Food and Gut Microbiota-Derived Metabolites in Nonalcoholic Fatty Liver Disease
Abstract
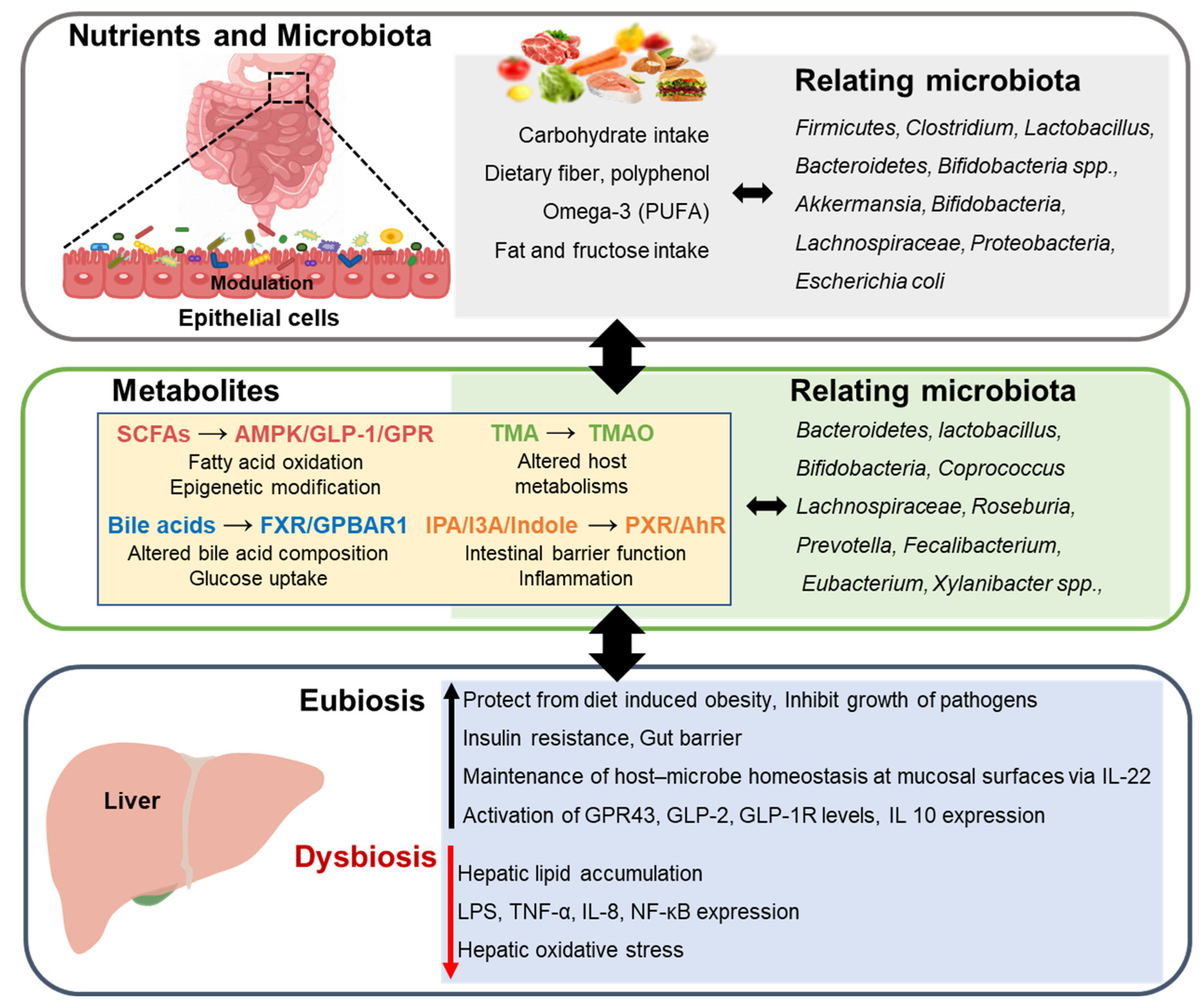
:1. Introduction
2. Food and Gut Microbiota
3. Nonalcoholic Fatty Liver Disease
4. Nutrients Associated with Microbiota in Nonalcoholic Fatty Liver Disease
4.1. Bile Acid
4.2. Short-Chain Fatty Acid
4.3. Amino Acid and Tryptophan
4.4. Choline
4.5. Polyphenol
4.6. ω-3 Polyunsaturated Fatty Acid
4.7. Others
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Moco, S.; Martin, F.P.; Rezzi, S. Metabolomics view on gut microbiome modulation by polyphenol-rich foods. J. Proteome Res. 2012, 11, 4781–4790. [Google Scholar] [CrossRef]
- David, L.A.; Maurice, C.F.; Carmody, R.N.; Gootenberg, D.B.; Button, J.E.; Wolfe, B.E.; Ling, A.V.; Devlin, A.S.; Varma, Y.; Fischbach, M.A.; et al. Diet rapidly and reproducibly alters the human gut microbiome. Nature 2014, 505, 559–563. [Google Scholar] [CrossRef]
- Mokhtari, Z.; Gibson, D.L.; Hekmatdoost, A. Nonalcoholic fatty liver disease, the gut microbiome, and diet. Adv. Nutr. 2017, 8, 240–252. [Google Scholar] [CrossRef]
- Schwenger, K.J.; Clermont-Dejean, N.; Allard, J.P. The role of the gut microbiome in chronic liver disease: The clinical evidence revised. JHEP Rep. 2019, 1, 214–226. [Google Scholar] [CrossRef]
- Smirnova, E.; Muthiah, M.D.; Narayan, N.; Siddiqui, M.S.; Puri, P.; Luketic, V.A.; Contos, M.J.; Idowu, M.; Chuang, J.C.; Billin, A.N.; et al. Metabolic reprogramming of the intestinal microbiome with functional bile acid changes underlie the development of NAFLD. Hepatology 2022. [Google Scholar] [CrossRef]
- Song, Q.; Zhang, X. The Role of Gut-Liver Axis in Gut Microbiome Dysbiosis Associated NAFLD and NAFLD-HCC. Biomedicines 2022, 10, 524. [Google Scholar] [CrossRef]
- Yadav, M.; Verma, M.K.; Chauhan, N.S. A review of metabolic potential of human gut microbiome in human nutrition. Arch. Microbiol. 2018, 200, 203–217. [Google Scholar] [CrossRef]
- Makki, K.; Deehan, E.C.; Walter, J.; Bäckhed, F. The impact of dietary fiber on gut microbiota in host health and disease. Cell Host Microbe 2018, 23, 705–715. [Google Scholar] [CrossRef]
- Hacquard, S.; Garrido-Oter, R.; González, A.; Spaepen, S.; Ackermann, G.; Lebeis, S.; McHardy, A.C.; Dangl, J.L.; Knight, R.; Ley, R. Microbiota and host nutrition across plant and animal kingdoms. Cell Host Microbe 2015, 17, 603–616. [Google Scholar] [CrossRef]
- Frame, L.A.; Costa, E.; Jackson, S.A. Current explorations of nutrition and the gut microbiome: A comprehensive evaluation of the review literature. Nutr. Rev. 2020, 78, 798–812. [Google Scholar] [CrossRef]
- Chalasani, N.; Younossi, Z.; Lavine, J.E.; Charlton, M.; Cusi, K.; Rinella, M.; Harrison, S.A.; Brunt, E.M.; Sanyal, A.J. The diagnosis and management of nonalcoholic fatty liver disease: Practice guidance from the American Association for the Study of Liver Diseases. Hepatology 2018, 67, 328–357. [Google Scholar] [CrossRef]
- Mouzaki, M.; Allard, J.P. The role of nutrients in the development, progression, and treatment of nonalcoholic fatty liver disease. J. Clin. Gastroenterol. 2012, 46, 457–467. [Google Scholar] [CrossRef]
- Stachowska, E.; Ryterska, K.; Maciejewska, D.; Banaszczak, M.; Milkiewicz, P.; Milkiewicz, M.; Gutowska, I.; Ossowski, P.; Kaczorowska, M.; Jamiol-Milc, D.; et al. Nutritional Strategies for the Individualized Treatment of Non-Alcoholic Fatty Liver Disease (NAFLD) Based on the Nutrient-Induced Insulin Output Ratio (NIOR). Int. J. Mol. Sci. 2016, 17, 1192. [Google Scholar] [CrossRef]
- Parry, S.A.; Hodson, L. Influence of dietary macronutrients on liver fat accumulation and metabolism. J. Investig. Med. 2017, 65, 1102–1115. [Google Scholar] [CrossRef]
- Bruzzese, E.; Volpicelli, M.; Squaglia, M.; Tartaglione, A.; Guarino, A. Impact of prebiotics on human health. Dig. Liver Dis. 2006, 38, S283–S287. [Google Scholar] [CrossRef]
- Valdes, A.M.; Walter, J.; Segal, E.; Spector, T.D. Role of the gut microbiota in nutrition and health. Bmj 2018, 361, k2179. [Google Scholar] [CrossRef]
- Qin, J.; Li, R.; Raes, J.; Arumugam, M.; Burgdorf, K.S.; Manichanh, C.; Nielsen, T.; Pons, N.; Levenez, F.; Yamada, T. A human gut microbial gene catalogue established by metagenomic sequencing. Nature 2010, 464, 59–65. [Google Scholar] [CrossRef]
- Blandino, G.; Inturri, R.; Lazzara, F.; Di Rosa, M.; Malaguarnera, L. Impact of gut microbiota on diabetes mellitus. Diabetes Metab. 2016, 42, 303–315. [Google Scholar] [CrossRef]
- Huttenhower, C.; Gevers, D.; Knight, R.; Abubucker, S.; Badger, J.H.; Chinwalla, A.T.; Creasy, H.H.; Earl, A.M.; FitzGerald, M.G.; Fulton, R.S. Structure, function and diversity of the healthy human microbiome. Nature 2012, 486, 207. [Google Scholar]
- Meelu, P.; Marin, R.; Clish, C.; Zella, G.; Cox, S.; Yajnik, V.; Nguyen, D.D.; Korzenik, J.R. Impaired innate immune function associated with fecal supernatant from Crohn’s disease patients: Insights into potential pathogenic role of the microbiome. Inflamm. Bowel Dis. 2014, 20, 1139–1146. [Google Scholar] [CrossRef]
