Effects of Daqu Attributes on Distribution and Assembly Patterns of Microbial Communities and Their Metabolic Function of Artificial Pit Mud
Abstract
:1. Introduction
2. Materials and Methods
2.1. The Manufacture of Fortified Daqu
2.2. The Artificial Pit Mud Manufacturing and Sample Collection
2.3. Analysis of Microbial Community
2.3.1. Fluorescence In Situ Hybridization (FISH)
2.3.2. High-Throughput Sequencing
2.4. Analysis of Metabolites
2.4.1. Analysis of Organic Acids
2.4.2. Analysis of Volatile Compounds
2.5. Detection of Physicochemical Properties
2.6. Statistical Analysis
3. Results and Discussion
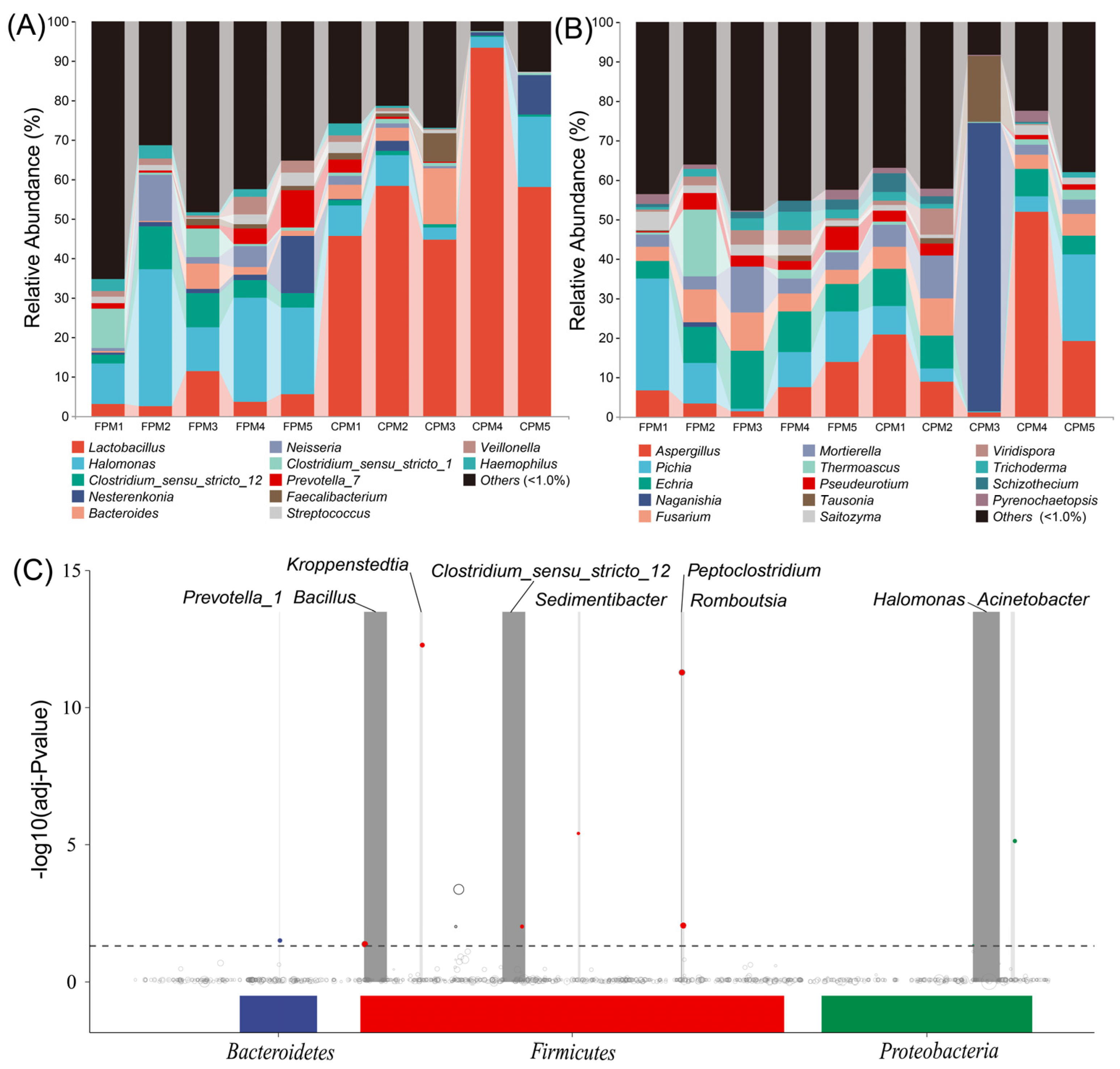
3.1. Effects of Daqu Attributes on Core Microbes of APM
3.2. Effects of Daqu Attributes on the Microbial Community of APM
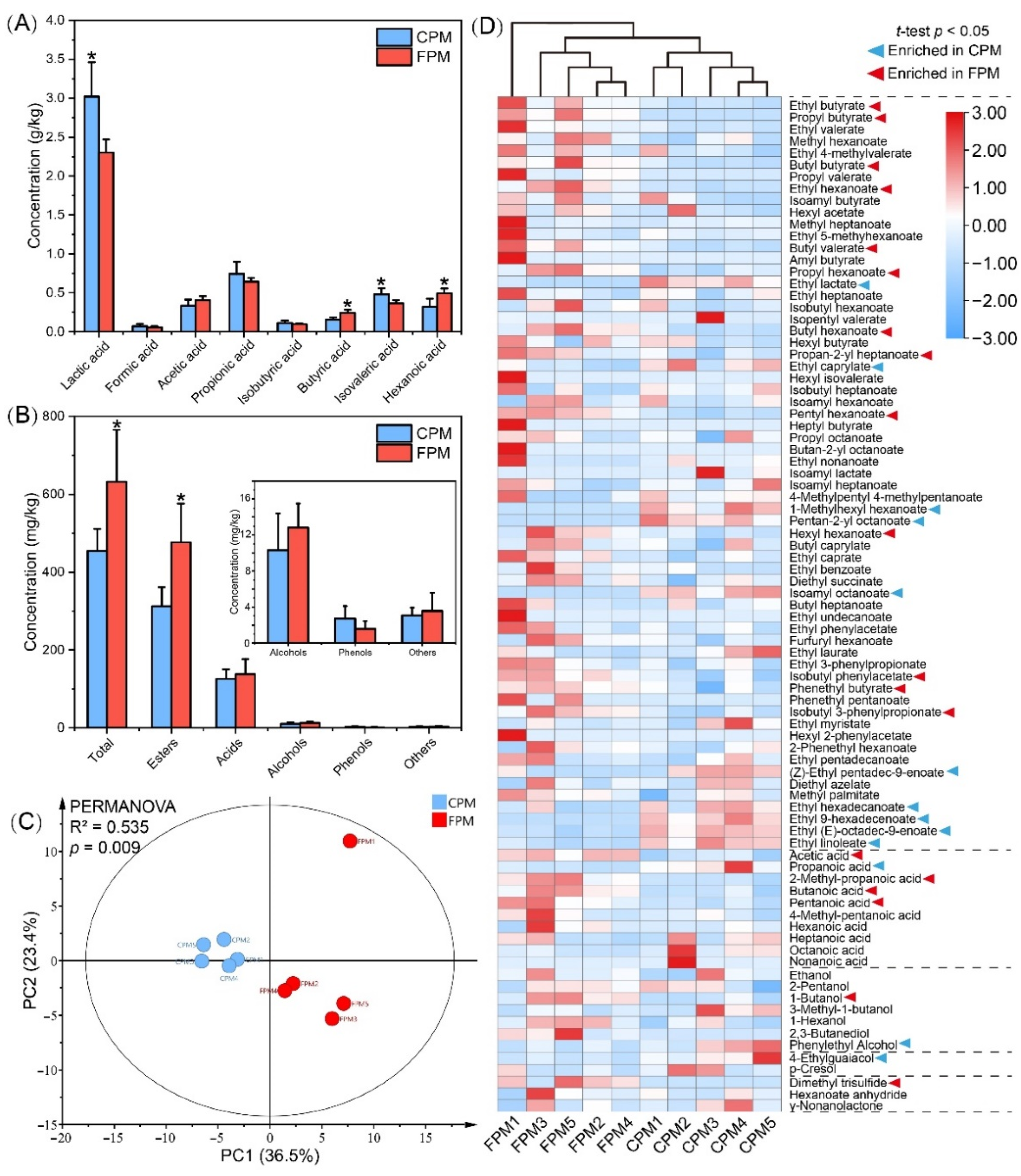
3.3. Effects of Daqu Attributes on Metabolic Profile of APM
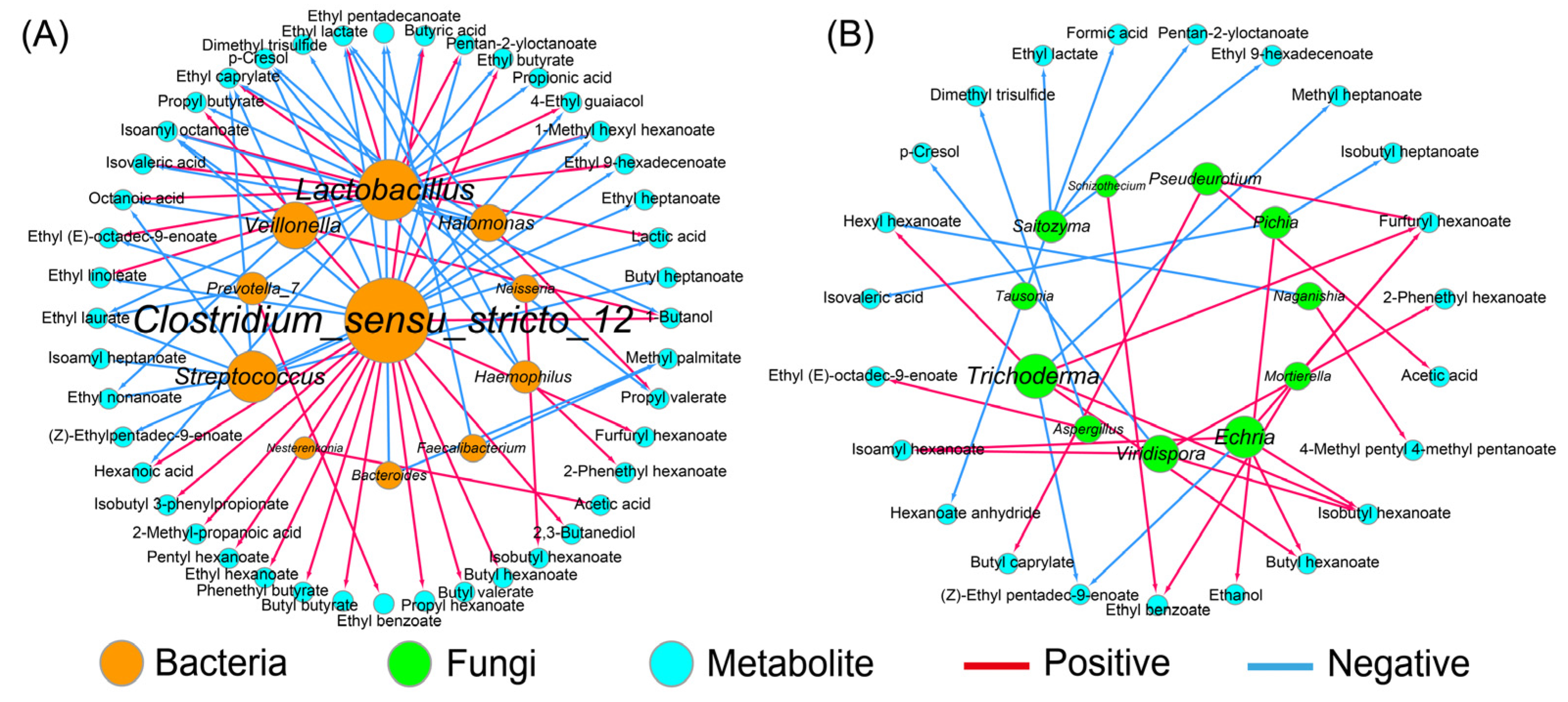
3.4. Correlation Analysis between Dominant Microbes and Metabolites
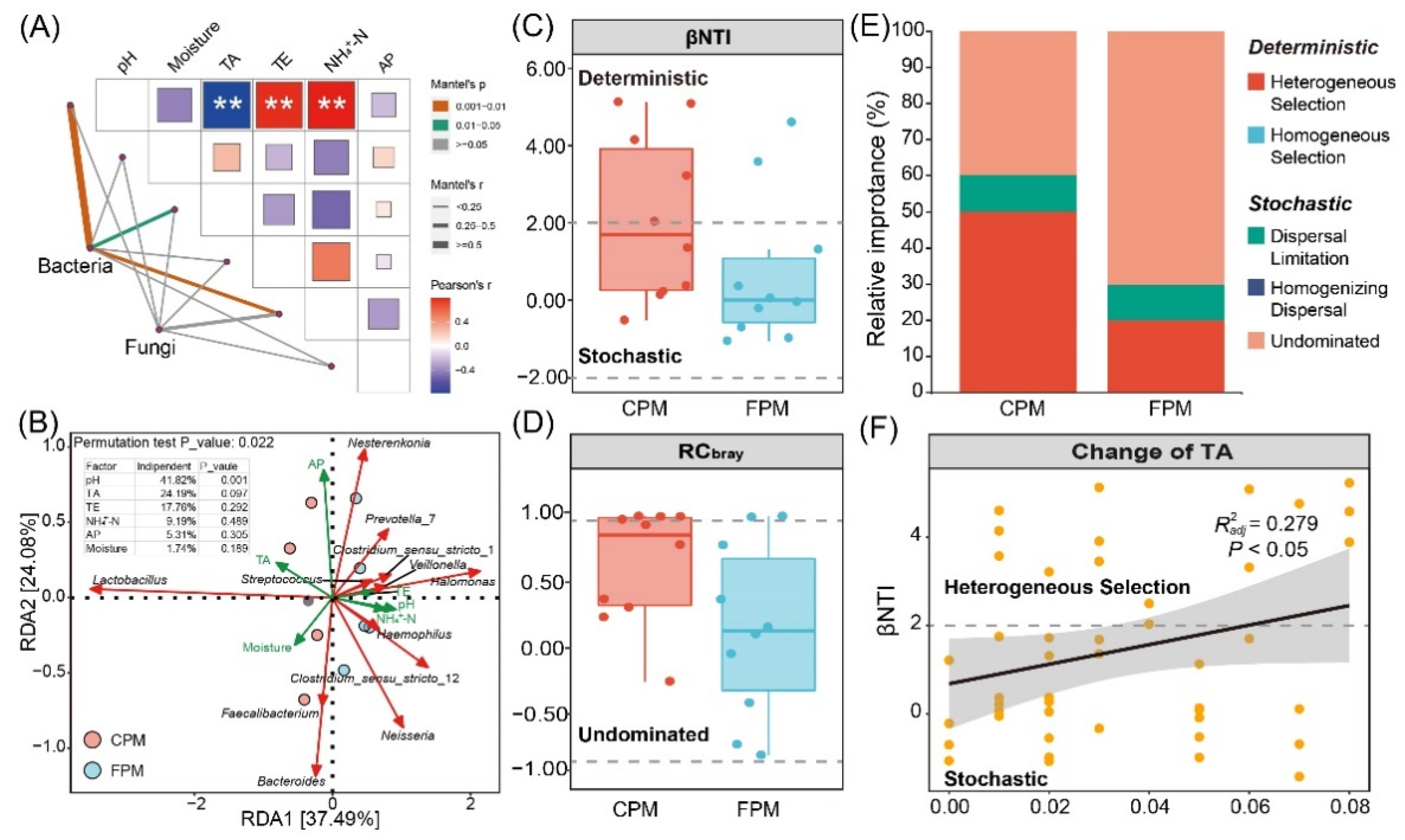
3.5. Driving Factors for Variation and Assembly of Bacterial Community
3.6. Co-Occurrence Network and Metabolic Functional of Bacterial Community
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Liu, H.; Sun, B. Effect of fermentation processing on the flavor of Baijiu. J. Agric. Food Chem. 2018, 66, 5425–5432. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.S.; Chen, H.; Wu, Y.S.; Zhao, D.R. Uncover the flavor code of strong-aroma baijiu: Research progress on the revelation of aroma compounds in strong-aroma baijiu by means of modern separation technology and molecular sensory evaluation. J. Food Compos. Anal. 2022, 109, 104499. [Google Scholar] [CrossRef]
- Ding, X.F.; Wu, C.D.; Huang, J.; Li, H.; Zhou, R.Q. Eubacterial and archaeal community characteristics in the man-made pit mud revealed by combined PCR-DGGE and FISH analyses. Food Res. Int. 2014, 62, 1047–1053. [Google Scholar] [CrossRef]
- Qian, W.; Lu, Z.M.; Chai, L.J.; Zhang, X.J.; Li, Q.; Wang, S.T.; Shen, C.H.; Shi, J.S.; Xu, Z.H. Cooperation within the microbial consortia of fermented grains and pit mud drives organic acid synthesis in strong-flavor Baijiu production. Food Res. Int. 2021, 147, 110449. [Google Scholar] [CrossRef]
