 | 0.061 | 1.352 | 0.030 | 1.387 | 0.030 | 1.216 | 0.061 | 0.729 | 0.665 |
| Catechol | 1.268 | 0.030 | 0.804 | 0.312 | 1.399 | 0.030 | 0.029 | 0.312 | 0.230 | 0.312 | 1.273 | 0.061 |
| Chlorogenic acid * | 0.105 | 0.885 | 1.429 | 0.030 | 1.333 | 0.030 | 1.294 | 0.030 | 0.363 | 0.194 | 1.190 | 0.030 |
| Chrysoeriol | 1.339 | 0.030 | 1.513 | 0.030 | 1.316 | 0.030 | 0.260 | 1.000 | 0.942 | 0.194 | 0.099 | 0.665 |
| Cinnamaldehyde | 1.346 | 0.021 | 1.434 | 0.030 | 1.451 | 0.021 | 1.125 | 0.021 | 1.210 | 0.021 | NA | NA |
| cis-Aconitic acid | 0.602 | 0.312 | 1.254 | 0.061 | 1.235 | 0.030 | 1.029 | 0.061 | 0.973 | 0.061 | 1.397 | 0.030 |
| Citramalic acid | 1.182 | 0.030 | 0.130 | 1.000 | 0.804 | 0.194 | 1.148 | 0.030 | 0.803 | 0.112 | 1.593 | 0.030 |
| Citric acid | 1.341 | 0.030 | 0.043 | 0.665 | 1.468 | 0.030 | 1.261 | 0.030 | 1.437 | 0.030 | 0.247 | 1.000 |
| Costunolide | 1.164 | 0.030 | 0.871 | 0.194 | 1.099 | 0.030 | 1.134 | 0.030 | 1.153 | 0.030 | 1.396 | 0.030 |
| Cucurbitacin E | 1.247 | 0.021 | NA | NA | 1.240 | 0.021 | 1.270 | 0.021 | 1.261 | 0.021 | 1.450 | 0.030 |
| D-Maltose | 1.125 | 0.021 | NA | NA | 1.283 | 0.021 | 1.145 | 0.021 | 1.304 | 0.021 | 1.570 | 0.030 |
| Ellagic acid | 1.211 | 0.030 | 1.426 | 0.030 | 1.279 | 0.030 | 0.410 | 0.312 | 0.008 | 1.000 | 1.388 | 0.030 |
| Epicatechin * | 1.209 | 0.030 | 0.471 | 0.885 | 0.981 | 0.112 | 1.145 | 0.061 | 0.078 | 0.312 | 0.104 | 0.312 |
| Estragole | 1.151 | 0.030 | 1.044 | 0.112 | 1.278 | 0.030 | 0.524 | 0.665 | 0.757 | 0.470 | 1.313 | 0.030 |
| Eugenol | 1.374 | 0.021 | 0.222 | 0.312 | 1.480 | 0.021 | 1.081 | 0.069 | 1.163 | 0.069 | NA | NA |
| Fraxetin | 0.978 | 0.030 | 1.377 | 0.030 | 1.203 | 0.030 | 1.220 | 0.030 | 0.371 | 0.470 | 0.023 | 1.000 |
| Fructose-1P | 1.272 | 0.021 | 0.300 | 0.665 | 1.353 | 0.021 | 1.222 | 0.021 | 1.335 | 0.021 | NA | NA |
| gamma-Aminobutyric acid | 1.281 | 0.030 | 1.414 | 0.030 | 1.191 | 0.030 | 1.378 | 0.030 | 1.407 | 0.030 | 0.763 | 0.194 |
| Garbanzol * | 0.507 | 0.312 | 1.523 | 0.030 | 0.884 | 0.312 | 1.100 | 0.061 | 1.218 | 0.061 | 1.217 | 0.030 |
| Genistein | 1.173 | 0.030 | 1.440 | 0.030 | 1.260 | 0.030 | 1.355 | 0.030 | 1.481 | 0.030 | 1.571 | 0.030 |
| Genistin | 0.799 | 0.194 | 1.448 | 0.030 | 1.349 | 0.030 | 1.156 | 0.030 | 0.353 | 0.312 | 1.104 | 0.030 |
| Glycylleucine | 1.236 | 0.030 | 1.358 | 0.030 | 1.393 | 0.030 | 0.690 | 0.312 | 1.265 | 0.030 | 1.576 | 0.030 |
| Herniarin | 1.280 | 0.030 | 1.042 | 0.194 | 1.414 | 0.030 | 0.312 | 1.000 | 0.643 | 0.312 | 1.397 | 0.030 |
