Lipidomics and Transcriptome Reveal the Effects of Feeding Systems on Fatty Acids in Yak’s Meat
Abstract
:1. Introduction
2. Materials and Methods
2.1. Animals and Sample Collection
2.2. Determination of Fatty Acids
2.3. RNA Extraction and Sequencing
2.4. Lipids Extraction and MS Data
2.5. Quantitative Real-Time PCR (qPCR)
2.6. Statistical Analysis
3. Results
3.1. Content of Fatty Acids
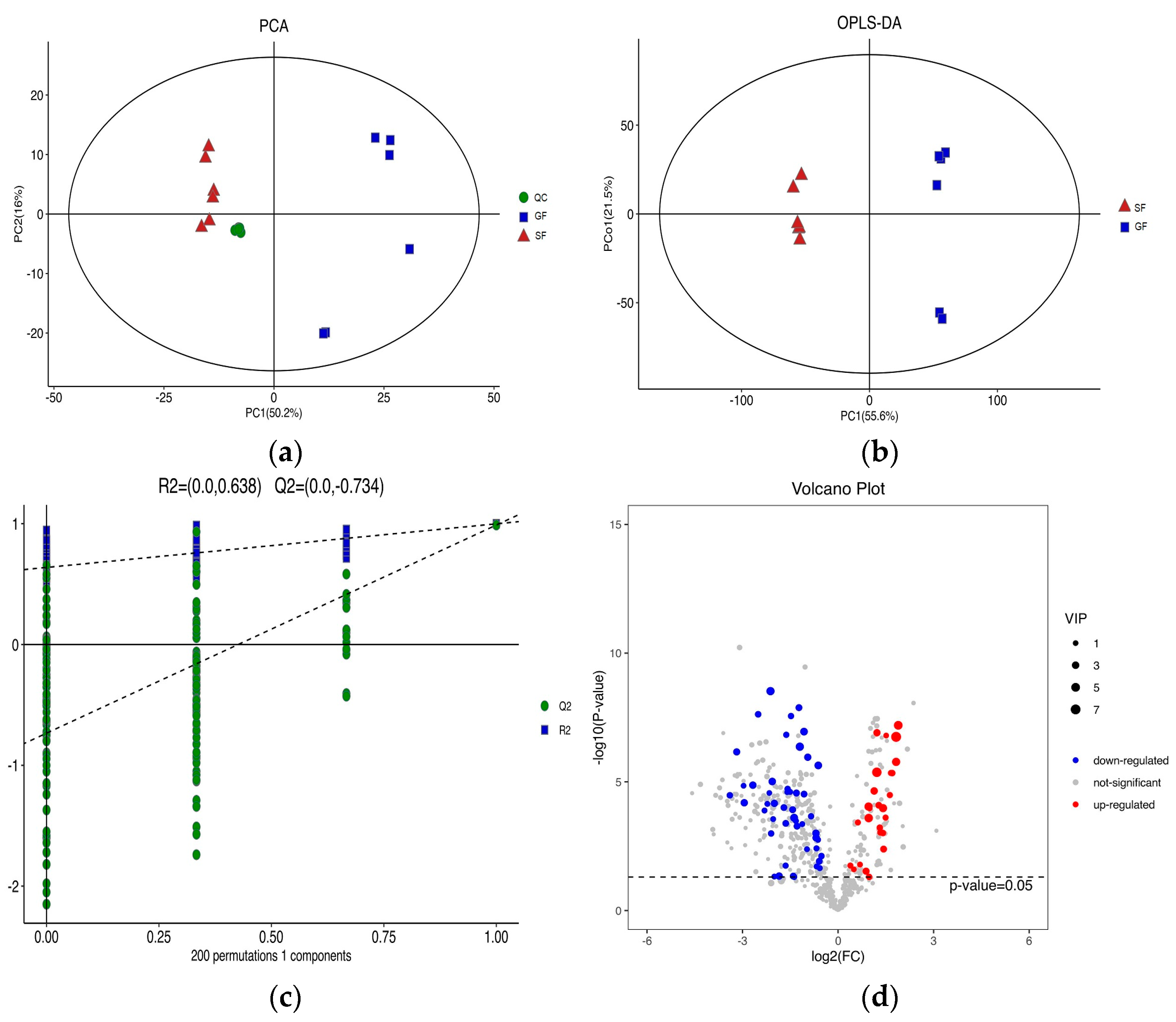
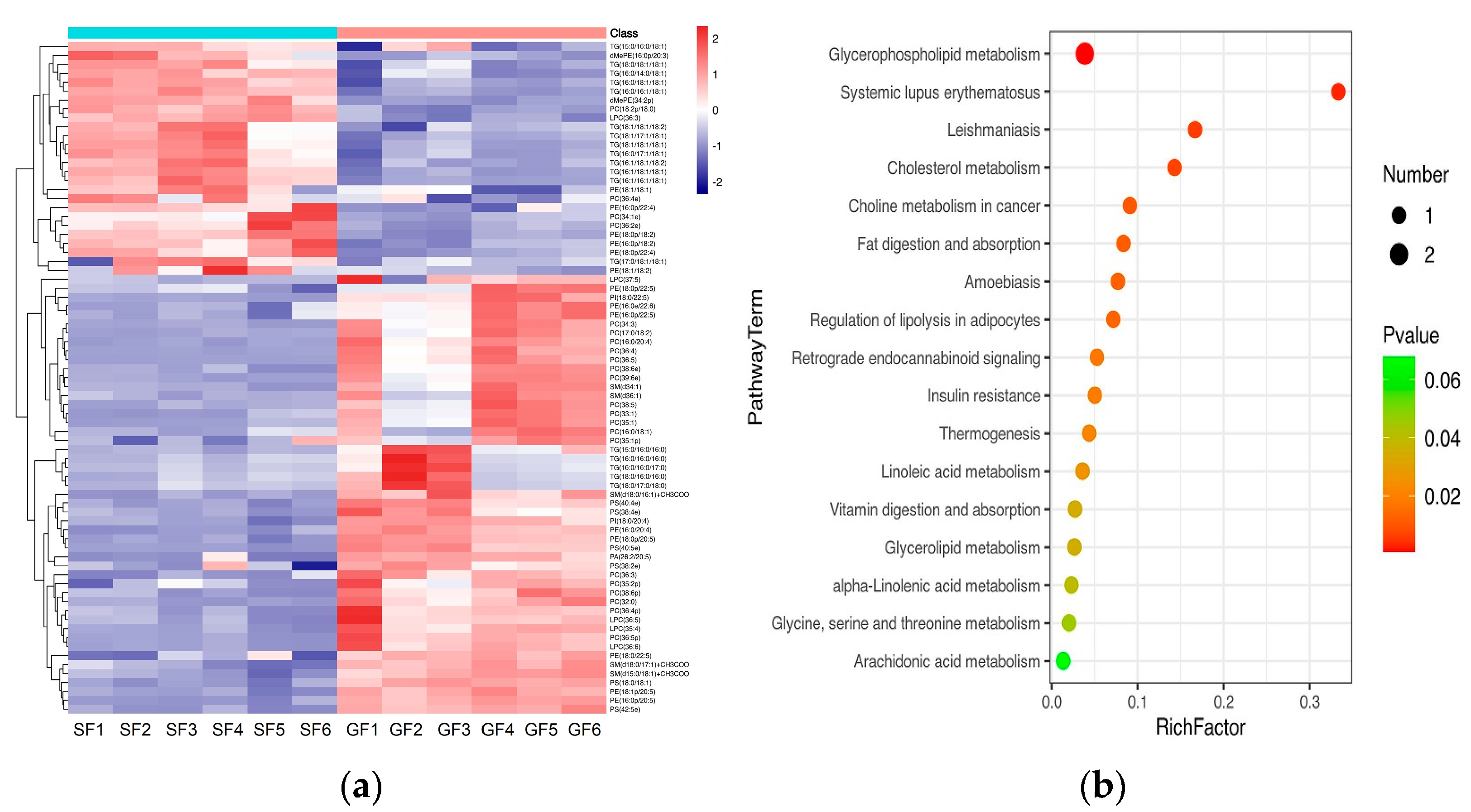
3.2. Lipidomics Results
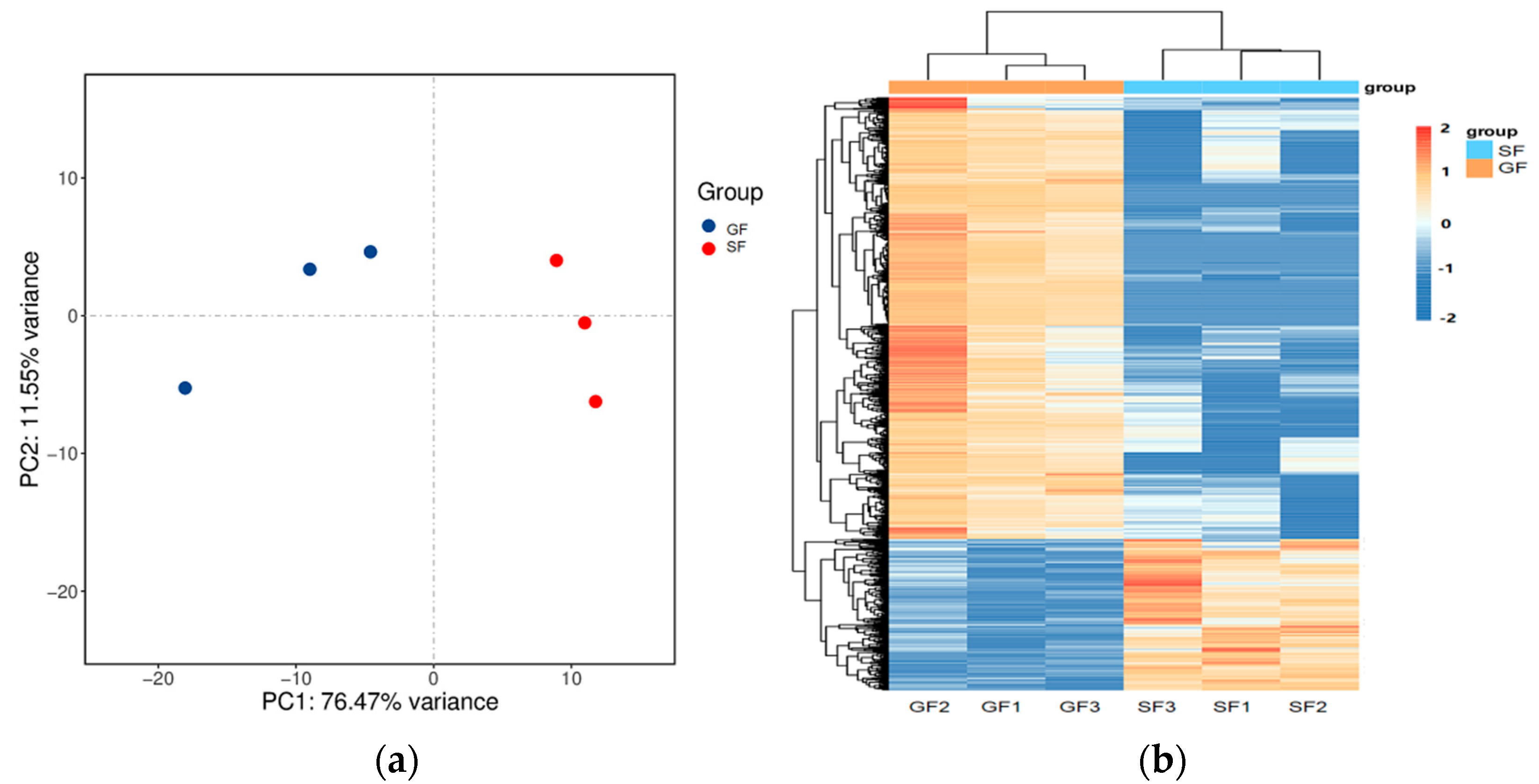
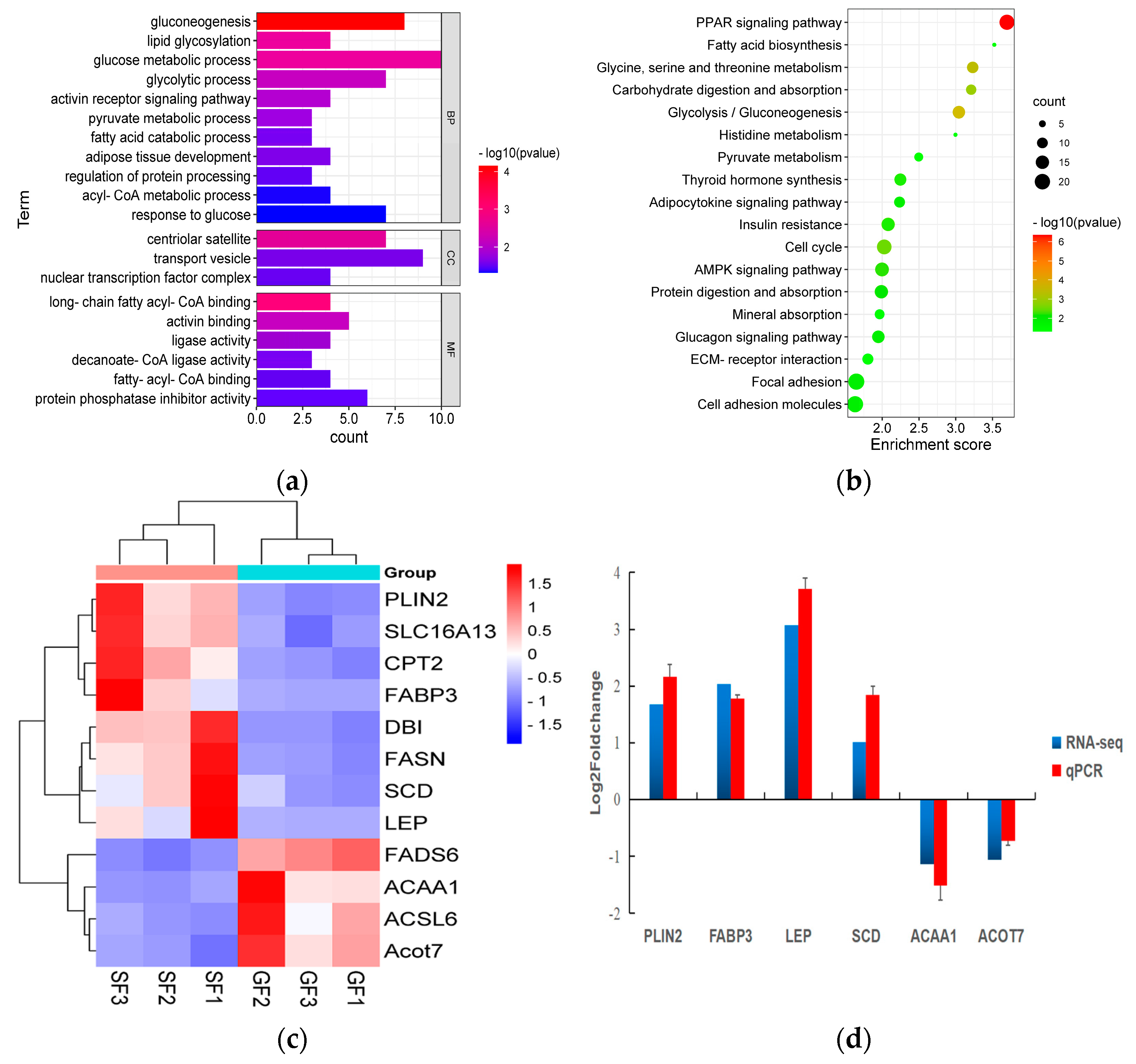
3.3. Transcriptome Results
3.4. Verification of mRNA Sequencing Using qPCR
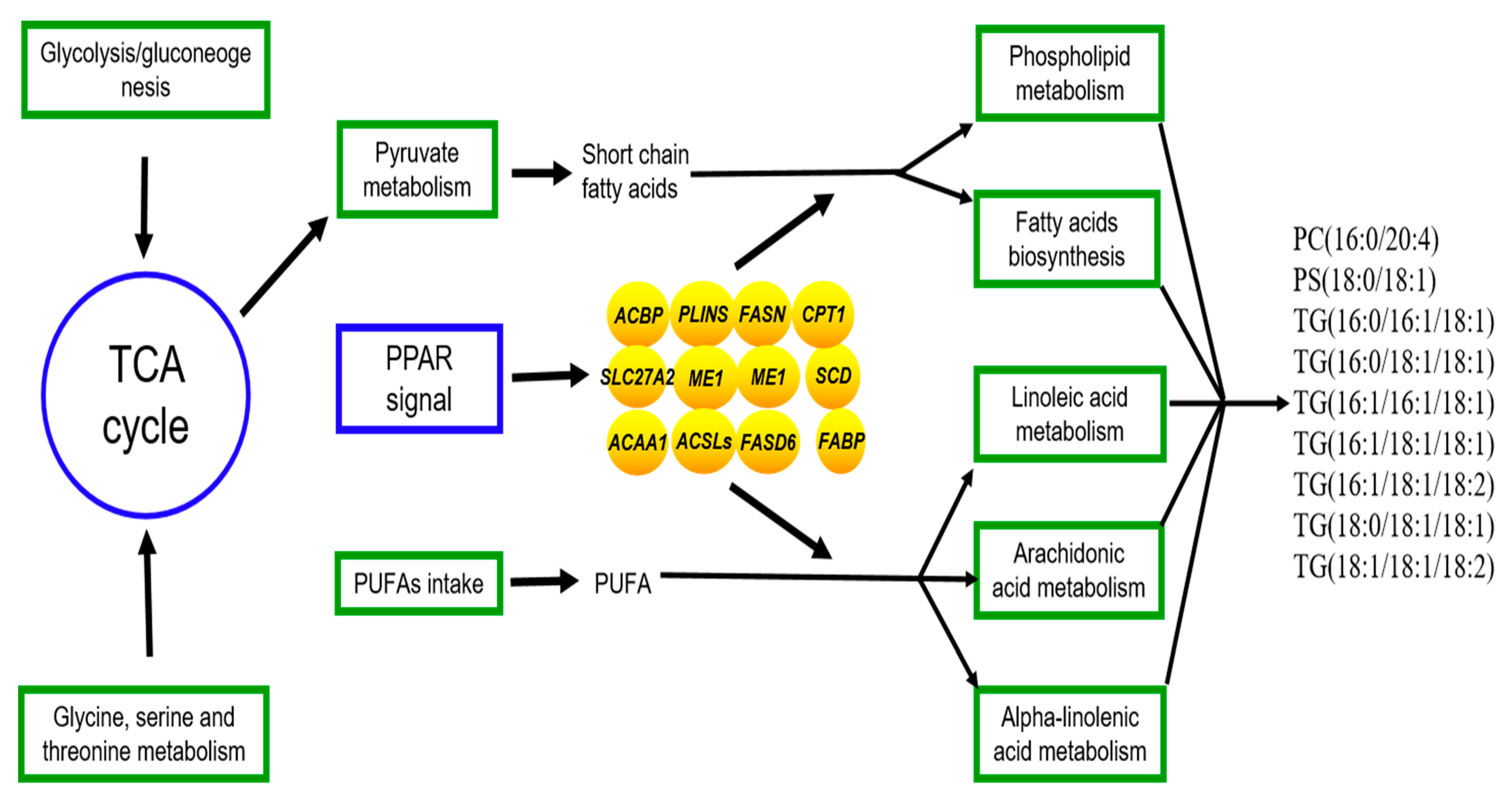
3.5. Joint Analysis of the Lipidomics and Transcriptome
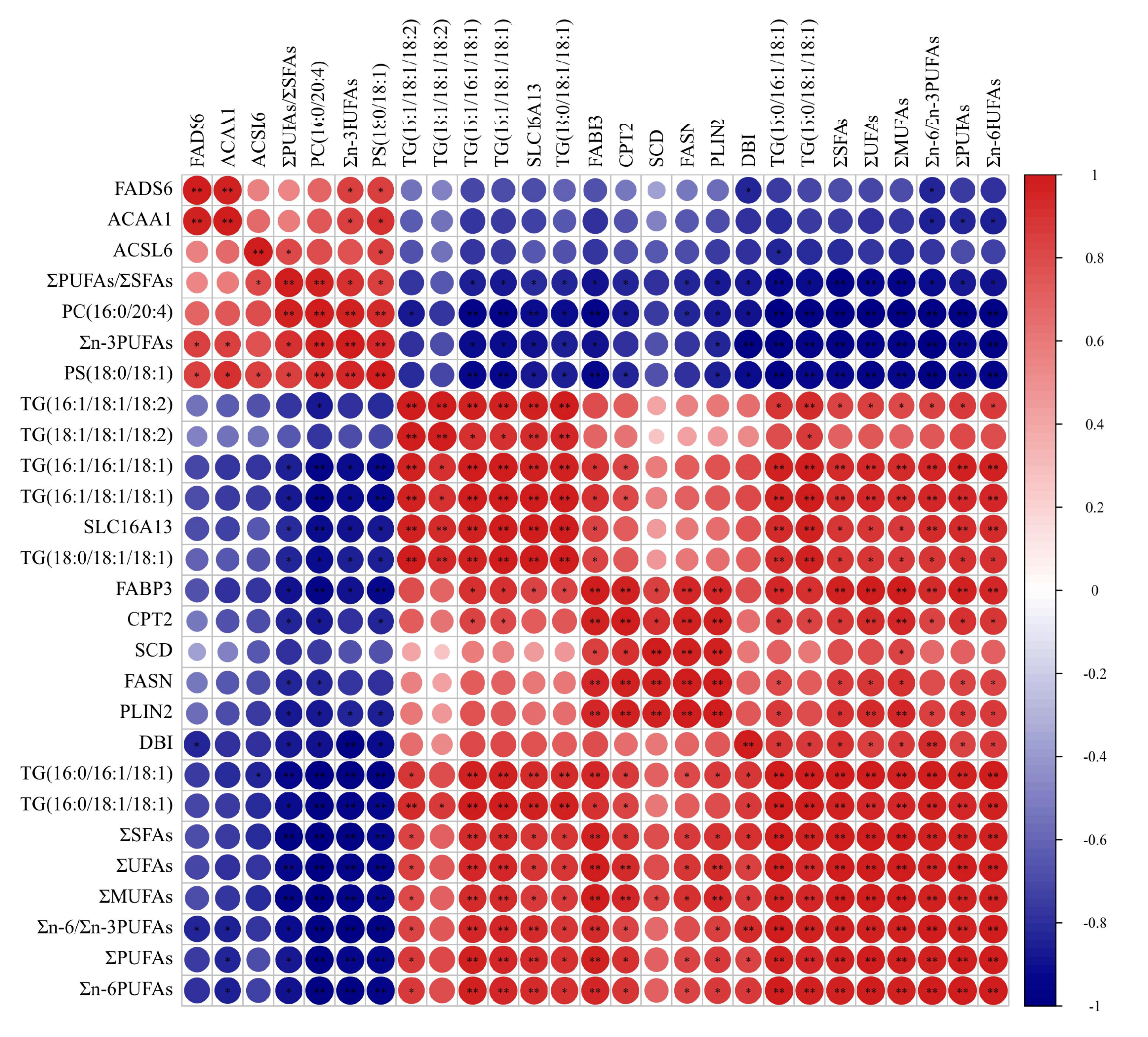
3.6. Correlation of Fatty Acids, DEGs and SDLs
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Jin, C.L.; Gao, C.Q.; Wang, Q.; Zhang, Z.M.; Xu, Y.L.; Li, H.C.; Yan, H.C.; Wang, X.Q. Effects of pioglitazone hydrochloride and vitamin E on meat quality, antioxidant status and fatty acid profiles in finishing pigs. Meat Sci. 2018, 145, 340–346. [Google Scholar] [CrossRef] [PubMed]
- Pan, L.; Chen, L.; Lv, J.; Pang, Y.; Guo, Y.; Pei, P.; Du, H.D.; Yang, L.; Millwood, I.Y.; Walters, R.G.; et al. Association of Red Meat Consumption, Metabolic Markers, and Risk of Cardiovascular Diseases. Front. Nutr. 2022, 9, 833271. [Google Scholar] [CrossRef] [PubMed]
- Webb, E.C.; O’Neill, H.A. The animal fat paradox and meat quality. Meat Sci. 2008, 80, 28–36. [Google Scholar] [CrossRef]
- Wood, J.D.; Enser, M.; Fisher, A.V.; Nute, G.R.; Sheard, P.R. Fat deposition, fatty acid composition and meat quality: A review. Meat Sci. 2008, 78, 343–358. [Google Scholar] [CrossRef]
- Chen, L.H.; Ekine-Dzivenu, C.; Vinsky, M.; Basarab, J.; Aalhus, J.; Dugan, M.E.R.; Fitzsimmons, C.; Stothard, P.; Li, C.X. Genome-wide association and genomic prediction of breeding values for fatty acid composition in subcutaneous adipose and longissimus lumborum muscle of beef cattle. BMC Genet. 2015, 16, 135. [Google Scholar] [CrossRef]
- Ruxton, C.H.S.; Reed, S.C.; Simpson, J.A.; Millington, K.J. The health benefits of omega-3 polyunsaturated fatty acids: A review of the evidence. J. Hum. Nutr. Diet. 2007, 20, 275–285. [Google Scholar] [CrossRef] [PubMed]
- Pewan, S.B.; Otto, J.R.; Huerlimann, R.; Budd, A.M.; Mwangi, F.W.; Edmunds, R.C.; Holman, B.W.B.; Henry, M.L.E.; Kinobe, R.T.; Adegboye, O.A.; et al. Genetics of Omega-3 Long-Chain Polyunsaturated Fatty Acid Metabolism and Meat Eating Quality in Tattykeel Australian White Lambs. Genes 2020, 11, 587. [Google Scholar] [CrossRef] [PubMed]
- Simopoulos, A.P. The importance of the ratio of omega-6/omega-3 essential fatty acids. Biomed. Pharmacother. 2002, 56, 365–379. [Google Scholar] [CrossRef]
- Dugan, M.E.R.; Kramer, J.G.K.; Robertson, W.M.; Meadus, W.J.; Aldai, N.; Rolland, D.C. Comparing subcutaneous adipose tissue in beef and muskox with emphasis on trans 18:1 and conjugated linoleic acids. Lipids 2007, 42, 509–518. [Google Scholar] [CrossRef]