- Wang, X.S.; Du, H.; Xu, Y. Source tracking of prokaryotic communities in fermented grain of Chinese strong-flavor liquor. Int. J. Food Microbiol. 2017, 244, 27–35. [Google Scholar] [CrossRef]
- Gao, J.J.; Liu, G.Y.; Li, A.J.; Liang, C.C.; Ren, C.; Xu, Y. Domination of pit mud microbes in the formation of diverse flavour compounds during Chinese strong aroma-type Baijiu fermentation. LWT 2021, 137, 110442. [Google Scholar] [CrossRef]
- Wang, C.D.; Chen, Q.; Wang, Q.; Li, C.H.; Leng, Y.Y.; Li, S.G.; Zhou, X.W.; Han, W.J.; Li, J.G.; Zhang, X.H.; et al. Long-term batch brewing accumulates adaptive microbes, which comprehensively produce more flavorful Chinese liquors. Food Res. Int. 2014, 62, 894–901. [Google Scholar] [CrossRef]
- Tao, Y.; Li, J.B.; Rui, J.P.; Xu, Z.C.; Zhou, Y.; Hu, X.H.; Wang, X.; Liu, H.M.; Li, D.P.; Li, X.Z. Prokaryotic communities in pit mud from different-aged cellars used for the production of Chinese strong-flavored liquor. Appl. Environ. Microbiol. 2014, 80, 2254–2260. [Google Scholar] [CrossRef]
- Liu, M.K.; Tang, Y.M.; Guo, X.J.; Zhao, K.; Penttinen, P.; Tian, X.H.; Zhang, X.Y.; Ren, D.Q.; Zhang, X.P. Structural and functional changes in prokaryotic communities in artificial pit mud during Chinese baijiu production. mSystems 2020, 5, e00829-19. [Google Scholar] [CrossRef]
- Lu, S.W.; Wei, C.C.; Li, Z.H.; He, X.H.; Tao, Y. Effects of a new caproic acid-producing bacteria on prokaryotic community structure and the content of acid and ester during the culture of artificial pit mud. Chin. J. Appl. Environ. Biol. 2020, 26, 144–151. [Google Scholar] [CrossRef]
- Zhang, L.Q.; Zhou, R.Q.; Niu, M.C.; Zheng, J.; Wu, C.D. Difference of microbial community stressed in artificial pit muds for luzhou-flavour liquor brewing revealed by multiphase culture-independent technology. J. Appl. Microbiol. 2015, 119, 1345–1356. [Google Scholar] [CrossRef] [PubMed]
- Jiang, Y.H.; Wang, X.; Wang, J.M.; Huang, H.T.; Luo, A.M. Comparison of physicochemical properties of artificial pit mud with different formulas. Liquor-Mak. Sci. Technol. 2021, 5, 70–76. [Google Scholar] [CrossRef]
