Microbial Diversity and Volatile Flavor Changes during Gayangju Fermentation, a Traditional Korean House Rice Wine
Abstract
:1. Introduction
2. Materials and Methods
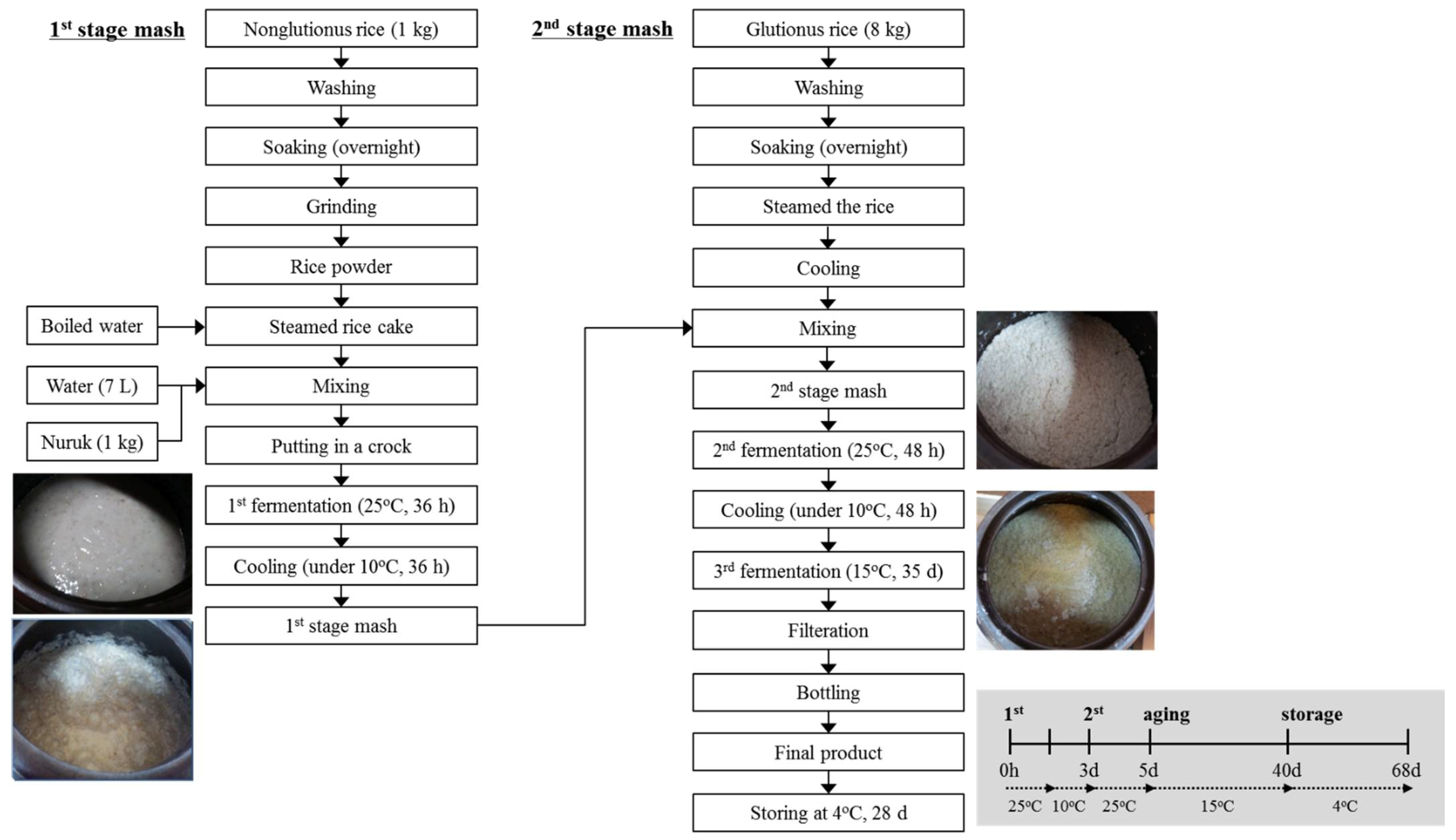
2.1. Gayangju Fermentation
2.2. Physicochemical Analysis
2.3. Free Amino Acids Analysis
2.4. Microbiological Analysis
2.5. Volatile Flavor Analysis
2.6. Statistical Analysis
3. Results and Discussion
3.1. Physicochemical Change during Gayangju Fermentation
3.2. Microbial Change during Makgeolli Fermentation
3.3. Volatile Compounds Change in Gayangju
4. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Bae, S.M. The Technology of Korean Traditional Liquor Making; WooKok Publishing Company: Seoul, Korea, 2002. [Google Scholar]
- Kim, S.A.; Yun, S.J.; Jeon, S.H.; Kim, N.H.; Kim, H.W.; Cho, T.J.; Lee, S.H.; Hwang, I.G.; Rhee, M.S. Microbial composition of turbid rice wine (Makgeolli) at different stages of production in a real processing line. Food Control 2015, 53, 1–8. [Google Scholar] [CrossRef]
- Bal, J.; Yun, S.-H.; Choi, M.-S.; Yeo, S.-H.; Kim, J.-M.; Kim, D.-H. Pyrosequencing reveals bacterial diversity in Korean traditional wheat-based nuruk. J. Microbiol. 2015, 53, 812–819. [Google Scholar] [CrossRef] [PubMed]
- Bal, J.; Yun, S.-H.; Yeo, S.-H.; Kim, J.-M.; Kim, D.-H. Metagenomic analysis of fungal diversity in Korean traditional wheat-based fermentation starter nuruk. Food Microbiol. 2016, 60, 73–83. [Google Scholar] [CrossRef]
- Chai, C.; Lim, G.S.; Kim, Y.J.; Oh, S.W. Microbial community changes in Makgeolli during brewing. J. Inst. Brew. 2015, 121, 304–308. [Google Scholar] [CrossRef]
- Lee, T.-S.; Choi, J.-Y. Volatile flavor components in Takju fermented with mashed glutinous rice and barley rice. Korean J. Food Sci. Technol. 1998, 30, 638–643. [Google Scholar]
- Lee, T.-S.; Choi, J.-Y. Volatile Flavor Components in Mash of Takju prepared by using Aspergillus kawachii Nuruks. Korean J. Food Sci. Technol. 2005, 37, 944–950. [Google Scholar]