- Aldai, N.; Najera, A.I.; Dugan, M.E.; Celaya, R.; Osoro, K. Characterisation of intramuscular, intermuscular and subcutaneous adipose tissues in yearling bulls of different genetic groups. Meat Sci. 2007, 76, 682–691. [Google Scholar] [CrossRef] [Green Version]
- Wang, Z.Z.; Zhu, B.; Niu, H.; Zhang, W.G.; Xu, L.; Xu, L.; Chen, Y.; Zhang, L.P.; Gao, X.; Gao, H.J.; et al. Genome wide association study identifies SNPs associated with fatty acid composition in Chinese Wagyu cattle. J. Anim. Sci. Biotechnol. 2019, 10, 27. [Google Scholar] [CrossRef] [PubMed]
- Baik, M.; Jeong, J.Y.; Vu, T.T.T.; Piao, M.Y.; Kang, H.J. Effects of castration on the adiposity and expression of lipid metabolism genes in various fat depots of Korean cattle. Livest. Sci. 2014, 168, 168–176. [Google Scholar] [CrossRef]
- Gui, L.; Hong, J.; Raza, S.H.A.; Zan, L. Genetic variants in SIRT3 transcriptional regulatory region affect promoter activity and fat deposition in three cattle breeds. Mol. Cell. Probe. 2017, 32, 40–45. [Google Scholar] [CrossRef]
- Juárez, M.; Lam, S.; Bohrer, B.M.; Dugan, M.E.R.; Vahmani, P.; Aalhus, J.; Juárez, A.; López-Campos, O.; Prieto, N.; Segura, J. Enhancing the Nutritional Value of Red Meat through Genetic and Feeding Strategies. Foods 2021, 10, 872. [Google Scholar] [CrossRef] [PubMed]
- Ramos, Z.; Barbieri, I.D.; Lier, E.V.; Montossi, F. Carcass and meat quality traits of grazing lambs are affected by supplementation during early post-weaning. Small Rumin. Res. 2020, 184, 106047. [Google Scholar] [CrossRef]
- Wang, L.L.; Han, L.; Ma, X.L.; Yu, Q.L.; Zhao, S.N. Effect of mitochondrial apoptotic activation through the mitochondrial membrane permeability transition pore on yak meat tenderness during postmortem aging. Food Chem. 2017, 234, 323–331. [Google Scholar] [CrossRef]
- Bai, W.L.; Yin, R.H.; Zhao, S.J.; Li, C.; Ma, Z.J.; Yin, R.L.; Luo, G.B.; Zhao, Z.H. A PCR Assay for Sex Determination of Yak (biNBos grunniensb/iN) Meat by Amplification of the Male-Specific biNSRYb/iN Gene; FAO: Rome, Italy, 2010; Volume 21, pp. 726–731. [Google Scholar]
- Lang, Y.M.; Sha, K.; Zhang, R.; Xie, P.; Luo, X.; Sun, B.Z.; Li, H.P.; Zhang, L.; Zhang, S.S.; Liu, X. Effect of electrical stimulation and hot boning on the eating quality of Gannan yak longissimus lumborum. Meat Sci. 2016, 112, 3–8. [Google Scholar] [CrossRef]
- Zhang, S.G.; Liu, T.; Brown, M.A.; Wu, J.P. Comparison of longissimus dorsi Fatty Acids Profiles in Gansu Black Yak and Chinese Yellow Cattle Steers and Heifers. Korean J. Food Sci. Anim. Resour. 2015, 35, 286–292. [Google Scholar] [CrossRef]
- Qin, W.; Liang, C.N.; Guo, X.; Chu, M.; Pei, J.; Bao, P.J.; Wu, X.Y.; Li, T.K.; Yan, P. PPARα signal pathway gene expression is associated with fatty acid content in yak and cattle longissimus dorsi muscle. Genet. Mol. Res. 2015, 14, 14469–14478. [Google Scholar] [CrossRef]