- Zoetendal, E.G.; Rajilić-Stojanović, M.; de Vos, W.M. High-throughput diversity and functionality analysis of the gastrointestinal tract microbiota. Gut 2008, 57, 1605–1615. [Google Scholar] [CrossRef]
- Cani, P.D.; Delzenne, N.M.; Amar, J.; Burcelin, R. Role of gut microflora in the development of obesity and insulin resistance following high-fat diet feeding. Pathol. Biol. 2008, 56, 305–309. [Google Scholar] [CrossRef]
- Martinez, F.D. The human microbiome. Early life determinant of health outcomes. Ann. Am. Thorac. Soc. 2014, 11, S7–S12. [Google Scholar] [CrossRef]
- Turnbaugh, P.J.; Ridaura, V.K.; Faith, J.J.; Rey, F.E.; Knight, R.; Gordon, J.I. The effect of diet on the human gut microbiome: A metagenomic analysis in humanized gnotobiotic mice. Sci. Transl. Med. 2009, 1, ra14–ra16. [Google Scholar] [CrossRef]
- Wen, L.; Ley, R.E.; Volchkov, P.Y.; Stranges, P.B.; Avanesyan, L.; Stonebraker, A.C.; Hu, C.; Wong, F.S.; Szot, G.L.; Bluestone, J.A. Innate immunity and intestinal microbiota in the development of Type 1 diabetes. Nature 2008, 455, 1109–1113. [Google Scholar] [CrossRef]
- Bäckhed, F.; Manchester, J.K.; Semenkovich, C.F.; Gordon, J.I. Mechanisms underlying the resistance to diet-induced obesity in germ-free mice. Proc. Natl. Acad. Sci. USA 2007, 104, 979–984. [Google Scholar] [CrossRef]
- Friedman, M. Analysis, nutrition, and health benefits of tryptophan. Int. J. Tryptophan Res. 2018, 11, 1178646918802282. [Google Scholar] [CrossRef]
- Cao, W.; Wang, C.; Chin, Y.; Chen, X.; Gao, Y.; Yuan, S.; Xue, C.; Wang, Y.; Tang, Q. DHA-phospholipids (DHA-PL) and EPA-phospholipids (EPA-PL) prevent intestinal dysfunction induced by chronic stress. Food Funct. 2019, 10, 277–288. [Google Scholar] [CrossRef]
- Costantini, L.; Molinari, R.; Farinon, B.; Merendino, N. Impact of omega-3 fatty acids on the gut microbiota. Int. J. Mol. Sci. 2017, 18, 2645. [Google Scholar] [CrossRef]
- Coker, J.K.; Moyne, O.; Rodionov, D.A.; Zengler, K. Carbohydrates great and small, from dietary fiber to sialic acids: How glycans influence the gut microbiome and affect human health. Gut Microbes 2021, 13, 1–18. [Google Scholar] [CrossRef]
- Samuel, B.S.; Shaito, A.; Motoike, T.; Rey, F.E.; Backhed, F.; Manchester, J.K.; Hammer, R.E.; Williams, S.C.; Crowley, J.; Yanagisawa, M. Effects of the gut microbiota on host adiposity are modulated by the short-chain fatty-acid binding G protein-coupled receptor, Gpr41. Proc. Natl. Acad. Sci. USA 2008, 105, 16767–16772. [Google Scholar] [CrossRef] [Green Version]
- De Filippo, C.; Cavalieri, D.; Di Paola, M.; Ramazzotti, M.; Poullet, J.B.; Massart, S.; Collini, S.; Pieraccini, G.; Lionetti, P. Impact of diet in shaping gut microbiota revealed by a comparative study in children from Europe and rural Africa. Proc. Natl. Acad. Sci. USA 2010, 107, 14691–14696. [Google Scholar] [CrossRef]
- Zmora, N.; Suez, J.; Elinav, E. You are what you eat: Diet, health and the gut microbiota. Nat. Rev. Gastroenterol. Hepatol. 2019, 16, 35–56. [Google Scholar] [CrossRef]
- Brown, K.; DeCoffe, D.; Molcan, E.; Gibson, D.L. Diet-induced dysbiosis of the intestinal microbiota and the effects on immunity and disease. Nutrients 2012, 4, 1095–1119. [Google Scholar] [CrossRef]
- Cani, P.D.; Delzenne, N.M. Involvement of the gut microbiota in the development of low grade inflammation associated with obesity: Focus on this neglected partner. Acta Gastroenterol Belg 2010, 73, 267–269. [Google Scholar]
- Zhu, W.; Gregory, J.C.; Org, E.; Buffa, J.A.; Gupta, N.; Wang, Z.; Li, L.; Fu, X.; Wu, Y.; Mehrabian, M. Gut microbial metabolite TMAO enhances platelet hyperreactivity and thrombosis risk. Cell 2016, 165, 111–124. [Google Scholar] [CrossRef]
- Barrea, L.; Annunziata, G.; Muscogiuri, G.; Di Somma, C.; Laudisio, D.; Maisto, M.; De Alteriis, G.; Tenore, G.C.; Colao, A.; Savastano, S. Trimethylamine-N-oxide (TMAO) as novel potential biomarker of early predictors of metabolic syndrome. Nutrients 2018, 10, 1971. [Google Scholar] [CrossRef]
- Zhang, C.; Zhang, M.; Wang, S.; Han, R.; Cao, Y.; Hua, W.; Mao, Y.; Zhang, X.; Pang, X.; Wei, C. Interactions between gut microbiota, host genetics and diet relevant to development of metabolic syndromes in mice. ISME J. 2010, 4, 232–241. [Google Scholar] [CrossRef]
- Sun, G.; Yin, Z.; Liu, N.; Bian, X.; Yu, R.; Su, X.; Zhang, B.; Wang, Y. Gut microbial metabolite TMAO contributes to renal dysfunction in a mouse model of diet-induced obesity. Biochem. Biophys. Res. Commun. 2017, 493, 964–970. [Google Scholar] [CrossRef]
- Chang, C.-J.; Lin, C.-S.; Lu, C.-C.; Martel, J.; Ko, Y.-F.; Ojcius, D.M.; Tseng, S.-F.; Wu, T.-R.; Chen, Y.-Y.M.; Young, J.D. Ganoderma lucidum reduces obesity in mice by modulating the composition of the gut microbiota. Nat. Commun. 2015, 6, 7489. [Google Scholar] [CrossRef]
- Terai, S.; Buchanan-Hughes, A.; Ng, A.; Lee, I.-H.; Hasegawa, K. Comorbidities and healthcare costs and resource use of patients with nonalcoholic fatty liver disease (NAFLD) and nonalcoholic steatohepatitis (NASH) in the Japan medical data vision database. J. Gastroenterol. 2021, 56, 274–284. [Google Scholar] [CrossRef]
- Friedman, S.L.; Neuschwander-Tetri, B.A.; Rinella, M.; Sanyal, A.J. Mechanisms of NAFLD development and therapeutic strategies. Nat. Med. 2018, 24, 908–922. [Google Scholar] [CrossRef]