- Zou, W.; Zhao, C.Q.; Luo, H.B. Diversity and function of the microbial community in Chinese strong-flavor baijiu ecosystem: A review. Front. Microbiol. 2018, 9, 671. [Google Scholar] [CrossRef] [PubMed]
- Liu, M.K.; Tang, Y.M.; Zhao, K.; Liu, Y.; Guo, X.J.; Ren, D.Q.; Yao, W.C.; Tian, X.H.; Gu, Y.F.; Yi, B.; et al. Determination of the fungal community of pit mud in fermentation cellars for Chinese strong-flavor liquor, using DGGE and Illumina Miseq sequencing. Food Res. Int. 2017, 91, 80–87. [Google Scholar] [CrossRef] [PubMed]
- Li, H.; Huang, J.; Liu, X.P.; Zhou, R.Q.; Ding, X.F.; Xiang, Q.Y.; Zhang, L.Q.; Wu, C.D. Characterization of interphase microbial community in Luzhou-flavored liquor manufacturing pits of various ages by polyphasic detection methods. J. Microbiol. Biotechnol. 2016, 27, 130–140. [Google Scholar] [CrossRef]
- Ding, X.F.; Wu, C.D.; Huang, J.; Zhou, R.Q. Interphase microbial community characteristics in the fermentation cellar of Chinese Luzhou-flavor liquor determined by PLFA and DGGE profiles. Food Res. Int. 2015, 72, 16–24. [Google Scholar] [CrossRef]
- Chen, S.Q.; Huang, J.; Qin, H.; Zhou, R.Q.; Yang, Y.; Qiu, C.F.; Zhang, S.Y. Characterizing the interaction relationship of the microbial communities between Zaopei and pit mud disturbing by Daqu. Food Sci. Biotechnol. 2021, 30, 1357–1367. [Google Scholar] [CrossRef]
- He, G.Q.; Huang, J.; Zhou, R.Q.; Wu, C.D.; Jin, Y. Effect of fortified Daqu on the microbial community and flavor in Chinese strong-flavor liquor brewing process. Front. Microbiol. 2019, 10, 56. [Google Scholar] [CrossRef]
- He, G.Q.; Huang, J.; Wu, C.D.; Jin, Y.; Zhou, R.Q. Bioturbation effect of fortified Daqu on microbial community and flavor metabolite in Chinese strong-flavor liquor brewing microecosystem. Food Res. Int. 2020, 129, 108851. [Google Scholar] [CrossRef]
- Mu, Y.; Huang, J.; Zhou, R.Q.; Mao, F.J.; Pan, Q.L.; Chen, S.Q.; Lu, Z.M.; Du, L.Q.; Xie, F. Exploring the response patterns of strong-flavor baijiu brewing microecosystem to fortified Daqu under different pit age. Food Res. Int. 2022, 155, 111062. [Google Scholar] [CrossRef]