- Hou, L.; Chai, S.T.; Liu, S.J.; Cui, Z.H.; Zhang, X.W.; Zhao, Y.P. Comparative Studies on Beef Amino Acid Composition and Fatty Acid Composition of Qinghai Yak and Qinchuan Cattle. Meat Res. 2013, 27, 30–36. [Google Scholar]
- Hu, R.; Zou, H.W.; Wang, Z.S.; Cao, B.H.; Peng, Q.H.; Jing, X.P.; Wang, Y.X.; Shao, Y.S.; Pei, Z.X.; Zhang, X.F.; et al. Nutritional Interventions Improved Rumen Functions and Promoted Compensatory Growth of Growth-Retarded Yaks as Revealed by Integrated Transcripts and Microbiome Analyses. Front. Microbiol. 2019, 10, 318. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wen, W.T.; Luo, X.L.; Xia, B.X.; Guan, J.Q.; Nie, Y.Y.; Li, L.; Duan, J.Y.; Suman, S.P.; Sun, Q. Post-mortem oxidative stability of three yak (Bos grunniens) muscles as influenced by animal age. Meat Sci. 2015, 105, 121–125. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Ren, Z.; Ma, P.; Wang, Z.; Niu, D.; Fu, H.; Elser, J.J. Effects of grassland degradation on ecological stoichiometry of soil ecosystems on the Qinghai-Tibet Plateau. Sci. Total Environ. 2020, 722, 137910. [Google Scholar] [CrossRef] [PubMed]
- Dai, Q.; Ma, J.; Cao, G.; Hu, R.; Zhu, Y.; Li, G.; Zou, H.W.; Wang, Z.S.; Peng, Q.H.; Xue, B.; et al. Comparative study of growth performance, nutrient digestibility, and ruminal and fecal bacterial community between yaks and cattle-yaks raised by stall-feeding. AMB Express 2021, 11, 98. [Google Scholar] [CrossRef] [PubMed]
- Sato, Y.; Takebe, H.; Tominaga, K.; Oishi, K.; Kumagai, H.; Yoshida, T. Taxonomic and functional characterization of the rumen microbiome of Japanese Black cattle revealed by 16S rRNA gene amplicon and metagenome shotgun sequencing. FEMS Microbiol. Ecol. 2021, 97, fiab152. [Google Scholar] [CrossRef] [PubMed]
- Wang, B.; Zhao, X.G.; Li, Z.; Luo, H.L.; Zhang, H.; Guo, Y.P.; Zhang, C.; Ma, Q. Changes of Metabolites and Gene Expression under Different Feeding Systems Associated with Lipid Metabolism in Lamb Meat. Foods 2021, 10, 2612. [Google Scholar] [CrossRef]
- Berton, M.P.; Fonseca, L.F.S.; Gimenez, D.F.J.; Utembergue, B.L.; Cesar, A.S.M.; Coutinho, L.L.; Lemos, M.V.A.D.; Aboujaoude, C.; Pereira, A.S.C.; Silva, R.M.D.O.; et al. Gene expression profile of intramuscular muscle in Nellore cattle with extreme values of fatty acid. BMC Genom. 2016, 17, 972. [Google Scholar] [CrossRef]
- Zhang, H.M.; Xia, H.L.; Jiang, H.R.; Mao, Y.J.; Qu, K.X.; Huang, B.Z.; Gong, Y.C.; Yang, Z.P. Longissimus dorsi muscle transcriptomic analysis of Yunling and Chinese simmental cattle differing in intramuscular fat content and fatty acid composition. Genome 2018, 61, 549–558. [Google Scholar] [CrossRef]
- Xu, Z.Y.; Chen, W.T.; Wang, L.Y.; Zhou, Y.B.; Nong, Q.Y.; Valencak, T.G.; Wang, Y.Z.; Xie, J.T.; Shan, T.Z. Cold Exposure Affects Lipid Metabolism, Fatty Acids Composition and Transcription in Pig Skeletal Muscle. Front. Physiol. 2021, 12, 748801. [Google Scholar] [CrossRef]
- Xie, Y.C.; Liu, Z.H.; Guo, J.T.; Su, X.; Zhao, C.; Zhang, C.Y.; Qin, Q.; Dai, D.L.; Zhao, Y.H.; Wang, Z.Y.; et al. MicroRNA-mRNA Regulatory Networking Fine-Tunes Polyunsaturated Fatty Acid Synthesis and Metabolism in the Inner Mongolia Cashmere Goat. Front. Genet. 2021, 12, 649015. [Google Scholar] [CrossRef]