- Larsen, O.F.A.; van der Grint, M.; Wiegers, C.; van de Burgwal, L.H.M. The Gut Microbiota: Master of Puppets Connecting the Epidemiology of Infectious, Autoimmune, and Metabolic Disease. Front. Microbiol. 2022, 13, 902106. [Google Scholar] [CrossRef]
- Burz, S.D.; Monnoye, M.; Philippe, C.; Farin, W.; Ratziu, V.; Strozzi, F.; Paillarse, J.-M.; Chêne, L.; Blottière, H.M.; Gérard, P. Fecal Microbiota Transplant from Human to Mice Gives Insights into the Role of the Gut Microbiota in Non-Alcoholic Fatty Liver Disease (NAFLD). Microorganisms 2021, 9, 199. [Google Scholar] [CrossRef]
- Raman, M.; Ahmed, I.; Gillevet, P.M.; Probert, C.S.; Ratcliffe, N.M.; Smith, S.; Greenwood, R.; Sikaroodi, M.; Lam, V.; Crotty, P. Fecal microbiome and volatile organic compound metabolome in obese humans with nonalcoholic fatty liver disease. Clin. Gastroenterol. Hepatol. 2013, 11, 868–875. e863. [Google Scholar] [CrossRef]
- Zhu, L.; Baker, S.S.; Gill, C.; Liu, W.; Alkhouri, R.; Baker, R.D.; Gill, S.R. Characterization of gut microbiomes in nonalcoholic steatohepatitis (NASH) patients: A connection between endogenous alcohol and NASH. Hepatology 2013, 57, 601–609. [Google Scholar] [CrossRef]
- Berlanga, A.; Guiu-Jurado, E.; Porras, J.A.; Auguet, T. Molecular pathways in non-alcoholic fatty liver disease. Clin. Exp. Gastroenterol. 2014, 7, 221. [Google Scholar]
- Caussy, C.; Tripathi, A.; Humphrey, G.; Bassirian, S.; Singh, S.; Faulkner, C.; Bettencourt, R.; Rizo, E.; Richards, L.; Xu, Z.Z. A gut microbiome signature for cirrhosis due to nonalcoholic fatty liver disease. Nat. Commun. 2019, 10, 1406. [Google Scholar] [CrossRef]
- Leong, P.K.; Ko, K.M. Schisandrin B: A double-edged sword in nonalcoholic fatty liver disease. Oxidative medicine and cellular longevity 2016, 2016, 6171658. [Google Scholar] [CrossRef]
- Gupta, H.; Youn, G.S.; Shin, M.J.; Suk, K.T. Role of Gut Microbiota in Hepatocarcinogenesis. Microorganisms 2019, 7, 121. [Google Scholar] [CrossRef]
- Aravalli, R.N.; Cressman, E.N.; Steer, C.J. Cellular and molecular mechanisms of hepatocellular carcinoma: An update. Arch. Toxicol. 2013, 87, 227–247. [Google Scholar] [CrossRef]
- Henao-Mejia, J.; Elinav, E.; Jin, C.; Hao, L.; Mehal, W.Z.; Strowig, T.; Thaiss, C.A.; Kau, A.L.; Eisenbarth, S.C.; Jurczak, M.J. Inflammasome-mediated dysbiosis regulates progression of NAFLD and obesity. Nature 2012, 482, 179–185. [Google Scholar] [CrossRef]
- Zeng, H.; Liu, J.; Jackson, M.I.; Zhao, F.-Q.; Yan, L.; Combs Jr, G.F. Fatty liver accompanies an increase in lactobacillus species in the hind gut of C57BL/6 mice fed a high-fat diet. J. Nutr. 2013, 143, 627–631. [Google Scholar] [CrossRef] [Green Version]
- Cano, P.G.; Santacruz, A.; Trejo, F.M.; Sanz, Y. Bifidobacterium CECT 7765 improves metabolic and immunological alterations associated with obesity in high-fat diet-fed mice. Obesity 2013, 21, 2310–2321. [Google Scholar] [CrossRef]
- Buzzetti, E.; Pinzani, M.; Tsochatzis, E.A. The multiple-hit pathogenesis of non-alcoholic fatty liver disease (NAFLD). Metabolism 2016, 65, 1038–1048. [Google Scholar] [CrossRef]
- Leung, C.; Rivera, L.; Furness, J.B.; Angus, P.W. The role of the gut microbiota in NAFLD. Nat. Rev. Gastroenterol. Hepatol. 2016, 13, 412–425. [Google Scholar] [CrossRef]
- Romero-Gómez, M.; Zelber-Sagi, S.; Trenell, M. Treatment of NAFLD with diet, physical activity and exercise. J. Hepatol. 2017, 67, 829–846. [Google Scholar] [CrossRef]
- Cheng, Y.; Zhang, K.; Chen, Y.; Li, Y.; Li, Y.; Fu, K.; Feng, R. Associations between dietary nutrient intakes and hepatic lipid contents in NAFLD patients quantified by 1H-MRS and dual-Echo MRI. Nutrients 2016, 8, 527. [Google Scholar] [CrossRef]
- Kolodziejczyk, A.A.; Zheng, D.; Shibolet, O.; Elinav, E. The role of the microbiome in NAFLD and NASH. EMBO Mol. Med. 2019, 11. [Google Scholar] [CrossRef]
- Maslowski, K.M.; Vieira, A.T.; Ng, A.; Kranich, J.; Sierro, F.; Yu, D.; Schilter, H.C.; Rolph, M.S.; Mackay, F.; Artis, D. Regulation of inflammatory responses by gut microbiota and chemoattractant receptor GPR43. Nature 2009, 461, 1282–1286. [Google Scholar] [CrossRef]
- Daliri, E.B.-M.; Lee, B.H. New perspectives on probiotics in health and disease. Food Sci. Hum. Wellness 2015, 4, 56–65. [Google Scholar] [CrossRef]
- Daliri, E.B.-M.; Wei, S.; Oh, D.H.; Lee, B.H. The human microbiome and metabolomics: Current concepts and applications. Crit. Rev. Food Sci. Nutr. 2017, 57, 3565–3576. [Google Scholar] [CrossRef]
- de Diego-Cabero, N.; Mereu, A.; Menoyo, D.; Holst, J.J.; Ipharraguerre, I.R. Bile acid mediated effects on gut integrity and performance of early-weaned piglets. BMC Vet. Res. 2015, 11, 111. [Google Scholar] [CrossRef] [Green Version]
- Adom, D.; Nie, D. Regulation of autophagy by short chain fatty acids in colon cancer cells. In Autophagy–A Double-Edged Sword–Cell Survival or Death; Intechopen: London, UK, 2013; pp. 235–247. [Google Scholar]
- Den Besten, G.; Bleeker, A.; Gerding, A.; van Eunen, K.; Havinga, R.; van Dijk, T.H.; Oosterveer, M.H.; Jonker, J.W.; Groen, A.K.; Reijngoud, D.-J. Short-chain fatty acids protect against high-fat diet–induced obesity via a PPARγ-dependent switch from lipogenesis to fat oxidation. Diabetes 2015, 64, 2398–2408. [Google Scholar] [CrossRef]