| Hydroxypyruvic acid | 1.302 | 0.021 | 0.653 | 0.665 | 1.403 | 0.021 | 1.252 | 0.021 | 1.346 | 0.021 | NA | NA |
| Indoleacetic acid | 1.369 | 0.030 | 1.649 | 0.030 | 1.383 | 0.030 | 1.385 | 0.030 | 1.346 | 0.030 | 0.396 | 0.47 |
| Isopentenyl pyrophosphate | 1.360 | 0.021 | 1.421 | 0.030 | 1.448 | 0.021 | 1.168 | 0.021 | 1.275 | 0.021 | NA | NA |
| Kaempferide * | 0.988 | 0.021 | 0.830 | 0.312 | 0.610 | 0.453 | 0.504 | 0.301 | 0.415 | 0.301 | 1.088 | 0.021 |
| Kaempferol * | 1.342 | 0.030 | 0.118 | 0.312 | 0.125 | 0.470 | 0.796 | 0.194 | 0.252 | 0.885 | 0.291 | 1.000 |
| Lamiide | 1.353 | 0.030 | 0.255 | 0.665 | 1.482 | 0.030 | 0.467 | 0.312 | 0.743 | 0.312 | 0.660 | 0.665 |
| L-Arginine | 0.875 | 0.030 | 1.607 | 0.030 | 0.784 | 0.665 | 1.127 | 0.030 | 1.101 | 0.030 | NA | NA |
| L-Asparagine | 1.207 | 0.021 | 1.394 | 0.030 | 1.301 | 0.021 | 1.367 | 0.021 | 1.470 | 0.021 | NA | NA |
| L-Cystine | 1.323 | 0.021 | 0.665 | 0.470 | 1.425 | 0.021 | 0.941 | 0.021 | 1.012 | 0.021 | NA | NA |
| Leucopelargonidin * | 1.123 | 0.030 | NA | NA | 1.288 | 0.030 | 1.223 | 0.030 | 1.357 | 0.030 | 0.245 | 0.885 |
| L-Histidine | 1.144 | 0.021 | 1.207 | 0.112 | 1.233 | 0.021 | 1.151 | 0.021 | 1.238 | 0.021 | NA | NA |
| Limonene-1,2-diol | 1.174 | 0.021 | 0.647 | 0.470 | 1.265 | 0.021 | 1.200 | 0.021 | 1.291 | 0.021 | 0.859 | 0.112 |
| L-Isoleucine | 1.221 | 0.021 | 0.903 | 0.194 | 1.473 | 0.021 | 1.224 | 0.021 | 1.514 | 0.021 | 0.888 | 0.194 |
| L-Leucine | 1.231 | 0.021 | 1.444 | 0.030 | 1.326 | 0.021 | 1.220 | 0.021 | 1.312 | 0.021 | 0.173 | 0.665 |
| L-Lysine | 0.858 | 0.112 | 1.453 | 0.030 | 1.163 | 0.030 | 1.148 | 0.030 | 0.339 | 0.665 | 1.008 | 0.061 |
| L-Ribulose | 0.900 | 0.194 | 0.903 | 0.194 | 1.032 | 0.030 | 0.442 | 0.665 | 0.752 | 0.112 | NA | NA |
| Luteolin * | 1.141 | 0.030 | 0.182 | 0.665 | 0.783 | 0.194 | 1.291 | 0.030 | 0.580 | 0.885 | 0.389 | 1.000 |
| Malvidin 3-glucoside | 1.412 | 0.030 | 1.662 | 0.030 | 1.510 | 0.030 | 0.617 | 0.312 | 1.355 | 0.030 | 1.498 | 0.030 |
| meso-2,6-Diaminoheptanedioate | 1.368 | 0.030 | 1.585 | 0.030 | 1.384 | 0.030 | 1.396 | 0.030 | 1.453 | 0.030 | 0.336 | 0.470 |
| myo-Inositol | 1.236 | 0.030 | 0.591 | 0.312 | 1.388 | 0.030 | 1.321 | 0.030 | 1.465 | 0.030 | 0.247 | 1.000 |
| Myricetin * | 1.102 | 0.021 | 0.789 | 0.312 | 1.172 | 0.021 | 1.129 | 0.021 | 1.233 | 0.021 | NA | NA |
| Naringenin 7-O-beta-D-glucoside | 1.299 | 0.030 | 1.343 | 0.061 | 1.075 | 0.030 | 1.339 | 0.030 | 1.320 | 0.030 | 1.491 | 0.030 |
| Norsanguinarine | 1.159 | 0.030 | 0.046 | 0.665 | 1.034 | 0.061 | 1.015 | 0.061 | 0.724 | 0.194 | 1.138 | 0.312 |