- Wang, H.; Zhong, J.C.; Zhang, C.F.; Chai, Z.X.; Cao, H.W.; Wang, J.K.; Zhu, J.J.; Wang, J.B.; Ji, Q.M. The whole-transcriptome landscape of muscle and adipose tissues reveals the ceRNA regulation network related to intramuscular fat deposition in yak. BMC Genom. 2020, 21, 347. [Google Scholar] [CrossRef]
- Nicholson, J.K.; Lindon, J.C.; Holmes, E. “Metabonomics”: Understanding the metabolic responses of living systems to pathophysiological stimuli via multivariate statistical analysis of biological NMR spectroscopic data. Xenobiotica 1999, 29, 1181–1189. [Google Scholar] [CrossRef] [PubMed]
- Fischer, H.P. Mathematical modeling of complex biological systems: From parts lists to understanding systems behavior. Alcohol Res. Health 2008, 31, 49–59. [Google Scholar] [PubMed]
- Li, M.; Li, Q.; Kang, S.; Cao, X.; Zheng, Y.; Wu, J.; Wu, R.N.; Shao, J.H.; Yang, M.; Yue, X.Q. Characterization and comparison of lipids in bovine colostrum and mature milk based on UHPLC-QTOF-MS lipidomics. Food Res. Int. 2020, 10, 136. [Google Scholar] [CrossRef] [PubMed]
- Rico, J.E.; Samii, S.S.; Zang, Y.; Deme, P.; Haughey, N.J.; Grilli, E.; McFadden, J.W. Characterization of the Plasma Lipidome in Dairy Cattle Transitioning from Gestation to Lactation: Identifying Novel Biomarkers of Metabolic Impairment. Metabolites 2021, 11, 290. [Google Scholar] [CrossRef] [PubMed]
- Artegoitia, V.M.; Foote, A.P.; Lewis, R.M.; Freetly, H.C. Metabolomics Profile and Targeted Lipidomics in Multiple Tissues Associated with Feed Efficiency in Beef Steers. ACS Omega 2019, 4, 3973–3982. [Google Scholar] [CrossRef]
- Ueda, S.; Sasaki, R.; Nakabayashi, R.; Yamanoue, M.; Sirai, Y.; Iwamoto, E. Exploring the Lipids Involved in the Formation of Characteristic Lactones in Japanese Black Cattle. Metabolites 2021, 11, 203. [Google Scholar] [CrossRef]
- Jia, W.; Shi, Q.Y.; Shi, L. Effect of irradiation treatment on the lipid composition and nutritional quality of goat meat. Food Chem. 2021, 351, 129295. [Google Scholar] [CrossRef]
- Zhang, Z.W.; Liao, Q.C.; Sun, Y.; Pan, T.L.; Liu, S.Q.; Miao, W.W.; Li, Y.X.; Zhou, L.; Xu, G.X. Lipidomic and Transcriptomic Analysis of the Longissimus Muscle of Luchuan and Duroc Pigs. Front. Nutr. 2021, 8, 667622. [Google Scholar] [CrossRef]
- Zhou, J.W.; Zhang, Y.; Wu, J.J.; Qiao, M.; Xu, Z.; Peng, X.W.; Mei, S.Q. Proteomic and lipidomic analyses reveal saturated fatty acids, phosphatidylinositol, phosphatidylserine, and associated proteins contributing to intramuscular fat deposition. J. Proteom. 2021, 241, 104235. [Google Scholar] [CrossRef]
- Song, S.Z.; Wu, J.P.; Zhao, S.G.; Casper, D.P.; He, B.; Liu, T.; Gong, X.Y.; Liu, L.S. The effect of energy restriction on fatty acid profiles of longissimus dorsi and tissue adipose depots in sheep. J. Anim. Sci. 2017, 95, 3940–3948. [Google Scholar] [CrossRef] [PubMed]
