Enhancement of Polypeptide Yield Derived from Rapeseed Meal with Low-Intensity Alternating Magnetic Field
Abstract
:1. Introduction
2. Materials and Methods
2.1. Materials and Reagents
2.2. Selection of Bacterial Strain
2.2.1. Selection and isolation of strains with high proteinase
2.2.2. Preliminary Screening of Strains
2.2.3. Rescreening of Strains
2.3. Identification of the Selected Strain
2.3.1. Morphology
2.3.2. 16S rDNA Sequence Analysis
2.4. Optimization of Cultural Conditions
2.5. Traditional RSM Solid-State Fermentation
2.6. Low-Intensity Alternating Magnetic-Field-Assisted RSM Fermentation
2.6.1. Low-Intensity Alternating Magnetic Field Set-Up
2.6.2. Optimization of Magnetic Field Parameters
2.7. Determination of Polypeptides Concentration
2.8. Statistical Analysis
3. Results and Discussion
3.1. Selection of Strains with High Proteinase Activity
3.2. Morphology and Identification of Strain
3.3. Optimization of Cultural Conditions
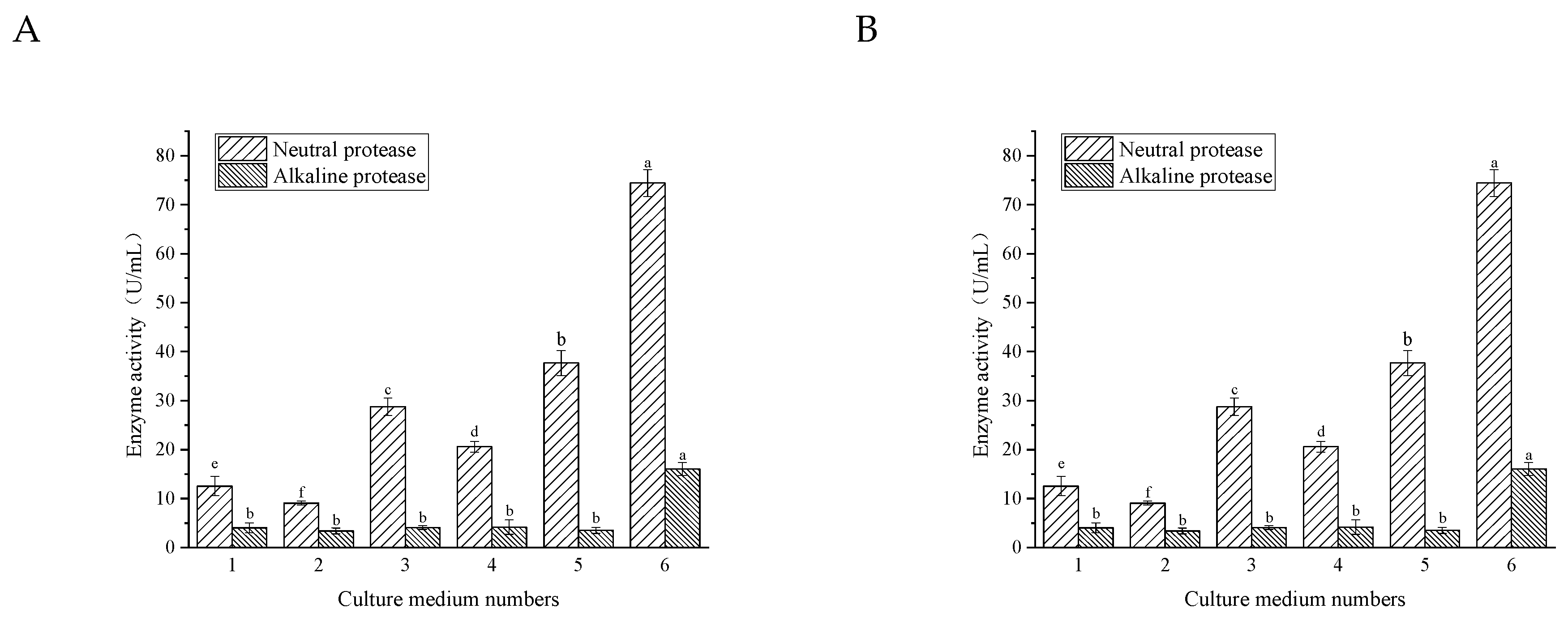
3.3.1. Culture Medium
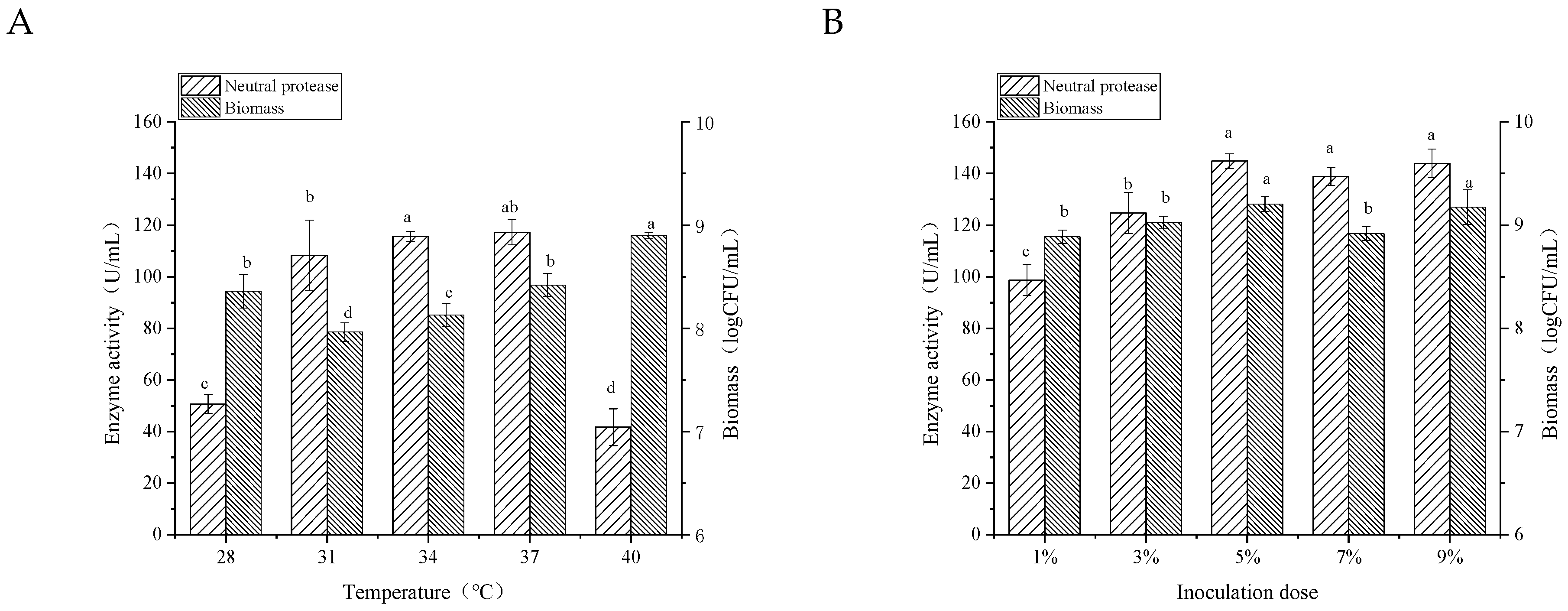
3.3.2. Incubation Temperature
3.3.3. Inoculation Dose and Initial pH in the Medium
3.3.4. Shaker Speed during Incubation
3.4. Optimization of the Traditional Fermentation Process
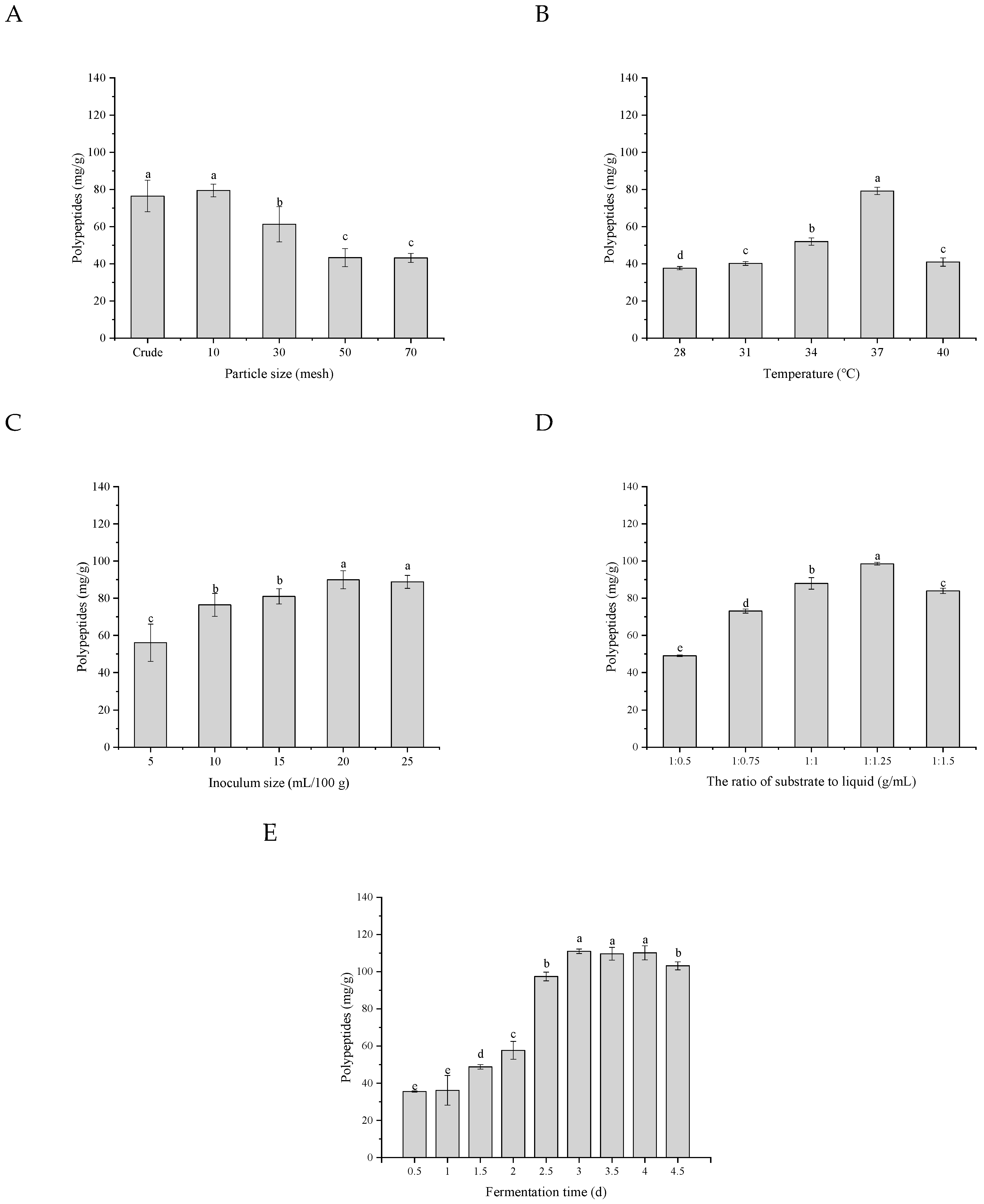
3.4.1. The Particle Size of the Substrate
3.4.2. Fermentation Temperature
3.4.3. Biomass Amounts and Substrate/Water Ratio
3.4.4. Fermentation Time
3.5. Effects of LF-MF Assisted Fermentation on Polypeptides
3.6. Possible Mechanisms
4. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Voća, N.; Pezo, L.; Jukić, Ž.; Lončar, B.; Šuput, D.; Krička, T. Estimation of the storage properties of rapeseeds using an artificial neural network. Ind. Crops Prod. 2022, 187, 115358. [Google Scholar] [CrossRef]
- Liu, S.; Raman, H.; Xiang, Y.; Zhao, C.; Huang, J.; Zhang, Y. De novo design of future rapeseed crops: Challenges and opportunities. Crop J. 2022, 10, 587–596. [Google Scholar] [CrossRef]
- Wang, R.; Shaarani, S.M.; Godoy, L.C.; Melikoglu, M.; Vergara, C.S.; Koutinas, A.; Webb, C. Bioconversion of rapeseed meal for the production of a generic microbial feedstock. Enzyme Microb. Technol. 2010, 47, 77–83. [Google Scholar] [CrossRef]
- Chmielewska, A.; Kozlowska, M.; Rachwal, D.; Wnukowski, P.; Amarowicz, R.; Nebesny, E.; Rosicka-Kaczmarek, J. Canola/rapeseed protein—Nutritional value, functionality and food application: A review. Crit. Rev. Food Sci. Nutr. 2021, 61, 3836–3856. [Google Scholar] [CrossRef]
- Olukomaiya, O.O.; Fernando, W.C.; Mereddy, R.; Li, X.; Sultanbawa, Y. Solid-state fermentation of canola meal with Aspergillus sojae, Aspergillus ficuum and their co-cultures: Effects on physicochemical, microbiological and functional properties. LWT-Food Sci. Technol. 2020, 127, 109362. [Google Scholar] [CrossRef]
- Wang, Y.; Li, Y.; Ruan, S.; Lu, F.; Tian, W.; Ma, H. Antihypertensive effect of rapeseed peptides and their potential in improving the effectiveness of captopril. J. Sci. Food Agric. 2021, 101, 3049–3055. [Google Scholar] [CrossRef]