- Xu, B.Y.; Xu, S.S.; Cai, J.; Sun, W.; Mu, D.D.; Wu, X.F.; Li, X.J. Analysis of the microbial community and the metabolic profile in medium-temperature Daqu after inoculation with Bacillus licheniformis and Bacillus velezensis. LWT 2022, 160, 113214. [Google Scholar] [CrossRef]
- Wang, X.J.; Zhu, H.M.; Ren, Z.Q.; Huang, Z.G.; Wei, C.H.; Deng, J. Characterization of microbial diversity and community structure in fermentation pit mud of different ages for production of strong-aroma Baijiu. Pol. J. Microbiol. 2020, 69, 151–164. [Google Scholar] [CrossRef]
- Wang, W.H.; Fan, G.S.; Li, X.T.; Fu, Z.L.; Liang, X.; Sun, B.G. Application of Wickerhamomyces anomalus in simulated solid-state fermentation for Baijiu production: Changes of microbial community structure and flavor metabolism. Front. Microbiol. 2020, 11, 598758. [Google Scholar] [CrossRef] [PubMed]
- Hu, X.L.; Yu, M.; Cao, Z.H.; Wang, K.L.; Tian, R.J.; Han, S.N.; Li, H.; Li, J.M.; Zeng, T.; Li, H. Spatial heterogeneity of prokaryotic microbial community diversity in degraded fermented pit for the production of strong-flavor Baijiu based on high throughput sequencing. Food Sci. 2021, 42, 86–93. [Google Scholar] [CrossRef]
- Hu, X.L.; Du, H.; Ren, C.; Xu, Y. Illuminating anaerobic microbial community and cooccurrence patterns across a quality gradient in Chinese liquor fermentation pit muds. Appl. Environ. Microbiol. 2016, 82, 2506–2515. [Google Scholar] [CrossRef]
- Tao, Y.; Wang, X.; Li, X.Z.; Wei, N.; Jin, H.; Xu, Z.C.; Tang, Q.L.; Zhu, X.Y. The functional potential and active populations of the pit mud microbiome for the production of Chinese strong-flavour liquor. Microb. Biotechnol. 2017, 10, 1603–1615. [Google Scholar] [CrossRef]
- He, G.Q.; Dong, Y.; Huang, J.; Wang, X.J.; Zhang, S.Y.; Wu, C.D.; Jin, Y.; Zhou, R.Q. Alteration of the microbial community for improving flavor character of Daqu by inoculation with Bacillus velezensis and Bacillus subtilis. LWT 2019, 111, 1–8. [Google Scholar] [CrossRef]