- Kramer, J.K.G.; Hernandez, M.; Cruz-Hernandez, C.; Kraft, J.; Dugan, M.E.R. Combining results of two GC separations partly achieves determination of all cis and trans 16:1, 18:1, 18:2 and 18:3 except CLA isomers of milk fat as demonstrated using ag-ion SPE fractionation. Lipids 2008, 43, 259–273. [Google Scholar] [CrossRef] [PubMed]
- Turner, T.; Aalhus, J.; Mapiye, C.; Rolland, D.; Larsen, I.; Basarab, J.; Baron, V.S.; McAllister, T.A.; Block, H.C.; Uttaro, B.; et al. Effects of diets supplemented with sunflower or flax seeds on quality and fatty acid profile of hamburgers made with perirenal or subcutaneous fat. Meat Sci. 2015, 99, 123–131. [Google Scholar] [CrossRef] [PubMed]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2-11 CT method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef] [PubMed]
- Andreo, N.; Bridi, A.M.; Soares, A.L.; Prohmann, P.E.F.; Peres, L.M.; Tarsitano, M.A.; Giangareli, B.D.L.; Takabayashi, A.A. Fatty acid profile of beef from immunocastrated (BOPRIVA®) Nellore bulls. Meat Sci. 2016, 117, 12–17. [Google Scholar] [CrossRef] [PubMed]
- Scollan, N.D.; Choi, N.J.; Kurt, E.; Fisher, A.V.; Enser, M.; Wood, J.D. Manipulating of fatty acid composition of muscle and adipose tissue in beef cattle. Br. J. Nutr. 2001, 85, 115–124. [Google Scholar] [CrossRef]
- WHO. Diet, Nutrition and the Prevention of Chronic Diseases. In Report of a Joint WHO/FAO Expert Consultation; WHO: Geneva, Switzerland, 2003; p. 916. [Google Scholar]
- Dewanckele, L.; Toral, P.G.; Vlaeminck, B.; Fievez, V. Invited review: Role of rumen biohydrogenation intermediates and rumen microbes in diet-induced milk fat depression: An update. J. Dairy Sci. 2020, 103, 7655–7681. [Google Scholar] [CrossRef]
- Wood, J.D.; Richardson, R.I.; Nute, G.R.; Fisher, A.V.; Campo, M.M.; Kasapidou, E.; Sheard, P.R.; Enser, M. Effects of fatty acids on meat quality: A review. Meat Sci. 2004, 66, 21–32. [Google Scholar] [CrossRef]
- Schäff, C.; Börner, S.; Hacke, S.; Kautzsch, U.; Albrecht, D.; Hammon, H.M.; Röntgen, M.; Kuhla, B. Increased anaplerosis, TCA cycling, and oxidative phosphorylation in the liver of dairy cows with intensive body fat mobilization during early lactation. J. Proteome Res. 2012, 11, 5503–5514. [Google Scholar] [CrossRef]
- Veshkini, A.M.; Hammon, H.; Sauerwein, H.; Tröscher, A.; Viala, D.; Delosière, M.; Ceciliani, F.; Déjean, S.; Bonnet, M. Longitudinal liver proteome profiling in dairy cows during the transition from gestation to lactation: Investigating metabolic adaptations and their interactions with fatty acids supplementation via repeated measurements ANOVA-simultaneous component analysis. J. Proteom. 2022, 252, 104435. [Google Scholar] [CrossRef]
- Busato, S.; Bionaz, M. When Two plus Two Is More than Four: Evidence for a Synergistic Effect of Fatty Acids on Peroxisome Proliferator—Activated Receptor Activity in a Bovine Hepatic Model. Genes 2021, 12, 1283. [Google Scholar] [CrossRef] [PubMed]