- Jung, H.; Lee, S.-J.; Lim, J.H.; Kim, B.-K.; Park, K.J. Chemical and sensory profiles of makgeolli, Korean commercial rice wine, from descriptive, chemical, and volatile compound analyses. Food Chem. 2014, 152, 624–632. [Google Scholar] [CrossRef] [PubMed]
- Song, Y.-R.; Jeong, D.-Y.; Baik, S.-H. Effects of indigenous yeasts on physicochemical and microbial properties of Korean soy sauce prepared by low-salt fermentation. Food Microbiol. 2015, 51, 171–178. [Google Scholar] [CrossRef] [PubMed]
- Izco, J.M.; Torre, P. Characterisation of volatile flavour compounds in Roncal cheese extracted by the ‘purge and trap’ method and analysed by GC–MS. Food Chem. 2000, 70, 409–417. [Google Scholar] [CrossRef]
- Hoenicke, K.; Simat, T.J.; Steinhart, H.; Christoph, N.; Geßner, M.; Köhler, H.-J. ‘Untypical aging off-flavor’ in wine: Formation of 2-aminoacetophenone and evaluation of its influencing factors. Anal. Chim. Acta 2002, 458, 29–37. [Google Scholar] [CrossRef]
- Jung, M.-J.; Nam, Y.-D.; Roh, S.W.; Bae, J.-W. Unexpected convergence of fungal and bacterial communities during fermentation of traditional Korean alcoholic beverages inoculated with various natural starters. Food Microbiol. 2012, 30, 112–123. [Google Scholar] [CrossRef] [PubMed]
- Song, S.H.; Lee, C.; Lee, S.; Park, J.M.; Lee, H.J.; Bai, D.H.; Yoon, S.S.; Choi, J.B.; Park, Y.S. Analysis of microflora profile in Korean traditional nuruk. J. Microbiol. Biotechnol. 2013, 23, 40–46. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chi, Z.; Chi, Z.; Liu, G.; Wang, F.; Ju, L.; Zhang, T. Saccharomycopsis fibuligera and its applications in biotechnology. Biotechnol. Adv. 2009, 27, 423–431. [Google Scholar] [CrossRef] [PubMed]
- Styger, G.; Prior, B.; Bauer, F. Wine flavor and aroma. J. Ind. Microbiol. Biotechnol. 2011, 38, 1145–1159. [Google Scholar] [CrossRef] [PubMed]
- Plata, C.; Millán, C.; Mauricio, J.; Ortega, J. Formation of ethyl acetate and isoamyl acetate by various species of wine yeasts. Food Microbiol. 2003, 20, 217–224. [Google Scholar] [CrossRef]