- Ding, Q.; Wu, R.A.; Yin, L.; Zhang, W.; He, R.; Zhang, T.; Jiang, H.; Luo, L.; Ma, H.; Dai, C. Antioxidation and memory protection effects of solid-state-fermented rapeseed meal peptides on D-galactose-induced memory impairment in aging-mice. J. Food Process Eng. 2019, 42, e13145. [Google Scholar] [CrossRef]
- Duan, X.; Zhang, M.; Chen, F. Prediction and analysis of antimicrobial peptides from rapeseed protein using in silico approach. J. Food Biochem. 2021, 45, e13598. [Google Scholar] [CrossRef]
- Olukomaiya, O.O.; Pan, L.; Zhang, D.; Mereddy, R.; Sultanbawa, Y.; Li, X. Performance and ileal amino acid digestibility in broilers fed diets containing solid-state fermented and enzyme-supplemented canola meals. Anim. Feed Sci. Technol. 2021, 275, 114876. [Google Scholar] [CrossRef]
- Shi, C.; He, J.; Yu, J.; Yu, B.; Huang, Z.; Mao, X.; Zheng, P.; Chen, D. Solid state fermentation of rapeseed cake with Aspergillus niger for degrading glucosinolates and upgrading nutritional value. J. Anim. Sci. Biotechnol. 2015, 6, 13. [Google Scholar] [CrossRef] [Green Version]
- Soccol, C.R.; Costa, E.S.F.d.; Letti, L.A.J.; Karp, S.G.; Woiciechowski, A.L.; Vandenberghe, L.P.d.S. Recent developments and innovations in solid state fermentation. Biotechnol. Res. Innov. 2017, 1, 52–71. [Google Scholar] [CrossRef]
- Ruan, S.; Luo, J.; Li, Y.; Wang, Y.; Huang, S.; Lu, F.; Ma, H. Ultrasound-assisted liquid-state fermentation of soybean meal with Bacillus subtilis: Effects on peptides content, ACE inhibitory activity and biomass. Process Biochem. 2020, 91, 73–82. [Google Scholar] [CrossRef]