| Palmitoleic acid | 1.354 | 0.030 | 0.610 | 1.000 | 1.460 | 0.030 | 1.114 | 0.030 | 1.200 | 0.030 | 0.973 | 0.030 |
| p-Coumaroyl quinic acid * | 1.310 | 0.030 | 1.279 | 0.030 | 1.350 | 0.030 | 1.147 | 0.061 | 1.537 | 0.030 | 1.079 | 0.030 |
| Pelargonic acid | 1.219 | 0.030 | 1.081 | 0.112 | 1.164 | 0.030 | 1.299 | 0.030 | 1.322 | 0.030 | 1.284 | 0.030 |
| Perillic acid | 1.002 | 0.112 | 1.306 | 0.061 | 0.988 | 0.312 | 1.204 | 0.030 | 1.184 | 0.061 | 0.010 | 0.665 |
| Procyanidin B2 | 1.123 | 0.030 | 1.312 | 0.030 | 0.968 | 0.030 | 0.577 | 0.312 | 1.056 | 0.112 | 1.491 | 0.030 |
| Pulegone | 1.311 | 0.030 | 0.485 | 0.665 | 1.255 | 0.030 | 1.291 | 0.030 | 1.230 | 0.030 | 0.441 | 0.885 |
| Qing Hau Sau | 1.078 | 0.030 | 1.089 | 0.061 | 1.182 | 0.030 | 1.173 | 0.030 | 1.268 | 0.030 | 0.396 | 0.470 |
| Quercetin * | 1.326 | 0.030 | 1.660 | 0.030 | 1.239 | 0.030 | 0.675 | 0.112 | 0.562 | 0.885 | 0.432 | 0.665 |
| Quercetin 3-O-glucoside * | 1.019 | 0.014 | 1.634 | 0.030 | 1.396 | 0.030 | 1.145 | 0.030 | 0.406 | 0.665 | 0.347 | 0.885 |
| Raucaffricine | 1.366 | 0.030 | 1.544 | 0.030 | 1.431 | 0.030 | 1.307 | 0.030 | 1.142 | 0.061 | 1.348 | 0.030 |
| Salicylic acid | 1.054 | 0.030 | 0.212 | 1.000 | 0.987 | 0.112 | 0.981 | 0.030 | 0.919 | 0.112 | 0.972 | 0.312 |
| Silibinin | 1.345 | 0.030 | 0.792 | 0.312 | 0.988 | 0.112 | 1.343 | 0.030 | 0.900 | 0.312 | 0.403 | 0.47 |
| Syringin | 1.181 | 0.021 | 1.482 | 0.030 | 1.272 | 0.021 | 1.349 | 0.021 | 1.451 | 0.021 | NA | NA |
| Taxifolin * | 1.136 | 0.030 | 0.592 | 0.312 | 1.287 | 0.030 | 0.411 | 1.000 | 0.736 | 0.194 | 1.474 | 0.030 |
| trans-Cinnamate | 1.295 | 0.030 | 1.438 | 0.030 | 1.403 | 0.030 | 0.509 | 0.665 | 0.748 | 0.112 | 1.316 | 0.030 |
| Uridine | 1.339 | 0.021 | 1.489 | 0.030 | 1.443 | 0.021 | 1.176 | 0.021 | 1.265 | 0.021 | NA | NA |
| Vaccenic acid | 1.095 | 0.021 | 0.988 | 0.061 | 1.179 | 0.021 | 1.328 | 0.021 | 1.429 | 0.021 | NA | NA |
| Xanthyletin | 1.184 | 0.030 | 0.572 | 0.665 | 1.345 | 0.030 | 0.993 | 0.061 | 1.144 | 0.030 | 1.085 | 0.112 |
| Sample | Raw Reads | Clean Reads | Clean Bases | Error (%) | Q20 (%) | Q30 (%) | GC Content (%) |
|---|---|---|---|---|---|---|---|
| T_C1 | 22,741,237 | 21,927,175 | 6.58 G | 0.02 | 98.16 | 94.26 | 44.30 |
| T_C2 | 22,753,116 | 21,910,412 | 6.57 G | 0.03 | 98.03 | 93.99 | 44.17 |
| T_C3 | 22,262,027 | 21,498,628 | 6.45 G | 0.02 | 98.22 | 94.48 | 44.39 |
| T_H1 | 22,897,426 | 22,263,553 | 6.68 G | 0.03 | 98.01 | 93.96 | 44.64 |
| T_H2 | 23,830,992 | 22,859,162 | 6.86 G | 0.02 | 98.08 | 94.17 | 44.61 |