- Callahan, B.J.; Mcmurdie, P.J.; Rosen, M.J.; Han, A.W.; Johnson, A.J.; Holmes, S.P. DADA2: High-resolution sample inference from Illumina amplicon data. Nat. Methods 2016, 13, 581–583. [Google Scholar] [CrossRef]
- Lai, J.S.; Zou, Y.; Zhang, J.L.; Peres-Neto, P.R. Generalizing hierarchical and variation partitioning in multiple regression and canonical analyses using the rdacca.hp R package. Methods Ecol. Evol. 2022, 13, 782–788. [Google Scholar] [CrossRef]
- Stegen, J.C.; Lin, X.J.; Fredrickson, J.K.; Chen, X.Y.; Kennedy, D.W.; Murray, C.J.; Rockhold, M.L.; Konopka, A. Quantifying community assembly processes and identifying features that impose them. ISME J. 2013, 7, 2069–2079. [Google Scholar] [CrossRef]
- Wen, T.; Xie, P.H.; Yang, S.D.; Niu, G.Q.; Liu, X.Y.; Ding, Z.X.; Xue, C.; Liu, Y.X.; Shen, Q.R.; Yuan, J. ggClusterNet: An R package for microbiome network analysis and modularity-based multiple network layouts. iMeta 2022, 1, e32. [Google Scholar] [CrossRef]
- Yuan, M.M.; Guo, X.; Wu, L.W.; Zhang, Y.; Xiao, N.J.; Ning, D.L.; Shi, Z.; Zhou, X.S.; Wu, L.W.; Yang, Y.F.; et al. Climate warming enhances microbial network complexity and stability. Nat. Clim. Chang. 2021, 11, 343–348. [Google Scholar] [CrossRef]
- Douglas, G.M.; Maffei, V.J.; Zaneveld, J.; Yurgel, S.N.; Brown, J.R.; Taylor, C.M.; Huttenhower, C.; Langille, M.G.I. PICRUSt2: An improved and extensible approach for metagenome inference. bioRxiv 2019. [Google Scholar] [CrossRef]
- Fu, J.X.; Chen, L.; Yang, S.Z.; Li, Y.Z.; Jin, L.; He, X.P.; He, L.; Ao, X.L.; Liu, S.L.; Liu, A.P.; et al. Metagenome and analysis of metabolic potential of the microbial community in pit mud used for Chinese strong-flavor liquor production. Food Res. Int. 2021, 143, 110294. [Google Scholar] [CrossRef] [PubMed]
- Chai, L.J.; Qian, W.; Zhong, X.Z.; Zhang, X.J.; Lu, Z.M.; Zhang, S.Y.; Wang, S.T.; Shen, C.H.; Shi, J.S.; Xu, Z.H. Mining the factors driving the evolution of the pit mud microbiome under the impact of long-term production of strong-flavor Baijiu. Appl. Environ. Microbiol. 2021, 87, e00885-21. [Google Scholar] [CrossRef]
- Gao, Z.Z.; Wu, Z.Y.; Zhang, W.X. Effect of pit mud on bacterial community and aroma components in yellow water and their changes during the fermentation of Chinese strong-flavor liquor. Foods 2020, 9, 372. [Google Scholar] [CrossRef]