- Wan, Y.; Zhang, J.; Han, H.; Li, L.; Liu, Y.; Gao, M. Citrinin-producing capacity of Monascus purpureus in response to low −frequency magnetic fields. Process Biochem. 2017, 53, 25–29. [Google Scholar] [CrossRef]
- Guo, L.; Azam, S.M.R.; Guo, Y.; Liu, D.; Ma, H. Germicidal efficacy of the pulsed magnetic field against pathogens and spoilage microorganisms in food processing: An overview. Food Control 2021, 136, 108496. [Google Scholar] [CrossRef]
- Canli, O.; Kurbanoglu, E.B. Application of low magnetic field on inulinase production by Geotrichum candidum under solid state fermentation using leek as substrate. Toxicol. Ind. Health 2012, 28, 894–900. [Google Scholar] [CrossRef]
- Bubanja, I.N.; Lončarević, B.; Lješević, M.; Beškoski, V.; Gojgić-Cvijović, G.; Velikić, Z.; Stanisavljev, D. The influence of low-frequency magnetic field regions on the Saccharomyces cerevisiae respiration and growth. Chem. Eng. Process. 2019, 143, 107593. [Google Scholar] [CrossRef]
- Hou, X.; Dai, C.; Tang, Y.; Xing, Z.; Mintah, B.K.; Dabbour, M.; Ding, Q.; He, R.; Ma, H. Thermophilic solid-state fermentation of rapeseed meal and analysis of microbial community diversity. LWT-Food Sci. Technol. 2019, 116, 108520. [Google Scholar] [CrossRef]
- Huang, G.; Chen, S.; Dai, C.; Sun, L.; Sun, W.; Tang, Y.; Xiong, F.; He, R.; Ma, H. Effects of ultrasound on microbial growth and enzyme activity. Ultrason. Sonochem. 2017, 37, 144–149. [Google Scholar] [CrossRef]