| T_H3 | 22,397,559 | 21,655,410 | 6.50 G | 0.02 | 98.14 | 94.29 | 44.79 |
| KEGG Pathways | ID | Input Number | Background Number | p-Value |
|---|---|---|---|---|
| Plant hormone signal transduction | ko04075 | 44 | 213 | 2.29 × 10−5 |
| Carotenoid biosynthesis | ko00906 | 16 | 41 | 2.87 × 10−5 |
| Phenylpropanoid biosynthesis | ko00940 | 29 | 132 | 2.30 × 10−4 |
| Plant–pathogen interaction | ko04626 | 33 | 177 | 1.14 × 10−3 |
| Flavonoid biosynthesis | ko00941 | 10 | 29 | 1.94 × 10−3 |
| Fatty acid elongation | ko00062 | 9 | 25 | 2.55 × 10−3 |
| Fatty acid biosynthesis | ko00061 | 14 | 54 | 2.64 × 10−3 |
| ABC transporters | ko02010 | 12 | 45 | 4.30 × 10−3 |
| Glycolysis/Gluconeogenesis | ko00010 | 27 | 158 | 8.49 × 10−3 |
| Galactose metabolism | ko00052 | 15 | 70 | 8.62 × 10−3 |
| Stilbenoid, diarylheptanoid and gingerol biosynthesis | ko00945 | 5 | 11 | 0.011 |
| Phenylalanine metabolism | ko00360 | 12 | 53 | 0.013 |
| Tryptophan metabolism | ko00380 | 9 | 37 | 0.020 |
| Limonene and pinene degradation | ko00903 | 4 | 10 | 0.032 |
| Sesquiterpenoid and triterpenoid biosynthesis | ko00909 | 6 | 22 | 0.036 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Lai, R.; Shen, C.; Feng, X.; Gao, M.; Zhang, Y.; Wei, X.; Chen, Y.; Cheng, C.; Wu, R. Integrated Metabolomic and Transcriptomic Analysis Reveals Differential Flavonoid Accumulation and Its Underlying Mechanism in Fruits of Distinct Canarium album Cultivars. Foods 2022, 11, 2527. https://doi.org/10.3390/foods11162527
Lai R, Shen C, Feng X, Gao M, Zhang Y, Wei X, Chen Y, Cheng C, Wu R. Integrated Metabolomic and Transcriptomic Analysis Reveals Differential Flavonoid Accumulation and Its Underlying Mechanism in Fruits of Distinct Canarium album Cultivars. Foods. 2022; 11(16):2527. https://doi.org/10.3390/foods11162527
Chicago/Turabian StyleLai, Ruilian, Chaogui Shen, Xin Feng, Minxia Gao, Yongyan Zhang, Xiaoxia Wei, Yiting Chen, Chunzhen Cheng, and Rujian Wu. 2022. "Integrated Metabolomic and Transcriptomic Analysis Reveals Differential Flavonoid Accumulation and Its Underlying Mechanism in Fruits of Distinct Canarium album Cultivars" Foods 11, no. 16: 2527. https://doi.org/10.3390/foods11162527
APA StyleLai, R., Shen, C., Feng, X., Gao, M., Zhang, Y., Wei, X., Chen, Y., Cheng, C., & Wu, R. (2022). Integrated Metabolomic and Transcriptomic Analysis Reveals Differential Flavonoid Accumulation and Its Underlying Mechanism in Fruits of Distinct Canarium album Cultivars. Foods, 11(16), 2527. https://doi.org/10.3390/foods11162527