- Zhang, Y.D.; Shen, Y.; Cheng, W.; Wang, X.; Xue, Y.S.; Chen, X.X.; Han, B.Z. Understanding the shifts of microbial community and metabolite profile from wheat to mature Daqu. Front. Microbiol. 2021, 12, 714726. [Google Scholar] [CrossRef]
- Liu, M.K.; Tang, Y.M.; Guo, X.J.; Zhao, K.; Tian, X.H.; Liu, Y.; Yao, W.C.; Deng, B.; Ren, D.Q.; Zhang, X.P. Deep sequencing reveals high bacterial diversity and phylogenetic novelty in pit mud from Luzhou Laojiao cellars for Chinese strong-flavor Baijiu. Food Res. Int. 2017, 102, 68–76. [Google Scholar] [CrossRef]
- Fan, W.L.; Qian, M.C. Characterization of aroma compounds of Chinese “Wuliangye” and “Jiannanchun” liquors by aroma extract dilution analysis. J. Agric. Food Chem. 2006, 54, 2695–2704. [Google Scholar] [CrossRef]
- Zhao, D.R.; Shi, D.M.; Sun, J.Y.; Li, A.J.; Sun, B.G.; Zhao, M.M.; Chen, F.; Sun, X.T.; Li, H.H.; Huang, M.Q.; et al. Characterization of key aroma compounds in Gujinggong Chinese Baijiu by gas chromatography-olfactometry, quantitative measurements, and sensory evaluation. Food Res. Int. 2018, 105, 616–627. [Google Scholar] [CrossRef]
- Cai, W.C.; Xue, Y.A.; Tang, F.X.; Wang, Y.R.; Yang, S.Y.; Liu, W.H.; Hou, Q.C.; Yang, X.Q.; Guo, Z.; Shan, C.H. The depth-depended fungal diversity and non-depth-depended aroma profiles of pit mud for strong-flavor baijiu. Front. Microbiol. 2022, 12, 789845. [Google Scholar] [CrossRef] [PubMed]
- Kredics, L.; Antal, Z.; Szekeres, A.; Hatvani, L.; Manczinger, L.; Vágvölgyi, C.; Nagy, E. Extracellular proteases of Trichoderma species. Acta Microbiol. Immunol. Hung. 2005, 52, 169–184. [Google Scholar] [CrossRef] [PubMed]
- Chase, J.M. Stochastic community assembly causes higher biodiversity in more productive environments. Science 2010, 328, 1388–1391. [Google Scholar] [CrossRef] [PubMed]
- Zheng, W.; Zhao, Z.Y.; Lv, F.L.; Wang, R.Z.; Zhao, Z.Y.; Li, Z.Y.; Zhai, B.N. Assembly of abundant and rare bacterial and fungal sub-communities in different soil aggregate sizes in an apple orchard treated with cover crop and fertilizer. Soil Biol. Biochem. 2021, 156, 108222. [Google Scholar] [CrossRef]