- Gao, Y.Y.; Cheng, G.; Cheng, Z.X.; Bao, C.; Yamada, T.; Cao, G.F.; Bao, S.Q.; Schreurs, N.M.; Zan, L.S.; Tong, B. Association of variants in FABP4, FASN, SCD, SREBP1 and TCAP genes with intramuscular fat, carcass traits and body size in Chinese Qinchuan cattle. Meat Sci. 2022, 192, 108882. [Google Scholar] [CrossRef] [PubMed]
- Du, L.L.; Li, K.N.; Chang, T.P.; An, B.X.; Liang, M.; Deng, T.Y.; Cao, S.; Du, Y.Y.; Cai, W.T.; Gao, X. Integrating genomics and transcriptomics to identify candidate genes for subcutaneous fat deposition in beef cattle. Genomics 2022, 114, 110406. [Google Scholar] [CrossRef] [PubMed]
- Ma, N.; Abaker, J.A.; Wei, G.; Chen, H.; Shen, X.; Chang, G. A high-concentrate diet induces an inflammatory response and oxidative stress and depresses milk fat synthesis in the mammary gland of dairy cows. J. Dairy Sci. 2022, 105, 5493–5505. [Google Scholar] [CrossRef]
- Raza, S.H.A.; Gui, L.S.; Khan, R.; Schreurs, N.M.; Wang, X.Y.; Wu, S.; Mei, C.G.; Wang, L.; Ma, X.Y.; Wei, D.W.; et al. Association between FASN gene polymorphisms ultrasound carcass traits and intramuscular fat in Qinchuan cattle. Gene 2017, 645, 55–59. [Google Scholar] [CrossRef]
- Laprairie, R.B.; Denovan-Wright, E.M.; Wright, J.M. Divergent evolution of 613 cis-acting peroxisome proliferator-activated receptor elements that differentially control the tandemly duplicated fatty acid-binding protein genes, fabp1b.1 and fabp1b.2, in zebrafish. Genome 2016, 59, 403–412. [Google Scholar] [CrossRef]
- Yadav, P.; Mukesh, M.; Kataria, R.S.; Yadav, A.; Mohanty, A.K.; Mishra, B.P. Semi-quantitative RT-PCR analysis of fat metabolism genes in mammary tissue of lactating and non-lactating water buffalo (Bubalus bubalis). Trop. Anim. Health Prod. 2012, 44, 693–696. [Google Scholar] [CrossRef]
- Rincon, G.; Islas-Trejo, A.; Castillo, A.R.; Bauman, D.E.; German, B.J.; Medrano, J.F. Polymorphisms in genes in the SREBP1 signalling pathway and SCD are associated with milk fatty acid composition in Holstein cattle. J. Dairy Res. 2012, 79, 66–75. [Google Scholar] [CrossRef]
- Schumann, T.; König, J.; von Loeffelholz, C.; Vatner, D.F.; Zhang, D.; Perry, R.J.; Bernier, M.; Chami, J.; Henke, C.; Kurzbach, A.; et al. Deletion of the diabetes candidate gene Slc16a13 in mice attenuates diet-induced ectopic lipid accumulation and insulin resistance. Commun. Biol. 2021, 4, 826. [Google Scholar] [CrossRef]
- Moallem, U. Invited review: Roles of dietary n-3 fatty acids in performance, milk fat composition, and reproductive and immune systems in dairy cattle. J. Dairy Sci. 2018, 101, 8641–8661. [Google Scholar] [CrossRef]
- Zhang, J.T.; Zhu, J.Q.; Zhu, Q.; Liu, H.; Gao, X.S.; Zhang, H.X. Fatty acid desaturase-6 (Fad6) is required for salt tolerance in Arabidopsis thaliana. Biochem. Bioph. Res. Commun. 2009, 390, 469–474. [Google Scholar] [CrossRef] [PubMed]
- Sun, J.; Li, J.Q.; Li, Y.N.; Du, J.L.; Zhao, N.N.; Mai, K.S.; Ai, Q.H. Regulation of ∆6Fads2 Gene Involved in LC-PUFA Subjected to Fatty Acid in Large Yellow Croaker (Larimichthys crocea) and Rainbow Trout (Oncorhynchus mykiss). Biomolecules 2022, 12, 659. [Google Scholar] [CrossRef] [PubMed]