- Kimura, I.; Ozawa, K.; Inoue, D.; Imamura, T.; Kimura, K.; Maeda, T.; Terasawa, K.; Kashihara, D.; Hirano, K.; Tani, T. The gut microbiota suppresses insulin-mediated fat accumulation via the short-chain fatty acid receptor GPR43. Nat. Commun. 2013, 4, 1829. [Google Scholar] [CrossRef]
- Sahuri-Arisoylu, M.; Brody, L.; Parkinson, J.; Parkes, H.; Navaratnam, N.; Miller, A.D.; Thomas, E.; Frost, G.; Bell, J. Reprogramming of hepatic fat accumulation and ‘browning’ of adipose tissue by the short-chain fatty acid acetate. Int. J. Obes. 2016, 40, 955–963. [Google Scholar] [CrossRef]
- Zhou, D.; Chen, Y.-W.; Zhao, Z.-H.; Yang, R.-X.; Xin, F.-Z.; Liu, X.-L.; Pan, Q.; Zhou, H.; Fan, J.-G. Sodium butyrate reduces high-fat diet-induced non-alcoholic steatohepatitis through upregulation of hepatic GLP-1R expression. Exp. Mol. Med. 2018, 50, 1–12. [Google Scholar] [CrossRef]
- Cani, P.D.; Possemiers, S.; Van de Wiele, T.; Guiot, Y.; Everard, A.; Rottier, O.; Geurts, L.; Naslain, D.; Neyrinck, A.; Lambert, D.M. Changes in gut microbiota control inflammation in obese mice through a mechanism involving GLP-2-driven improvement of gut permeability. Gut 2009, 58, 1091–1103. [Google Scholar] [CrossRef]
- Joyce, S.A.; Gahan, C.G. Bile acid modifications at the microbe-host interface: Potential for nutraceutical and pharmaceutical interventions in host health. Annu. Rev. Food Sci. Technol. 2016, 7, 313–333. [Google Scholar] [CrossRef]
- Fiorucci, S.; Distrutti, E. Bile acid-activated receptors, intestinal microbiota, and the treatment of metabolic disorders. Trends Mol. Med. 2015, 21, 702–714. [Google Scholar] [CrossRef]
- Esteller, A. Physiology of bile secretion. World J. Gastroenterol. WJG 2008, 14, 5641. [Google Scholar] [CrossRef]
- Zhu, C.; Fuchs, C.D.; Halilbasic, E.; Trauner, M. Bile acids in regulation of inflammation and immunity: Friend or foe. Clin. Exp. Rheumatol. 2016, 34, 25–31. [Google Scholar]
- Li, Y.C.; Wang, D.P.; Chiang, J. Regulation of cholesterol 7 alpha-hydroxylase in the liver. Cloning, sequencing, and regulation of cholesterol 7 alpha-hydroxylase mRNA. J. Biol. Chem. 1990, 265, 12012–12019. [Google Scholar] [CrossRef]
- Chiang, J.Y. Bile acid regulation of gene expression: Roles of nuclear hormone receptors. Endocr. Rev. 2002, 23, 443–463. [Google Scholar] [CrossRef] [Green Version]
- Chiang, J.Y. Bile acids: Regulation of synthesis: Thematic Review Series: Bile Acids. J. Lipid Res. 2009, 50, 1955–1966. [Google Scholar] [CrossRef]
- Monte, M.J.; Marin, J.J.; Antelo, A.; Vazquez-Tato, J. Bile acids: Chemistry, physiology, and pathophysiology. World J. Gastroenterol. WJG 2009, 15, 804. [Google Scholar] [CrossRef]
- Arab, J.P.; Karpen, S.J.; Dawson, P.A.; Arrese, M.; Trauner, M. Bile acids and nonalcoholic fatty liver disease: Molecular insights and therapeutic perspectives. Hepatology 2017, 65, 350–362. [Google Scholar] [CrossRef]
- Mouzaki, M.; Wang, A.Y.; Bandsma, R.; Comelli, E.M.; Arendt, B.M.; Zhang, L.; Fung, S.; Fischer, S.E.; McGilvray, I.G.; Allard, J.P. Bile acids and dysbiosis in non-alcoholic fatty liver disease. PLoS ONE 2016, 11, e0151829. [Google Scholar] [CrossRef]
- Ferslew, B.C.; Xie, G.; Johnston, C.K.; Su, M.; Stewart, P.W.; Jia, W.; Brouwer, K.L.; Barritt, A.S. Altered bile acid metabolome in patients with nonalcoholic steatohepatitis. Dig. Dis. Sci. 2015, 60, 3318–3328. [Google Scholar] [CrossRef]
- Chávez-Talavera, O.; Tailleux, A.; Lefebvre, P.; Staels, B. Bile acid control of metabolism and inflammation in obesity, type 2 diabetes, dyslipidemia, and nonalcoholic fatty liver disease. Gastroenterology 2017, 152, 1679–1694. e1673. [Google Scholar] [CrossRef]
- Houten, S.M.; Watanabe, M.; Auwerx, J. Endocrine functions of bile acids. EMBO J. 2006, 25, 1419–1425. [Google Scholar] [CrossRef]
- Nguyen, A.; Bouscarel, B. Bile acids and signal transduction: Role in glucose homeostasis. Cell. Signal. 2008, 20, 2180–2197. [Google Scholar] [CrossRef]
- Modica, S.; Murzilli, S.; Salvatore, L.; Schmidt, D.R.; Moschetta, A. Nuclear bile acid receptor FXR protects against intestinal tumorigenesis. Cancer Res. 2008, 68, 9589–9594. [Google Scholar] [CrossRef]
- Chiang, J.Y.; Ferrell, J.M. Bile acid metabolism in liver pathobiology. Gene Expr. 2018, 18, 71. [Google Scholar] [CrossRef] [Green Version]
- Jiang, C.; Xie, C.; Li, F.; Zhang, L.; Nichols, R.G.; Krausz, K.W.; Cai, J.; Qi, Y.; Fang, Z.-Z.; Takahashi, S. Intestinal farnesoid X receptor signaling promotes nonalcoholic fatty liver disease. J. Clin. Investig. 2015, 125, 386–402. [Google Scholar] [CrossRef]
- Louis, P.; Flint, H.J. Diversity, metabolism and microbial ecology of butyrate-producing bacteria from the human large intestine. FEMS Microbiol. Lett. 2009, 294, 1–8. [Google Scholar] [CrossRef]
- Silva, Y.P.; Bernardi, A.; Frozza, R.L. The role of short-chain fatty acids from gut microbiota in gut-brain communication. Front. Endocrinol. 2020, 11, 25. [Google Scholar] [CrossRef]