- Kang, B.-S.; Lee, J.-E.; Park, H.-J. Qualitative and Quantitative Prediction of Volatile Compounds from Initial Amino Acid Profiles in Korean Rice Wine (makgeolli) Model. J. Food Sci. 2014, 79, C1106–C1116. [Google Scholar] [CrossRef] [PubMed]
- Chuenchomrat, P.; Assavanig, A.; Lertsiri, S. Volatile flavour compounds analysis of solid state fermented Thai rice wine (Ou). ScienceAsia 2008, 34, 199–206. [Google Scholar] [CrossRef]
- Park, H.-J.; Lee, S.M.; Song, S.H.; Kim, Y.-S. Characterization of Volatile Components in Makgeolli, a Traditional Korean Rice Wine, with or without Pasteurization, During Storage. Molecules 2013, 18, 5317–5325. [Google Scholar] [CrossRef] [PubMed]
- Dragone, G.; Mussatto, S.I.; Oliveira, J.; Teixeira, J.A. Characterisation of volatile compounds in an alcoholic beverage produced by whey fermentation. Food Chem. 2009, 112, 929–935. [Google Scholar] [CrossRef]
- Lasekan, O.; Buettner, A.; Christlbauer, M. Investigation of important odorants of palm wine (Elaeis guineensis). Food Chem. 2007, 105, 15–23. [Google Scholar] [CrossRef]
 ) and total acidity (●); (B) contents of alcohol (
) and total acidity (●); (B) contents of alcohol ( ), reducing sugars (
), reducing sugars ( ), and total soluble solids (●).
), and total soluble solids (●).
 ) and total acidity (●); (B) contents of alcohol (
) and total acidity (●); (B) contents of alcohol ( ), reducing sugars (
), reducing sugars ( ), and total soluble solids (●).
), and total soluble solids (●).



 , GY), GY3, GY5, GY15, GY40, and GY68, indicate samples of different fermentation time of Gayangju. CO (
, GY), GY3, GY5, GY15, GY40, and GY68, indicate samples of different fermentation time of Gayangju. CO ( ) indicates commercial Makgeolli.
) indicates commercial Makgeolli.
 , GY), GY3, GY5, GY15, GY40, and GY68, indicate samples of different fermentation time of Gayangju. CO (
, GY), GY3, GY5, GY15, GY40, and GY68, indicate samples of different fermentation time of Gayangju. CO ( ) indicates commercial Makgeolli.