- Dulay, R.M.R.; Cabrera, E.C.; Kalaw, S.P.; Reyes, R.G. Optimization of submerged culture conditions for mycelial biomass production of fourteen Lentinus isolates from Luzon Island, Philippines. Biocatal. Agric. Biotechnol. 2021, 38, 102226. [Google Scholar] [CrossRef]
- Elisashvili, V. Submerged cultivation of medicinal mushrooms: Bioprocesses and products (review). Int. J. Med. Mushrooms 2012, 14, 211. [Google Scholar] [CrossRef]
- Rodrigues, T.H.S.; Pinto, G.A.S.; Gonçalves, L.R.B. Effects of inoculum concentration, temperature, and carbon sources on tannase production during solid state fermentation of cashew apple bagasse. Biotechnol. Bioprocess Eng. 2008, 13, 571–576. [Google Scholar] [CrossRef]
- Dai, C.; Ma, H.; He, R.; Huang, L.; Zhu, S.; Ding, Q.; Luo, L. Improvement of nutritional value and bioactivity of soybean meal by solid-state fermentation with Bacillus subtilis. LWT-Food Sci. Technol. 2017, 86, 1–7. [Google Scholar] [CrossRef]
- Lu, F.; Alenyorege, E.A.; Ouyang, N.; Zhou, A.; Ma, H. Simulated natural and high temperature solid-state fermentation of soybean meal: A comparative study regarding microorganisms, functional properties and structural characteristics. LWT-Food Sci. Technol. 2022, 159, 113125. [Google Scholar] [CrossRef]
- Ma, H.; Pan, Z.; Gao, M.; Luo, L. Efficacy in Microbial Sterilization of Pulsed Magnetic Field Treatment. Int. J. Food Eng. 2008, 4, 1–14. [Google Scholar] [CrossRef]
- Miñano, H.L.A.; Silva, A.C.d.S.; Souto, S.; Costa, E.J.X. Magnetic Fields in Food Processing Perspectives, Applications and Action Models. Processes 2020, 8, 814. [Google Scholar] [CrossRef]