- Schwiertz, A.; Taras, D.; Schäfer, K.; Beijer, S.; Bos, N.A.; Donus, C.; Hardt, P.D. Microbiota and SCFA in lean and overweight healthy subjects. Obesity 2010, 18, 190–195. [Google Scholar] [CrossRef]
- Ruppin, H.; Bar-Meir, S.; Soergel, K.H.; Wood, C.M.; Schmitt Jr, M.G. Absorption of short-chain fatty acids by the colon. Gastroenterology 1980, 78, 1500–1507. [Google Scholar] [CrossRef]
- Vinolo, M.A.; Rodrigues, H.G.; Nachbar, R.T.; Curi, R. Regulation of inflammation by short chain fatty acids. Nutrients 2011, 3, 858–876. [Google Scholar] [CrossRef]
- De Vadder, F.; Kovatcheva-Datchary, P.; Goncalves, D.; Vinera, J.; Zitoun, C.; Duchampt, A.; Bäckhed, F.; Mithieux, G. Microbiota-generated metabolites promote metabolic benefits via gut-brain neural circuits. Cell 2014, 156, 84–96. [Google Scholar] [CrossRef]
- Byndloss, M.X.; Olsan, E.E.; Rivera-Chávez, F.; Tiffany, C.R.; Cevallos, S.A.; Lokken, K.L.; Torres, T.P.; Byndloss, A.J.; Faber, F.; Gao, Y. Microbiota-activated PPAR-γ signaling inhibits dysbiotic Enterobacteriaceae expansion. Science 2017, 357, 570–575. [Google Scholar] [CrossRef]
- Da Zhou, Q.P.; Xin, F.-Z.; Zhang, R.-N.; He, C.-X.; Chen, G.-Y.; Liu, C.; Chen, Y.-W.; Fan, J.-G. Sodium butyrate attenuates high-fat diet-induced steatohepatitis in mice by improving gut microbiota and gastrointestinal barrier. World J. Gastroenterol. 2017, 23, 60. [Google Scholar] [CrossRef]
- Wu, G.; Meininger, C.J. Regulation of nitric oxide synthesis by dietary factors. Annu. Rev. Nutr. 2002, 22, 61–86. [Google Scholar] [CrossRef]
- Evenepoel, P.; Claus, D.; Geypens, B.; Hiele, M.; Geboes, K.; Rutgeerts, P.; Ghoos, Y. Amount and fate of egg protein escaping assimilation in the small intestine of humans. Am. J. Physiol. -Gastrointest. Liver Physiol. 1999, 277, G935–G943. [Google Scholar] [CrossRef]
- Smith, E.A.; Macfarlane, G.T. Enumeration of human colonic bacteria producing phenolic and indolic compounds: Effects of pH, carbohydrate availability and retention time on dissimilatory aromatic amino acid metabolism. J. Appl. Bacteriol. 1996, 81, 288–302. [Google Scholar] [CrossRef]
- Chen, J.; Vitetta, L. Gut microbiota metabolites in nafld pathogenesis and therapeutic implications. Int. J. Mol. Sci. 2020, 21, 5214. [Google Scholar] [CrossRef]
- Herrera, C.P.; Smith, K.; Atkinson, F.; Ruell, P.; Chow, C.M.; O’Connor, H.; Brand-Miller, J. High-glycaemic index and-glycaemic load meals increase the availability of tryptophan in healthy volunteers. Br. J. Nutr. 2011, 105, 1601–1606. [Google Scholar] [CrossRef]
- Roager, H.M.; Licht, T.R. Microbial tryptophan catabolites in health and disease. Nat. Commun. 2018, 9, 3294. [Google Scholar] [CrossRef]
- Gostner, J.M.; Geisler, S.; Stonig, M.; Mair, L.; Sperner-Unterweger, B.; Fuchs, D. Tryptophan metabolism and related pathways in psychoneuroimmunology: The impact of nutrition and lifestyle. Neuropsychobiology 2020, 79, 89–99. [Google Scholar] [CrossRef]
- Bansal, T.; Alaniz, R.C.; Wood, T.K.; Jayaraman, A. The bacterial signal indole increases epithelial-cell tight-junction resistance and attenuates indicators of inflammation. Proc. Natl. Acad. Sci. USA 2010, 107, 228–233. [Google Scholar] [CrossRef]
- Cherayil, B.J. Indoleamine 2, 3-dioxygenase in intestinal immunity and inflammation. Inflamm. Bowel Dis. 2009, 15, 1391–1396. [Google Scholar] [CrossRef]
- Laurans, L.; Venteclef, N.; Haddad, Y.; Chajadine, M.; Alzaid, F.; Metghalchi, S.; Sovran, B.; Denis, R.G.; Dairou, J.; Cardellini, M. Genetic deficiency of indoleamine 2, 3-dioxygenase promotes gut microbiota-mediated metabolic health. Nat. Med. 2018, 24, 1113–1120. [Google Scholar] [CrossRef]
- Ji, Y.; Yin, Y.; Li, Z.; Zhang, W. Gut microbiota-derived components and metabolites in the progression of non-alcoholic fatty liver disease (NAFLD). Nutrients 2019, 11, 1712. [Google Scholar] [CrossRef] [Green Version]
- Hou, Q.; Ye, L.; Liu, H.; Huang, L.; Yang, Q.; Turner, J.; Yu, Q. Lactobacillus accelerates ISCs regeneration to protect the integrity of intestinal mucosa through activation of STAT3 signaling pathway induced by LPLs secretion of IL-22. Cell Death Differ. 2018, 25, 1657–1670. [Google Scholar] [CrossRef]
- Ma, L.; Li, H.; Hu, J.; Zheng, J.; Zhou, J.; Botchlett, R.; Matthews, D.; Zeng, T.; Chen, L.; Xiao, X. Indole Alleviates Diet-Induced Hepatic Steatosis and Inflammation in a Manner Involving Myeloid Cell 6-Phosphofructo-2-Kinase/Fructose-2, 6-Biphosphatase 3. Hepatology 2020, 72, 1191–1203. [Google Scholar] [CrossRef]
- Kessel, A.; Toubi, E.; Pavlotzky, E.; Mogilner, J.; Coran, A.; Lurie, M.; Karry, R.; Sukhotnik, I. Treatment with glutamine is associated with down-regulation of Toll-like receptor-4 and myeloid differentiation factor 88 expression and decrease in intestinal mucosal injury caused by lipopolysaccharide endotoxaemia in a rat. Clin. Exp. Immunol. 2008, 151, 341–347. [Google Scholar] [CrossRef]
- Simon, J.; Martinez-Chantar, M.L.; Delgado, T.C. Glutamine, fatty liver disease and aging. Aging 2021, 13, 3165–3166. [Google Scholar] [CrossRef]
- Zhang, W.-X.; Zhou, L.-F.; Zhang, L.; Bao, L.; Wang, C.-C.; Meng, H.-Y.; Yin, W. Protective effects of glutamine preconditioning on ischemia-reperfusion injury in rats. Hepatobiliary Pancreat. Dis. Int. 2011, 10, 78–82. [Google Scholar] [CrossRef]