) indicates commercial Makgeolli.
| No. | Flavor Compounds | Relative Peak Area | |||||
|---|---|---|---|---|---|---|---|
| Commercial Makgeolli | Gayangju Makgeolli | ||||||
| 3 Days | 5 Days | 15 Days | 40 Days | 68 Days | |||
| F1 | 2,3-Butanediol | 0 | 1 a | 2.15 b | 5.79 c | 5.43 c | 9.41 d |
| F2 | 3-Methyl-1-butanol | 2.97 a | 1 b | 2.09 c | 4.59 d | 3.56 e | 7.29 f |
| F3 | 2-Methyl-1-butanol | 0 | 1 a | 2 a | 6.79 b | 59.35 c | 691.53 d |
| F4 | 2-Methyl-1-propanol | 5.79 | 0 | 0 | 0 | 0 | 0 |
| F5 | Ethyl tetradecanoate | 0 | 1 a | 1.59 b | 3.02 c | 4.53 d | 4.92 d |
| F6 | Ethyl dodecanoate | 0 | 1 a | 2.20 b | 5.26 c | 5.15 c | 5.54 d |
| F7 | Ethyl decanoate | 0 | 1 a | 2.36 b | 5.42 c | 4.45 d | 8.65 e |
| F8 | butanoic acid | 0 | 1 a | 3 b | 16.2 c | 4.05 b | 9.11 d |
| F9 | 2-Methylbutanoic acid | 0 | 1 a | 2.74 | 5.71 | 4.74 | 8.13 |
| F10 | 3-Methylbutanoic acid | 0 | 1 a | 2.67 | 1.9 | 5.37 | 8.63 |
| F11 | 2-Methylbutylaldehyde | 0.85 a | 1 a | 1.2 | 1.25 | 3 | 2.75 |
| F12 | 3-Methylbutylaldehyde | 1.60 a | 1 b | 1.67 a | 1.87 a | 0.23 b | 2.04 a |
| F13 | 2-Methylthio-1-propanol | 0 | 1 a | 2.51 b | 5.45 c | 4.89 c | 6.59 d |
| F14 | Ethyl acetate | 0.69 a | 1 a | 1.2 a | 1.7 b | 2 b | 2.2 b |
| F15 | Ethyl caprylate | 0.1 b | 1 a | 1.96 c | 2.38 d | 4.29 e | 6.19 f |
| F16 | 2-phenylethylethanlol | 0 | 1 a | 2.23 b | 5.11 d | 4.32 c | 8.49 e |
| F17 | Ethyl pentadecanoate | 0 | 1 a | 1.22 a | 2.81 b | 2.69 b | 3.12 c |
| F18 | 3-Mehtylbutyl acetate | 0.13 a | 1 a | 1.74 b | 2.74 c | 4.07 d | 8.15 e |
| F19 | 2-Ethyl-1-hexene | 0.07 a | 1 a | 0.8 a | 6.07 b | 8.11 c | 9.43 d |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Song, Y.-R.; Lim, B.-U.; Baik, S.-H. Microbial Diversity and Volatile Flavor Changes during Gayangju Fermentation, a Traditional Korean House Rice Wine. Foods 2022, 11, 2604. https://doi.org/10.3390/foods11172604
Song Y-R, Lim B-U, Baik S-H. Microbial Diversity and Volatile Flavor Changes during Gayangju Fermentation, a Traditional Korean House Rice Wine. Foods. 2022; 11(17):2604. https://doi.org/10.3390/foods11172604
Chicago/Turabian StyleSong, Young-Ran, Byeong-Uk Lim, and Sang-Ho Baik. 2022. "Microbial Diversity and Volatile Flavor Changes during Gayangju Fermentation, a Traditional Korean House Rice Wine" Foods 11, no. 17: 2604. https://doi.org/10.3390/foods11172604
APA StyleSong, Y.-R., Lim, B.-U., & Baik, S.-H. (2022). Microbial Diversity and Volatile Flavor Changes during Gayangju Fermentation, a Traditional Korean House Rice Wine. Foods, 11(17), 2604. https://doi.org/10.3390/foods11172604