- Zablotskii, V.; Polyakova, T.; Lunov, O.; Dejneka, A. How a High-Gradient Magnetic Field Could Affect Cell Life. Sci. Rep. 2016, 6, 37407. [Google Scholar] [CrossRef]
- Konopacki, M.; Rakoczy, R. The analysis of rotating magnetic field as a trigger of Gram-positive and Gram-negative bacteria growth. Biochem. Eng. J. 2019, 141, 259–267. [Google Scholar] [CrossRef]
- Zhang, J.; Zeng, D.; Xu, C.; Gao, M. Effect of low-frequency magnetic field on formation of pigments of Monascus purpureus. Eur. Food Res. Technol. 2014, 240, 577–582. [Google Scholar] [CrossRef]
- Pilla, A.; Fitzsimmons, R.; Muehsam, D.; Wu, J.; Rohde, C.; Casper, D. Electromagnetic fields as first messenger in biological signaling: Application to calmodulin-dependent signaling in tissue repair. Biochim. Biophys. Acta 2011, 1810, 1236–1245. [Google Scholar] [CrossRef]
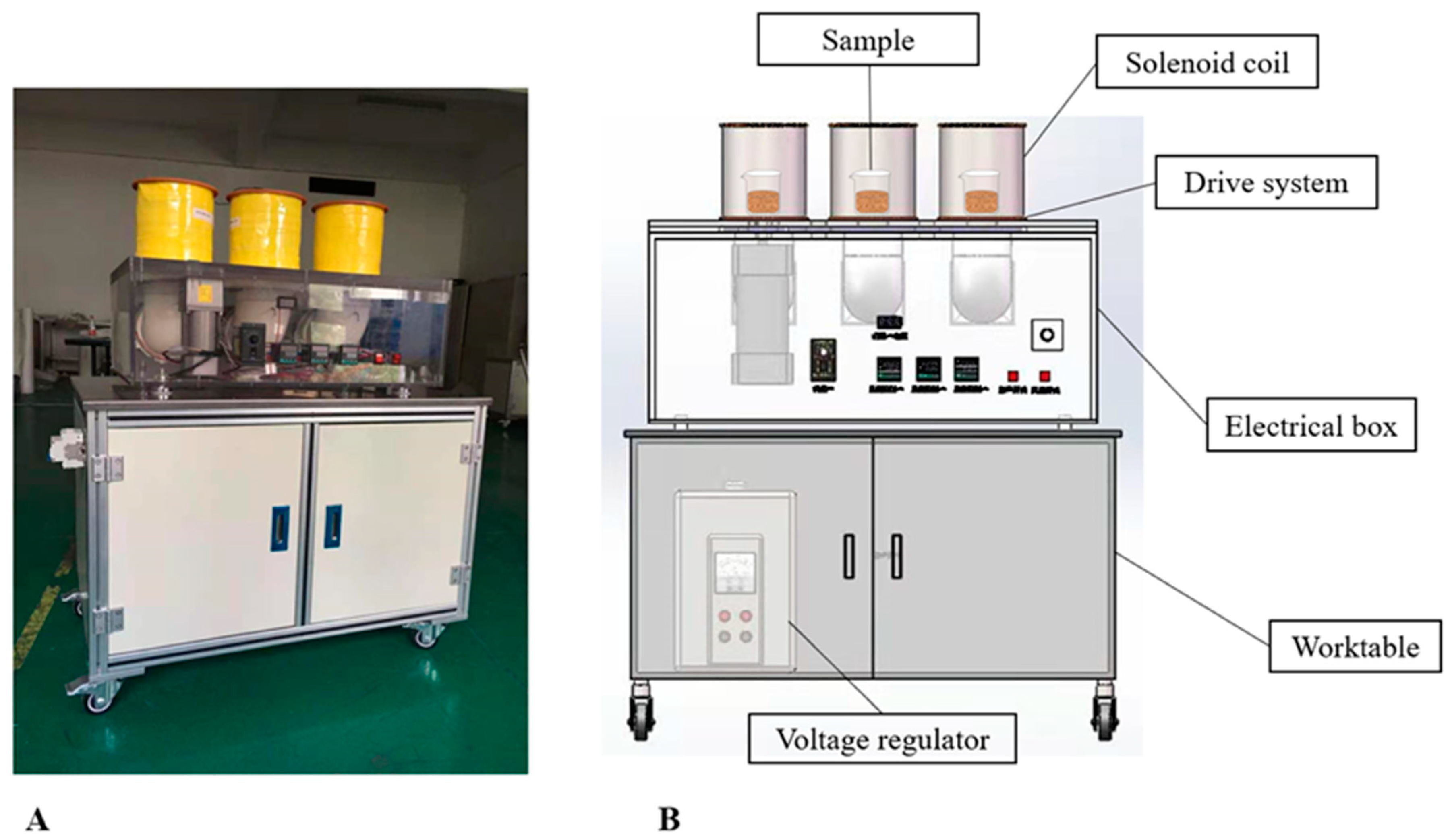



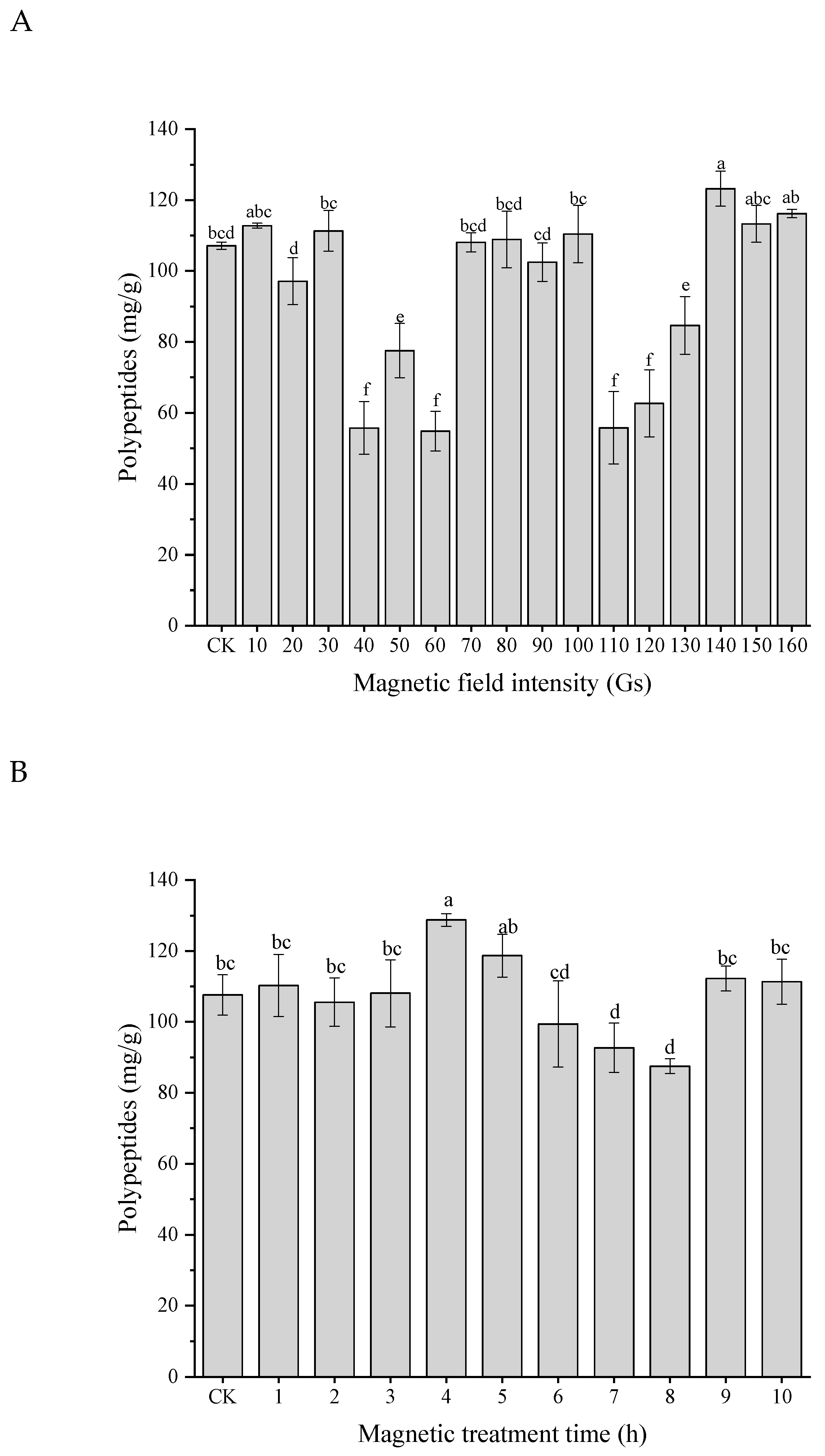
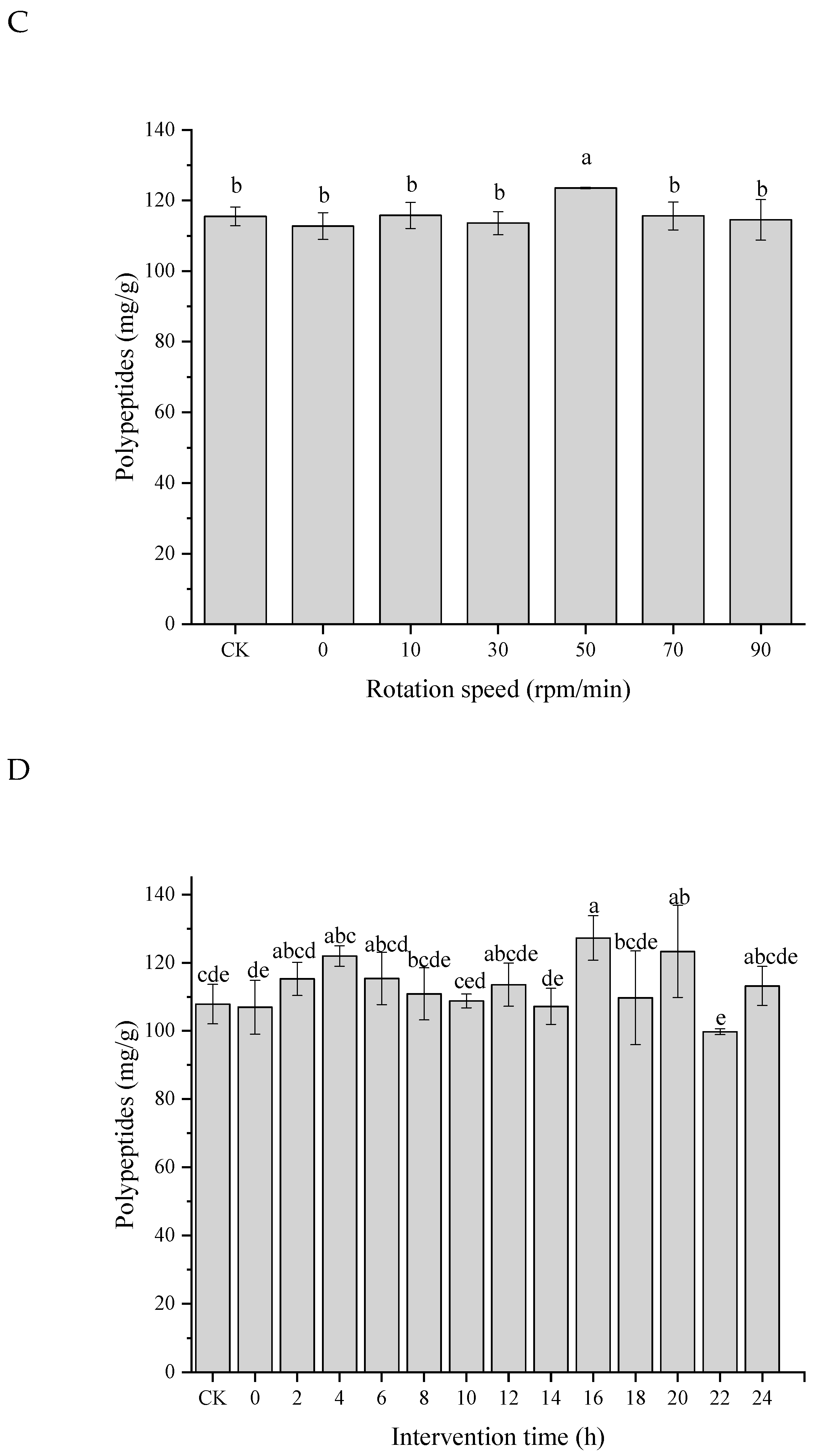
| Single Factor | Treatments | Fixed Parameters |
|---|---|---|
| Magnetic field intensity (Gs) | CK, 10, 20, 30, 40, 50, 60, 70, 80, 90, 100,110, 120, 130, 140, 150, 160 | LF-MF exposure time: 1 h; Rotation speed: 50 rpm/min; Intervention time: 4 h |
| LF-MF exposure time (h) | CK, 1, 2, 3, 4, 5, 6, 7, 8, 9, 10 | Magnetic field intensity: 140 Gs; Rotation speed: 50 rpm/min; Intervention time: 4 h |