- Neu, J.; DeMarco, V.; Li, N. Glutamine: Clinical applications and mechanisms of action. Curr. Opin. Clin. Nutr. Metab. Care 2002, 5, 69–75. [Google Scholar] [CrossRef]
- Corbin, K.D.; Zeisel, S.H. Choline metabolism provides novel insights into nonalcoholic fatty liver disease and its progression. Curr. Opin. Gastroenterol. 2012, 28, 159–165. [Google Scholar] [CrossRef]
- Guerrerio, A.L.; Colvin, R.M.; Schwartz, A.K.; Molleston, J.P.; Murray, K.F.; Diehl, A.; Mohan, P.; Schwimmer, J.B.; Lavine, J.E.; Torbenson, M.S.; et al. Choline intake in a large cohort of patients with nonalcoholic fatty liver disease. Am. J. Clin. Nutr. 2012, 95, 892–900. [Google Scholar] [CrossRef] [PubMed]
- Yao, Z.M.; Vance, D.E. Reduction in VLDL, but not HDL, in plasma of rats deficient in choline. Biochem Cell Biol. 1990, 68, 552–558. [Google Scholar] [CrossRef] [PubMed]
- Schneider, K.M.; Mohs, A.; Kilic, K.; Candels, L.S.; Elfers, C.; Bennek, E.; Schneider, L.B.; Heymann, F.; Gassler, N.; Penders, J.; et al. Intestinal Microbiota Protects against MCD Diet-Induced Steatohepatitis. Int. J. Mol. Sci. 2019, 20, 308. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ye, J.Z.; Li, Y.T.; Wu, W.R.; Shi, D.; Fang, D.Q.; Yang, L.Y.; Bian, X.Y.; Wu, J.J.; Wang, Q.; Jiang, X.W.; et al. Dynamic alterations in the gut microbiota and metabolome during the development of methionine-choline-deficient diet-induced nonalcoholic steatohepatitis. World J. Gastroenterol. 2018, 24, 2468–2481. [Google Scholar] [CrossRef]
- Baker, J.R.; Chaykin, S. The biosynthesis of trimethylamine-N-oxide. J. Biol. Chem. 1962, 237, 1309–1313. [Google Scholar] [CrossRef]
- Lang, D.H.; Yeung, C.K.; Peter, R.M.; Ibarra, C.; Gasser, R.; Itagaki, K.; Philpot, R.M.; Rettie, A.E. Isoform specificity of trimethylamine N-oxygenation by human flavin-containing monooxygenase (FMO) and P450 enzymes: Selective catalysis by FMO3. Biochem. Pharmacol. 1998, 56, 1005–1012. [Google Scholar] [CrossRef]
- Romano, K.A.; Martinez-Del Campo, A.; Kasahara, K.; Chittim, C.L.; Vivas, E.I.; Amador-Noguez, D.; Balskus, E.P.; Rey, F.E. Metabolic, Epigenetic, and Transgenerational Effects of Gut Bacterial Choline Consumption. Cell Host Microbe 2017, 22, 279–290.e277. [Google Scholar] [CrossRef]
- Spencer, M.D.; Hamp, T.J.; Reid, R.W.; Fischer, L.M.; Zeisel, S.H.; Fodor, A.A. Association between composition of the human gastrointestinal microbiome and development of fatty liver with choline deficiency. Gastroenterology 2011, 140, 976–986. [Google Scholar] [CrossRef]
- Chen, Y.M.; Liu, Y.; Zhou, R.F.; Chen, X.L.; Wang, C.; Tan, X.Y.; Wang, L.J.; Zheng, R.D.; Zhang, H.W.; Ling, W.H.; et al. Associations of gut-flora-dependent metabolite trimethylamine-N-oxide, betaine and choline with non-alcoholic fatty liver disease in adults. Sci. Rep. 2016, 6, 19076. [Google Scholar] [CrossRef]
- Gao, X.; Liu, X.; Xu, J.; Xue, C.; Xue, Y.; Wang, Y. Dietary trimethylamine N-oxide exacerbates impaired glucose tolerance in mice fed a high fat diet. J. Biosci. Bioeng. 2014, 118, 476–481. [Google Scholar] [CrossRef] [PubMed]
- Shabbir, U.; Rubab, M.; Daliri, E.B.-M.; Chelliah, R.; Javed, A.; Oh, D.-H. Curcumin, Quercetin, Catechins and Metabolic Diseases: The Role of Gut Microbiota. Nutrients 2021, 13, 206. [Google Scholar] [CrossRef] [PubMed]
- Ozdal, T.; Sela, D.A.; Xiao, J.; Boyacioglu, D.; Chen, F.; Capanoglu, E. The reciprocal interactions between polyphenols and gut microbiota and effects on bioaccessibility. Nutrients 2016, 8, 78. [Google Scholar] [CrossRef] [PubMed]
- Ding, S.; Xu, S.; Ma, Y.; Liu, G.; Jang, H.; Fang, J. Modulatory mechanisms of the NLRP3 inflammasomes in diabetes. Biomolecules 2019, 9, 850. [Google Scholar] [CrossRef] [Green Version]
- Ma, J.; Zheng, Y.; Tang, W.; Yan, W.; Nie, H.; Fang, J.; Liu, G. Dietary polyphenols in lipid metabolism: A role of gut microbiome. Anim. Nutr. 2020, 6, 404–409. [Google Scholar] [CrossRef]
- Bian, Y.; Wei, J.; Zhao, C.; Li, G. Natural polyphenols targeting senescence: A novel prevention and therapy strategy for cancer. Int. J. Mol. Sci. 2020, 21, 684. [Google Scholar] [CrossRef]
- Alves-Santos, A.M.; Sugizaki, C.S.A.; Lima, G.C.; Naves, M.M.V. Prebiotic effect of dietary polyphenols: A systematic review. J. Funct. Foods 2020, 74, 104169. [Google Scholar] [CrossRef]
- Moorthy, M.; Chaiyakunapruk, N.; Jacob, S.A.; Palanisamy, U.D. Prebiotic potential of polyphenols, its effect on gut microbiota and anthropometric/clinical markers: A systematic review of randomised controlled trials. Trends Food Sci. Technol. 2020, 99, 634–649. [Google Scholar] [CrossRef]
- Roberfroid, M.; Gibson, G.R.; Hoyles, L.; McCartney, A.L.; Rastall, R.; Rowland, I.; Wolvers, D.; Watzl, B.; Szajewska, H.; Stahl, B. Prebiotic effects: Metabolic and health benefits. Br. J. Nutr. 2010, 104, S1–S63. [Google Scholar] [CrossRef]
- Rafiei, H.; Omidian, K.; Bandy, B. Dietary polyphenols protect against oleic acid-induced steatosis in an in vitro model of NAFLD by modulating lipid metabolism and improving mitochondrial function. Nutrients 2019, 11, 541. [Google Scholar] [CrossRef]