| Item | TMR | Natural Grass |
|---|---|---|
| Ingredient (%) | ||
| Corn | 19.20 | - |
| Wheat bran | 9.20 | - |
| Whole corn silage | 32.00 | - |
| Oat Hay | 28.00 | - |
| Rapeseed meal | 8.10 | - |
| NaHCO3 | 1.00 | - |
| NaCl | 1.50 | - |
| Premix | 1.00 | - |
| Total | 100.00 | - |
| Common nutrition (%) | ||
| Crude fat | 4.52 | 2.63 |
| Crude protein | 16.96 | 11.93 |
| Neutral detergent fiber | 23.24 | 76.14 |
| Acid detergent fiber | 13.84 | 10.09 |
| Calcium | 0.79 | 5.22 |
| Phosphorus | 0.37 | 0.07 |
| Fatty acids (%) | ||
| C16:0 | 0.68 | 0.30 |
| C18:0 | 0.21 | 0.09 |
| C18:1 | 0.18 | 0.07 |
| C18:2n6 | 0.58 | 0.29 |
| C18:3n3 | 1.42 | 0.68 |
| Variable | Absolute Content (mg/100g) | Relative Content (%) | ||||
|---|---|---|---|---|---|---|
| GF Yaks (n = 6, Mean ± SEM) | SF Yaks (n = 6, Mean ± SEM) | F | GF Yaks (n = 6, Mean ± SEM) | SF Yaks (n = 6, Mean ± SEM) | F | |
| C4:0 | 2.12 ± 0.08 | 3.75 ± 0.27 | ** | 0.15 ± 0.01 | 0.17 ± 0.01 | ns |
| C6:0 | 0.36 ± 0.02 | 0.60 ± 0.05 | ** | 0.03 ± 0.001 | 0.03 ± 0.002 | ns |
| C8:0 | 0.53 ± 0.02 | 0.13 ± 0.02 | ** | 0.04 ± 0.001 | 0.01 ± 0.001 | ** |
| C10:0 | 0.44 ± 0.02 | 0.61 ± 0.03 | ** | 0.03 ± 0.001 | 0.03 ± 0.001 | ns |
| C11:0 | 0.06 ± 0.004 | 0.09 ± 0.01 | ** | 0.005 ± 0.0002 | 0.004 ± 0.0003 | ns |
| C12:0 | 0.37 ± 0.01 | 0.53 ± 0.05 | ** | 0.03 ± 0.001 | 0.03 ± 0.002 | ns |
| C13:0 | 0.79 ± 0.03 | 1.35 ± 0.17 | ** | 0.06 ± 0.002 | 0.06 ± 0.01 | ns |
| C14:0 | 7.67 ± 0.27 | 10.66 ± 0.36 | ** | 0.56 ± 0.02 | 0.50 ± 0.02 | ns |
| C14:0iso | 0.40 ± 0.05 | 0.44 ± 0.02 | ns | 0.03 ± 0.005 | 0.02 ± 0.001 | ns |
| C15:0 | 9.00 ± 0.34 | 7.48 ± 0.65 | ns | 0.65 ± 0.02 | 0.35 ± 0.03 | ** |
| C15:0iso | 1.13 ± 0.06 | 1.31 ± 0.05 | ns | 0.08 ± 0.01 | 0.06 ± 0.003 | ** |
| C15:0ai | 1.08 ± 0.05 | 0.88 ± 0.03 | * | 0.08 ± 0.003 | 0.04 ± 0.001 | ** |
| C16:0 | 173.53 ± 3.86 | 335.64 ± 6.60 | ** | 12.61 ± 0.15 | 15.62 ± 0.36 | ** |
| C16:0iso | 1.76 ± 0.03 | 1.68 ± 0.04 | ns | 0.13 ± 0.004 | 0.08 ± 0.002 | ** |
| C17:0 | 22.02 ± 1.55 | 16.79 ± 2.16 | ns | 1.59 ± 0.09 | 0.78 ± 0.09 | ** |
| C17:0iso | 1.74 ± 0.07 | 1.91 ± 0.06 | ns | 0.13 ± 0.01 | 0.09 ± 0.003 | ** |
| C17:0ai | 2.06 ± 0.08 | 2.18 ± 0.12 | ns | 0.15 ± 0.004 | 0.10 ± 0.01 | ** |
| C18:0 | 326.20 ± 10.16 | 637.71 ± 9.09 | ** | 23.69 ± 0.39 | 29.66 ± 0.21 | ** |
| C18:0iso | 3.81 ± 2.52 | 1.37 ± 0.08 | ns | 0.29 ± 0.19 | 0.06 ± 0.003 | ns |
| C20:0 | 1.53 ± 0.09 | 2.17 ± 0.30 | ns | 0.11 ± 0.01 | 0.10 ± 0.01 | ns |
| C21:0 | 4.73 ± 0.28 | 7.03 ± 0.58 | ** | 0.34 ± 0.02 | 0.33 ± 0.03 | ns |
| C22:0 | 2.23 ± 0.22 | 3.04 ± 0.21 | * | 0.16 ± 0.02 | 0.14 ± 0.01 | ns |
| C23:0 | 9.42 ± 0.31 | 12.59 ± 2.01 | ns | 0.68 ± 0.02 | 0.58 ± 0.09 | ns |
| C24:0 | 68.21 ± 3.57 | 94.62 ± 6.14 | ** | 4.95 ± 0.21 | 4.39 ± 0.24 | ns |
| C14:1 | 17.45 ± 1.26 | 1.16 ± 0.13 | ** | 1.27 ± 0.09 | 0.05 ± 0.01 | ** |
| c-C15:1 | 4.88 ± 0.28 | 3.13 ± 0.32 | ** | 0.35 ± 0.02 | 0.15 ± 0.02 | ** |
| C16:1 | 35.33 ± 1.52 | 39.10 ± 4.24 | ns | 2.57 ± 0.10 | 1.81 ± 0.18 | ** |