| Rotation speed (rpm/min) | CK, 0, 10, 30, 50, 70 | Magnetic field intensity: 140 Gs; LF-MF exposure time: 4 h; Intervention time: 4 h |
| Intervention time (h) | CK, 0, 2, 4, 6, 8, 10, 12, 14, 16, 18, 20, 22, 24 | Magnetic field intensity: 140 Gs; LF-MF exposure time: 4 h; Rotation speed: 50 rpm/min; |
| Strains | Protease | Amylase | Cellulase |
|---|---|---|---|
| B1 | 3.25 ± 0.26 cd | 2.07 ± 0.20 cd | 2.03 ± 0.09 bc |
| B2 | 3.26 ± 0.21 cd | 2.18 ± 0.18 c | 2.00 ± 0.09 bcd |
| B5 | 3.57 ± 0.14 bc | 2.38 ± 0.13 b | 1.88 ± 0.19 d |
| B9 | 3.17 ± 0.17 d | 1.79 ± 0.10 e | 1.93 ± 0.15 cd |
| B10 | 3.60 ± 0.40 b | 1.96 ± 0.09 de | 2.09 ± 0.07 bc |
| B13 | 4.00 ± 0.34 a | 2.50 ± 0.08 ab | 2.18 ± 0.10 a |
| B14 | 3.93 ± 0.23 a | 2.63 ± 0.11 a | 2.06 ± 0.06 bc |
| B15 | 3.42 ± 0.23 bcd | 2.49 ± 0.29 ab | 1.86 ± 0.17 d |
| B16 | 3.49 ± 0.31 bc | 2.56 ± 0.12 ab | 1.98 ± 0.11 cd |
| BY1 | 3.35 ± 0.25 bcd | 1.90 ± 0.11 de | 2.16 ± 0.08 a |
| BY3 | 3.44 ± 0.31 bc | 2.16 ± 0.14 c | 2.04 ± 0.13 bc |
| BY4 | 3.60 ± 0.22 b | 1.95 ± 0.01 de | 2.13 ± 0.05 b |
| Strains | Enzyme Activity (U/mL) | |||
|---|---|---|---|---|
| Neutral Protease | Alkaline Protease | Acid Proteinase | Total Protease | |
| B16 | 22.62 ± 0.91 a | 2.54 ± 0.51 a | 0.46 ± 0.26 c | 25.61 ± 1.68 |
| BY4 | 22.27 ± 2.00 a | 0.94 ± 0.16 def | 1.60 ± 0.13 ab | 24.81 ± 2.29 |
| BY1 | 21.56 ± 0.17 a | 2.62 ± 0.29 a | -- | 24.18 ± 0.46 |
| BY3 | 21.97 ± 2.44 a | 0.67 ± 0.23 ef | 0.96 ± 0.29 bc | 23.60 ± 2.96 |
| B15 | 4.42 ± 0.53 b | 1.37 ± 0.27 bcd | 0.69 ± 0.66 c | 6.48 ± 1.46 |