- Li, A.-N.; Li, S.; Zhang, Y.-J.; Xu, X.-R.; Chen, Y.-M.; Li, H.-B. Resources and biological activities of natural polyphenols. Nutrients 2014, 6, 6020–6047. [Google Scholar] [CrossRef] [PubMed]
- Lamuel-Raventos, R.M.; Onge, M.-P.S. Prebiotic nut compounds and human microbiota. Crit. Rev. Food Sci. Nutr. 2017, 57, 3154–3163. [Google Scholar] [CrossRef] [PubMed]
- Li, S.; Tan, H.Y.; Wang, N.; Cheung, F.; Hong, M.; Feng, Y. The potential and action mechanism of polyphenols in the treatment of liver diseases. Oxidative Med. Cell. Longev. 2018, 2018, 8394818. [Google Scholar] [CrossRef] [PubMed]
- Byelashov, O.A.; Sinclair, A.J.; Kaur, G. Dietary sources, current intakes, and nutritional role of omega-3 docosapentaenoic acid. Lipid Technol. 2015, 27, 79–82. [Google Scholar] [CrossRef] [Green Version]
- Calder, P.C. Marine omega-3 fatty acids and inflammatory processes: Effects, mechanisms and clinical relevance. Biochim. Et Biophys. Acta (BBA)-Mol. Cell Biol. Lipids 2015, 1851, 469–484. [Google Scholar] [CrossRef]
- Calder, P.C. Omega-3 polyunsaturated fatty acids and inflammatory processes: Nutrition or pharmacology? Br. J. Clin. Pharmacol. 2013, 75, 645–662. [Google Scholar] [CrossRef]
- Brown, T.J.; Brainard, J.; Song, F.; Wang, X.; Abdelhamid, A.; Hooper, L. Omega-3, omega-6, and total dietary polyunsaturated fat for prevention and treatment of type 2 diabetes mellitus: Systematic review and meta-analysis of randomised controlled trials. BMJ 2019, 366, l4697. [Google Scholar] [CrossRef]
- He, X.-X.; Wu, X.-L.; Chen, R.-P.; Chen, C.; Liu, X.-G.; Wu, B.-J.; Huang, Z.-M. Effectiveness of omega-3 polyunsaturated fatty acids in non-alcoholic fatty liver disease: A meta-analysis of randomized controlled trials. PLoS ONE 2016, 11, e0162368. [Google Scholar] [CrossRef]
- Simopoulos, A.P. An increase in the omega-6/omega-3 fatty acid ratio increases the risk for obesity. Nutrients 2016, 8, 128. [Google Scholar] [CrossRef]
- de Castro, G.S.; Calder, P.C. Non-alcoholic fatty liver disease and its treatment with n-3 polyunsaturated fatty acids. Clin. Nutr. 2018, 37, 37–55. [Google Scholar] [CrossRef]
- Liu, H.-Q.; Qiu, Y.; Mu, Y.; Zhang, X.-J.; Liu, L.; Hou, X.-H.; Zhang, L.; Xu, X.-N.; Ji, A.-L.; Cao, R. A high ratio of dietary n-3/n-6 polyunsaturated fatty acids improves obesity-linked inflammation and insulin resistance through suppressing activation of TLR4 in SD rats. Nutr. Res. 2013, 33, 849–858. [Google Scholar] [CrossRef] [PubMed]
- Druart, C.; Bindels, L.B.; Schmaltz, R.; Neyrinck, A.M.; Cani, P.D.; Walter, J.; Ramer-Tait, A.E.; Delzenne, N.M. Ability of the gut microbiota to produce PUFA-derived bacterial metabolites: Proof of concept in germ-free versus conventionalized mice. Mol. Nutr. Food Res. 2015, 59, 1603–1613. [Google Scholar] [CrossRef] [PubMed]
- Cieślak, A.; Trottier, J.; Verreault, M.; Milkiewicz, P.; Vohl, M.-C.; Barbier, O. N-3 polyunsaturated fatty acids stimulate bile acid detoxification in human cell models. Can. J. Gastroenterol. Hepatol. 2018, 2018, 6031074. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Z.; Hyun, J.E.; Thiesen, A.; Park, H.; Hotte, N.; Watanabe, H.; Higashiyama, T.; Madsen, K.L. Sex-Specific Differences in the Gut Microbiome in Response to Dietary Fiber Supplementation in IL-10-Deficient Mice. Nutrients 2020, 12, 2088. [Google Scholar] [CrossRef] [PubMed]
- Stephen, A.M.; Champ, M.M.-J.; Cloran, S.J.; Fleith, M.; van Lieshout, L.; Mejborn, H.; Burley, V.J. Dietary fibre in Europe: Current state of knowledge on definitions, sources, recommendations, intakes and relationships to health. Nutr. Res. Rev. 2017, 30, 149–190. [Google Scholar] [CrossRef]
- Den Besten, G.; Van Eunen, K.; Groen, A.K.; Venema, K.; Reijngoud, D.-J.; Bakker, B.M. The role of short-chain fatty acids in the interplay between diet, gut microbiota, and host energy metabolism. J. Lipid Res. 2013, 54, 2325–2340. [Google Scholar] [CrossRef]
- Murga-Garrido, S.M.; Hong, Q.; Cross, T.-W.L.; Hutchison, E.R.; Han, J.; Thomas, S.P.; Vivas, E.I.; Denu, J.; Ceschin, D.G.; Tang, Z.-Z. Gut microbiome variation modulates the effects of dietary fiber on host metabolism. Microbiome 2021, 9, 117. [Google Scholar] [CrossRef]
- Kuo, S.-M. The interplay between fiber and the intestinal microbiome in the inflammatory response. Adv. Nutr. 2013, 4, 16–28. [Google Scholar] [CrossRef]
- Choe, S.S.; Huh, J.Y.; Hwang, I.J.; Kim, J.I.; Kim, J.B. Adipose tissue remodeling: Its role in energy metabolism and metabolic disorders. Front. Endocrinol. 2016, 7, 30. [Google Scholar] [CrossRef]
- Argo, C.K.; Northup, P.G.; Al-Osaimi, A.M.; Caldwell, S.H. Systematic review of risk factors for fibrosis progression in non-alcoholic steatohepatitis. J. Hepatol. 2009, 51, 371–379. [Google Scholar] [CrossRef]
- Ma, J.; Fox, C.S.; Jacques, P.F.; Speliotes, E.K.; Hoffmann, U.; Smith, C.E.; Saltzman, E.; McKeown, N.M. Sugar-sweetened beverage, diet soda, and fatty liver disease in the Framingham Heart Study cohorts. J. Hepatol. 2015, 63, 462–469. [Google Scholar] [CrossRef] [PubMed]