| c-C17:1 | 14.55 ± 0.95 | 6.06 ± 0.81 | ** | 1.06 ± 0.06 | 0.29 ± 0.04 | ** |
| c-C18:1 | 287.11 ± 5.40 | 459.89 ± 11.29 | ** | 20.87 ± 0.26 | 21.38 ± 0.32 | ns |
| t-C18:1 | 5.81 ± 0.59 | 12.18 ± 1.41 | ** | 0.42 ± 0.04 | 0.57 ± 0.07 | ns |
| t11-C18:1 | 2.99 ± 0.07 | 3.99 ± 0.09 | ** | 0.22 ± 0.01 | 0.19 ± 0.004 | ** |
| c-C20:1 | 1.71 ± 0.05 | 5.37 ± 0.51 | ** | 0.12 ± 0.004 | 0.25 ± 0.03 | ** |
| C24:1 | 37.28 ± 1.06 | 53.25 ± 3.72 | ** | 2.71 ± 0.09 | 2.47 ± 0.14 | ns |
| c-C18:2n6 | 166.09 ± 4.86 | 270.36 ± 5.23 | ** | 12.07 ± 0.25 | 12.59 ± 0.36 | ** |
| t-C18:2n6 | 0.45 ± 0.04 | 0.58 ± 0.07 | ns | 0.03 ± 0.003 | 0.03 ± 0.003 | ns |
| c9, t11-C18:2 | 3.32 ± 0.14 | 3.84 ± 0.14 | * | 0.24 ± 0.01 | 0.18 ± 0.01 | ** |
| t11, c15-C18:2 | 5.14 ± 0.19 | 6.86 ± 0.24 | ** | 0.37 ± 0.01 | 0.32 ± 0.01 | * |
| c-C18:3n6 | 10.23 ± 0.30 | 5.76 ± 0.59 | ** | 0.74 ± 0.02 | 0.27 ± 0.03 | ** |
| C18:3n3 | 50.31 ± 0.93 | 33.25 ± 0.72 | ** | 3.66 ± 0.04 | 1.55 ± 0.04 | ** |
| c9, t11, t15-C18:3 | 0.64 ± 0.02 | 0.74 ± 0.04 | ns | 0.05 ± 0.002 | 0.03 ± 0.002 | ** |
| c-C20:2 | 5.94 ± 0.35 | 3.43 ± 0.60 | ** | 0.43 ± 0.02 | 0.16 ± 0.03 | ** |
| C20:4n6 | 41.63 ± 1.63 | 59.06 ± 2.50 | ** | 3.03 ± 0.13 | 2.74 ± 0.09 | ns |
| c-C20:3n3 | 0.24 ± 0.01 | 0.27 ± 0.04 | ns | 0.02 ± 0.001 | 0.01 ± 0.002 | ns |
| c-C20:5n3 | 41.49 ± 0.93 | 34.62 ± 1.09 | ** | 3.02 ± 0.09 | 1.61 ± 0.05 | ** |
| c-C22:6n3 | 2.21 ± 0.08 | 2.71 ± 0.27 | ns | 0.16 ± 0.01 | 0.13 ± 0.01 | * |
| ΣSFAs | 641.20 ± 16.10 | 1144.55 ± 14.32 | ** | 46.58 ± 0.45 | 53.23 ± 0.21 | ** |
| ΣMUFAs | 407.10 ± 6.21 | 584.14 ± 14.59 | ** | 29.60 ± 0.19 | 27.15 ± 0.32 | ** |
| ΣPUFAs | 327.70 ± 6.88 | 421.46 ± 4.30 | ** | 23.82 ± 0.37 | 19.62 ± 0.34 | ** |
| ΣUFAs | 734.80 ± 11.95 | 1005.60 ± 14.73 | ** | 53.42 ± 0.45 | 46.77 ± 0.21 | ** |
| Σn-3PUFAs | 94.25 ± 1.21 | 70.86 ± 1.58 | ** | 6.86 ± 0.10 | 3.30 ± 0.08 | ** |
| Σn-6PUFAs | 218.41 ± 6.19 | 335.75 ± 4.62 | ** | 15.88 ± 0.36 | 15.63 ± 0.32 | ns |
| Σn-6/Σn-3PUFAs | 2.32 ± 0.06 | 4.75 ± 0.15 | ** | 2.32 ± 0.06 | 4.75 ± 0.15 | ** |
| ΣPUFAs/ΣSFAs | 0.51 ± 0.01 | 0.37 ± 0.01 | ** | 0.51 ± 0.01 | 0.37 ± 0.01 | ** |
| ΣBIs | 12.09 ± 0.22 | 15.43 ± 0.30 | ** | 0.88 ± 0.02 | 0.72 ± 0.01 | ** |
| ΣBCFAs | 11.98 ± 2.59 | 9.77 ± 0.18 | ns | 0.88 ± 0.20 | 0.45 ± 0.01 | ns |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Xiong, L.; Pei, J.; Wang, X.; Guo, S.; Guo, X.; Yan, P. Lipidomics and Transcriptome Reveal the Effects of Feeding Systems on Fatty Acids in Yak’s Meat. Foods 2022, 11, 2582. https://doi.org/10.3390/foods11172582
Xiong L, Pei J, Wang X, Guo S, Guo X, Yan P. Lipidomics and Transcriptome Reveal the Effects of Feeding Systems on Fatty Acids in Yak’s Meat. Foods. 2022; 11(17):2582. https://doi.org/10.3390/foods11172582
Chicago/Turabian StyleXiong, Lin, Jie Pei, Xingdong Wang, Shaoke Guo, Xian Guo, and Ping Yan. 2022. "Lipidomics and Transcriptome Reveal the Effects of Feeding Systems on Fatty Acids in Yak’s Meat" Foods 11, no. 17: 2582. https://doi.org/10.3390/foods11172582