| B14 | 3.45 ± 0.23 bc | 2.01 ± 0.27 b | 1.83 ± 0.88 a | 7.29 ± 1.38 |
| B1 | 2.69 ± 1.31 bcd | 1.98 ± 0.55 b | -- | 4.67 ± 1.86 |
| B13 | 2.44 ± 0.38 cde | 0.80 ± 0.17 ef | 0.73 ± 0.11 c | 3.97 ± 0.66 |
| B9 | 1.30 ± 0.13 de | 1.16 ± 0.22 cde | -- | 2.46 ± 0.35 |
| B2 | 1.21 ± 0.21 de | 0.53 ± 0.20 f | 1.14 ± 0.15 abc | 2.88 ± 0.56 |
| B10 | 0.84 ± 0.26 de | 0.73 ± 0.19 ef | -- | 1.57 ± 0.45 |
| B5 | 0.63 ± 0.15 e | 1.57 ± 0.37 c | 0.54 ± 0.08 c | 2.74 ± 0.60 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Guo, L.; Guo, Y.; Wu, P.; Liu, S.; Gu, C.; Yolandani; Wu, M.; Ma, H.; He, R. Enhancement of Polypeptide Yield Derived from Rapeseed Meal with Low-Intensity Alternating Magnetic Field. Foods 2022, 11, 2952. https://doi.org/10.3390/foods11192952
Guo L, Guo Y, Wu P, Liu S, Gu C, Yolandani, Wu M, Ma H, He R. Enhancement of Polypeptide Yield Derived from Rapeseed Meal with Low-Intensity Alternating Magnetic Field. Foods. 2022; 11(19):2952. https://doi.org/10.3390/foods11192952
Chicago/Turabian StyleGuo, Lina, Yiting Guo, Ping Wu, Shiyi Liu, Chen Gu, Yolandani, Mengdi Wu, Haile Ma, and Ronghai He. 2022. "Enhancement of Polypeptide Yield Derived from Rapeseed Meal with Low-Intensity Alternating Magnetic Field" Foods 11, no. 19: 2952. https://doi.org/10.3390/foods11192952
APA StyleGuo, L., Guo, Y., Wu, P., Liu, S., Gu, C., Yolandani, Wu, M., Ma, H., & He, R. (2022). Enhancement of Polypeptide Yield Derived from Rapeseed Meal with Low-Intensity Alternating Magnetic Field. Foods, 11(19), 2952. https://doi.org/10.3390/foods11192952