- Do, M.H.; Lee, E.; Oh, M.-J.; Kim, Y.; Park, H.-Y. High-glucose or-fructose diet cause changes of the gut microbiota and metabolic disorders in mice without body weight change. Nutrients 2018, 10, 761. [Google Scholar] [CrossRef] [PubMed]
- Ishimoto, T.; Lanaspa, M.A.; Rivard, C.J.; Roncal-Jimenez, C.A.; Orlicky, D.J.; Cicerchi, C.; McMahan, R.H.; Abdelmalek, M.F.; Rosen, H.R.; Jackman, M.R. High-fat and high-sucrose (western) diet induces steatohepatitis that is dependent on fructokinase. Hepatology 2013, 58, 1632–1643. [Google Scholar] [CrossRef] [PubMed] [Green Version]
| Nutrient | Microbiota Changes | Altered Metabolites | Reference |
|---|---|---|---|
| Protein | (↑): Bacteroidetes, Lactobacillus | (↑): Sulfide, polysaccharide lyases, tryptophan catabolism | [26,27] |
| (↓): Firmicutes, Clostridium | |||
| Omega-3 | (↑): Bifidobacteria, Lachnospiraceae, Roseburia, Bacteroidetes | (↑): SCFAs | [28,29] |
| (↓): Enterobacteria, Faecalibacterium | (↓): IL-1β, IL-6, TNF-α | ||
| Fiber | (↑): Prevotella, Xylanibacter spp., Bifidobacterium, Roseburia, Faecalibacterium | (↑): SCFAs | [30,31,32] |
| Low-fiber | (↑): Akkermansia, Bacteroides caccae | [33] | |
| (↓): Escherichia coli | |||
| Fat intake | (↑): Firmicutes, Clostridium | (↑): LPS, Indoxyl sulfate, p-cresyl sulfate | [34,35] |
| (↓): Lactobacillus, Bacteroidetes, Bifidobacteria spp., Akkermansia | |||
| Fructose intake | (↑): Clostridium innocuum, Catenibacterium mitsuokai, Enterococcus spp. | TMAO | [36,37] |
| High-fat diet | (↑): Firmicutes, Proteobacteria | (↑): TMAO, LPS | [24,38,39,40] |
| (↓): Bacteroidetes, Bifidobacteria |
| Liver Disease | Microbiota Changes | Major Impacts | References |
|---|---|---|---|
| NAFLD | (↑): Proteobacteria, Firmicutes, Lactobacillus, Parabacteroides, Allisonella, C. coccoides (↓): Oscillibacter, Faecalibacterium, Anaerosporobacter | (↑): FFAs, triglycerides, de novo lipogenesis (↓): ApoB | [45,46,47] |
| NAFLD-associated Cirrhosis | (↑): Streptococcus, Lactococcus, Enterobacteriaceae (↓): Bacteroidetes, Bacillus, F. prausnitzii, Prevotella | (↑): PDGF, TGF-β, ECM | [48,49] |
| NAFLD-associated HCC | (↑): E. coli, Actinobacteria (↓): Parabacteroides, butyrate-producing genera | (↑): HBV, HCV, Wnt/β-catenin, JAK/STAT, Rb, p53, MAPK | [50,51] |
| Model | |||
| Methionine choline-deficient diet model, leptin receptor deficiency steatosis model, and the high-fat diet model. | (↑): Hepatic steatosis, hepatic inflammation, lipid biosynthesis (↑): Bacteroidetes, Prevotellaceae, Deferribacteres, Oscillibacter (↓): Lactobacillus, Bacteroidetes | [52] | |
| High-fat (45% energy) or low-fat (10% energy) diet for 10 weeks. | (↑): Body weight (by 34%) (↓): Hepatic steatosis, hepatic inflammation (↑): L. gasseri, L.s taiwanensis | [53] | |
| High-fat diet-induced and lean mice (7 weeks): supplemented with B. pseudocatenulatum | (↓): Insulin resistance, hepatic fat, serum inflammatory markers, body weight (↑): Bifidobacteria, Enterobacteriaceae | [54] | |
| Nutrients | Microbiota | Function | Reference |
|---|---|---|---|
| Bile acid | Bifidobacterium, Lactobacillus, Enterobacter, Clostridium, Bacteroides | (↑): Inhibit growth of pathogens, gut-derived hormone secretion, insulin resistance, gut barrier | [60,61,62,63] |
| SCFAs | Bifidobacterium sp., Roseburia, Clostridium, Faecalibacterium, Coprococcus | (↑): Protect from diet-induced obesity, (↓): Hepatic lipid accumulation | [64,65] |
| Acetate | Bacteroidetes | (↑): Insulin signaling, hepatic function, activation of GPR43 (↓): Fat accumulation, lipid storage | [66,67] |
| Propionate | Bacteroidetes, B. obeum, C. catus, R. inulinivorans, P. copri | (↑): Regulation of colonic T reg cell homeostasis, | |
| Butyrate | Clostridia, F. prausnitzii, Eubacterium, Roseburia, C. catus, A. hadrus | (↑): GLP-2, GLP-1R level, antiinflammation, gut–gut barrier function, T reg cell homeostasis, colonic suppression of colonic inflammation, activation of GPR43 | [60,68,69] |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Jeong, M.K.; Min, B.H.; Choi, Y.R.; Hyun, J.Y.; Park, H.J.; Eom, J.A.; Won, S.M.; Jeong, J.J.; Oh, K.K.; Gupta, H.; et al. Food and Gut Microbiota-Derived Metabolites in Nonalcoholic Fatty Liver Disease. Foods 2022, 11, 2703. https://doi.org/10.3390/foods11172703
Jeong MK, Min BH, Choi YR, Hyun JY, Park HJ, Eom JA, Won SM, Jeong JJ, Oh KK, Gupta H, et al. Food and Gut Microbiota-Derived Metabolites in Nonalcoholic Fatty Liver Disease. Foods. 2022; 11(17):2703. https://doi.org/10.3390/foods11172703
Chicago/Turabian StyleJeong, Min Kyo, Byeong Hyun Min, Ye Rin Choi, Ji Ye Hyun, Hee Jin Park, Jung A Eom, Sung Min Won, Jin Ju Jeong, Ki Kwang Oh, Haripriya Gupta, and et al. 2022. "Food and Gut Microbiota-Derived Metabolites in Nonalcoholic Fatty Liver Disease" Foods 11, no. 17: 2703. https://doi.org/10.3390/foods11172703
APA StyleJeong, M. K., Min, B. H., Choi, Y. R., Hyun, J. Y., Park, H. J., Eom, J. A., Won, S. M., Jeong, J. J., Oh, K. K., Gupta, H., Ganesan, R., Sharma, S. P., Yoon, S. J., Choi, M. R., Kim, D. J., & Suk, K. T. (2022). Food and Gut Microbiota-Derived Metabolites in Nonalcoholic Fatty Liver Disease. Foods, 11(17), 2703. https://doi.org/10.3390/foods11172703