- Jiao, S.; Lu, Y.H. Abundant fungi adapt to broader environmental gradients than rare fungi in agricultural fields. Glob. Chang. Biol. 2020, 26, 4506–4520. [Google Scholar] [CrossRef]
- Zhou, J.Z.; Ning, D.L. Stochastic community assembly: Does it matter in microbial ecology? Microbiol. Mol. Biol. Rev. 2017, 81, e00002-17. [Google Scholar] [CrossRef]
- She, Z.X.; Pan, X.; Wang, J.; Shao, R.; Wang, G.C.; Wang, S.P.; Yue, Z.B. Vertical environmental gradient drives prokaryotic microbial community assembly and species coexistence in a stratified acid mine drainage lake. Water Res. 2021, 206, 117739. [Google Scholar] [CrossRef]
- Zhang, H.M.; Meng, Y.J.; Wang, Y.L.; Zhou, Q.W.; Li, A.J.; Liu, G.Y.; Li, J.X.; Xing, X.H. Prokaryotic communities in multidimensional bottom-pit-mud from old and young pits used for the production of Chinese strong-flavor Baijiu. Food Chem. 2020, 312, 126084. [Google Scholar] [CrossRef]
- Hernandez, D.J.; David, A.S.; Menges, E.S.; Searcy, C.A.; Afkhami, M.E. Environmental stress destabilizes microbial networks. ISME J. 2021, 15, 1722–1734. [Google Scholar] [CrossRef]
- de Araújo, C.W.; Leitão, R.C.; Gehring, T.A.; Angenent, L.T.; Santaella, S.T. Anaerobic fermentation for n-caproic acid production: A review. Process Biochem. 2017, 54, 106–119. [Google Scholar] [CrossRef]
- Zhu, X.Y.; Tao, Y.; Liang, C.; Li, X.Z.; Wei, N.; Zhang, W.J.; Zhou, Y.; Yang, Y.F.; Bo, T. The synthesis of n-caproate from lactate: A new efficient process for medium-chain carboxylates production. Sci. Rep. 2015, 5, 14360. [Google Scholar] [CrossRef] [PubMed]
- Gao, M.; Lin, Y.J.; Wang, P.; Jin, Y.; Wang, Q.H.; Ma, H.Z.; Shen, Y.Q.; Le, Q.V.; Xia, C.L.; Lam, S.S. Production of medium-chain fatty acid caproate from Chinese liquor distillers’ grain using pit mud as the fermentation microbes. J. Hazard. Mater. 2021, 417, 126037. [Google Scholar] [CrossRef] [PubMed]
- Zhu, X.Y.; Zhou, Y.; Wang, Y.; Wu, T.T.; Li, X.Z.; Li, D.P.; Tao, Y. Production of high-concentration n-caproic acid from lactate through fermentation using a newly isolated Ruminococcaceae bacterium CPB6. Biotechnol. Biofuels 2017, 10, 102. [Google Scholar] [CrossRef] [PubMed]
- Candry, P.; Radić, L.; Favere, J.; Carvajal-Arroyo, J.M.; Rabaey, K.; Ganigué, R. Mildly acidic pH selects for chain elongation to caproic acid over alternative pathways during lactic acid fermentation. Water Res. 2020, 186, 116396. [Google Scholar] [CrossRef]

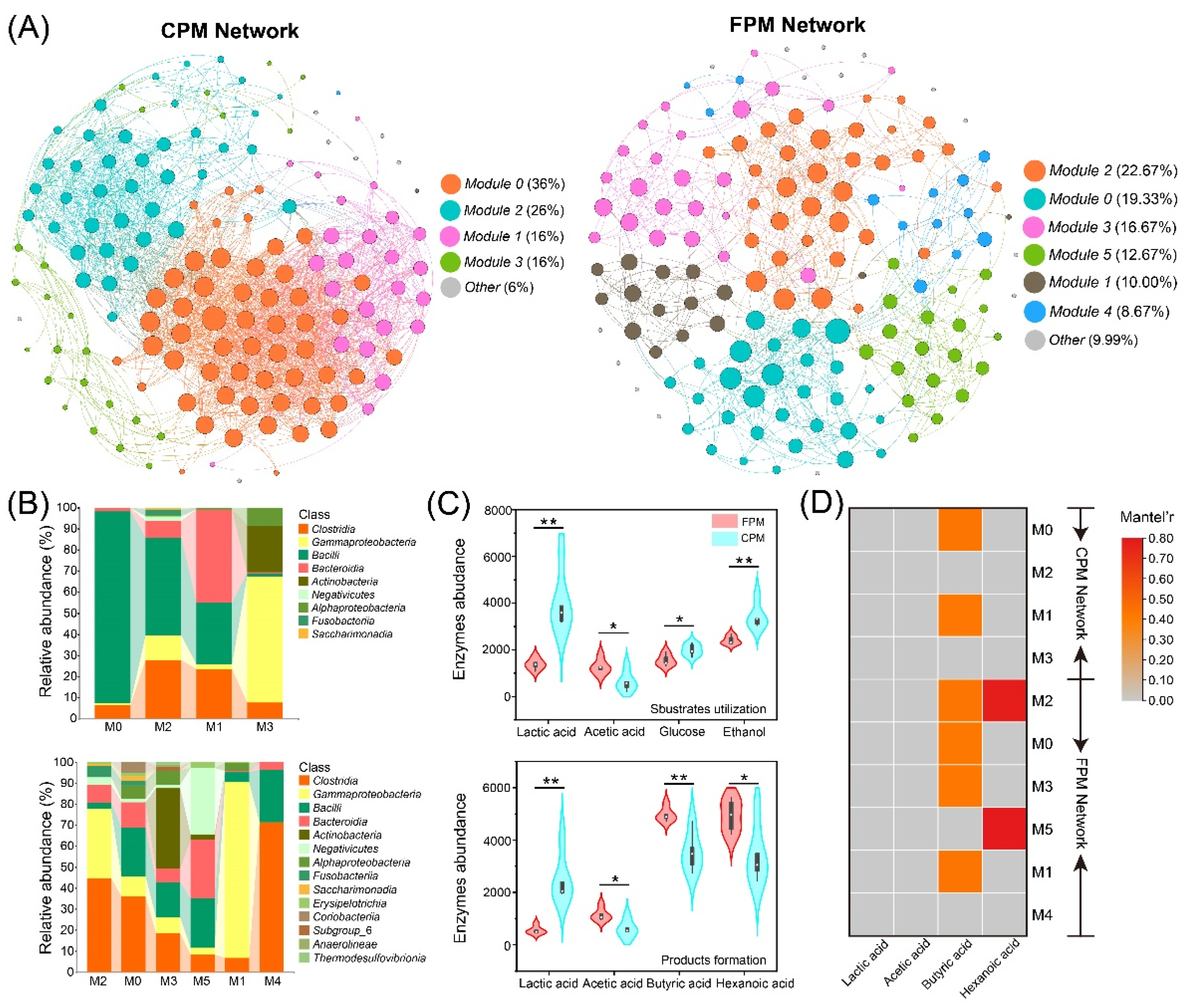
| Network Properties 1 | FPM | CPM |
|---|---|---|
| Node number | 150 | 150 |
| Edge number | 602 | 1493 |
| Proportion of positive edge | 64.12% | 91.06% |
| Proportion of negative edge | 35.88% | 8.91% |
| Average degree | 8.027 | 19.907 |
| Average path length | 3.553 | 2.945 |
| Average clustering coefficient | 0.236 | 0.135 |
| Diameter | 8.164 | 8.152 |
| Density | 0.054 | 0.134 |
| Modularity | 0.626 | 0.340 |
| Modules number | 21 | 13 |
| Vulnerability | 0.011 | 0.009 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Mu, Y.; Huang, J.; Zhou, R.; Zhang, S.; Qin, H.; Tang, H.; Pan, Q.; Tang, H. Effects of Daqu Attributes on Distribution and Assembly Patterns of Microbial Communities and Their Metabolic Function of Artificial Pit Mud. Foods 2022, 11, 2922. https://doi.org/10.3390/foods11182922
Mu Y, Huang J, Zhou R, Zhang S, Qin H, Tang H, Pan Q, Tang H. Effects of Daqu Attributes on Distribution and Assembly Patterns of Microbial Communities and Their Metabolic Function of Artificial Pit Mud. Foods. 2022; 11(18):2922. https://doi.org/10.3390/foods11182922
Chicago/Turabian StyleMu, Yu, Jun Huang, Rongqing Zhou, Suyi Zhang, Hui Qin, Hanlan Tang, Qianglin Pan, and Huifang Tang. 2022. "Effects of Daqu Attributes on Distribution and Assembly Patterns of Microbial Communities and Their Metabolic Function of Artificial Pit Mud" Foods 11, no. 18: 2922. https://doi.org/10.3390/foods11182922
APA StyleMu, Y., Huang, J., Zhou, R., Zhang, S., Qin, H., Tang, H., Pan, Q., & Tang, H. (2022). Effects of Daqu Attributes on Distribution and Assembly Patterns of Microbial Communities and Their Metabolic Function of Artificial Pit Mud. Foods, 11(18), 2922. https://doi.org/10.3390/foods11182922