Lactiplantibacillus plantarum LOC1 Isolated from Fresh Tea Leaves Modulates Macrophage Response to TLR4 Activation
Abstract
1. Introduction
2. Materials and Methods
2.1. Microorganisms
2.2. Cell Cultures
2.3. Cell Stimulation with Lactobacilli and LPS
2.4. Microarray Analysis
2.5. Two-Step Real-Time Quantitative PCR
2.6. L. plantarum LOC1 Genome Analysis
2.7. Statistical Analysis
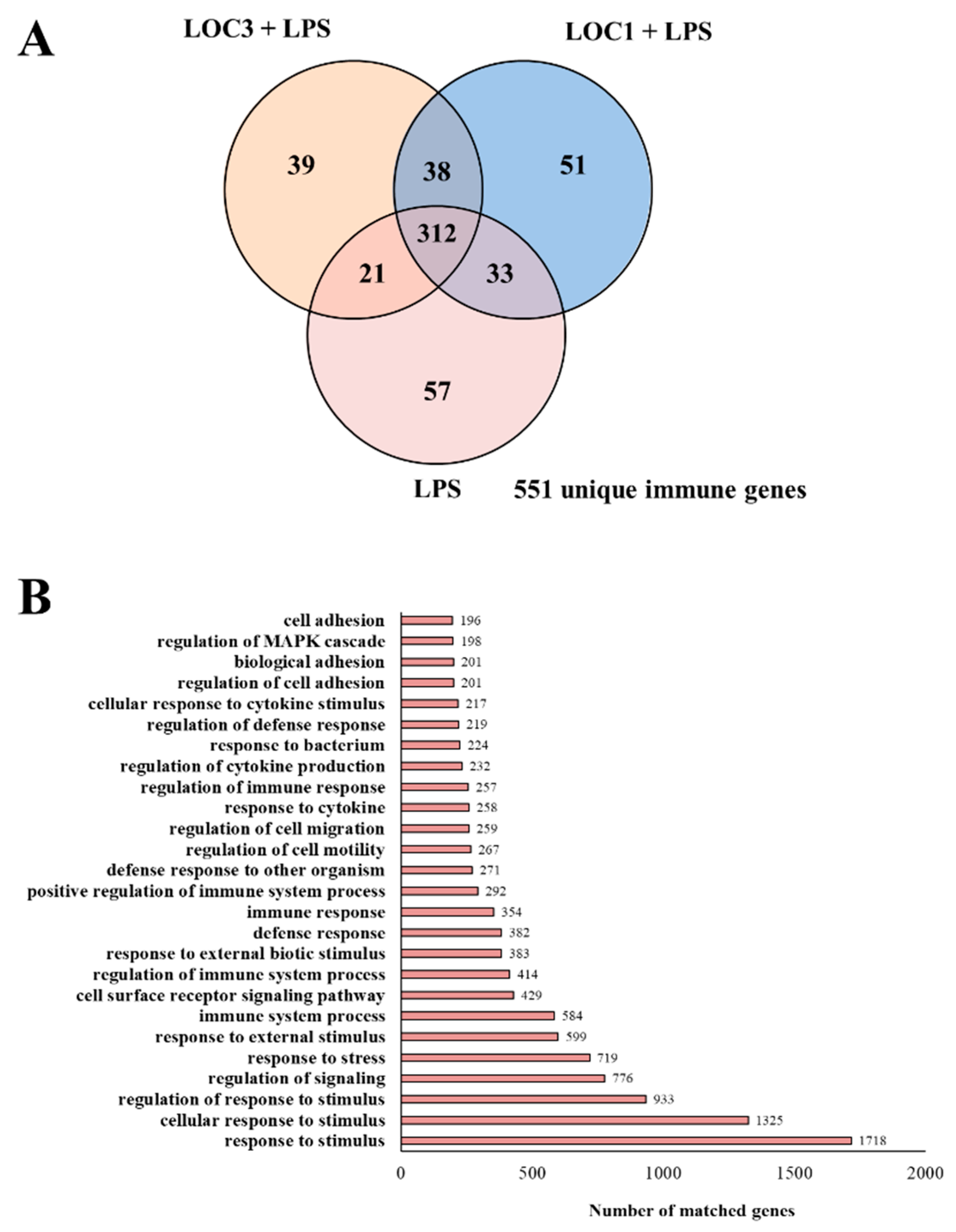
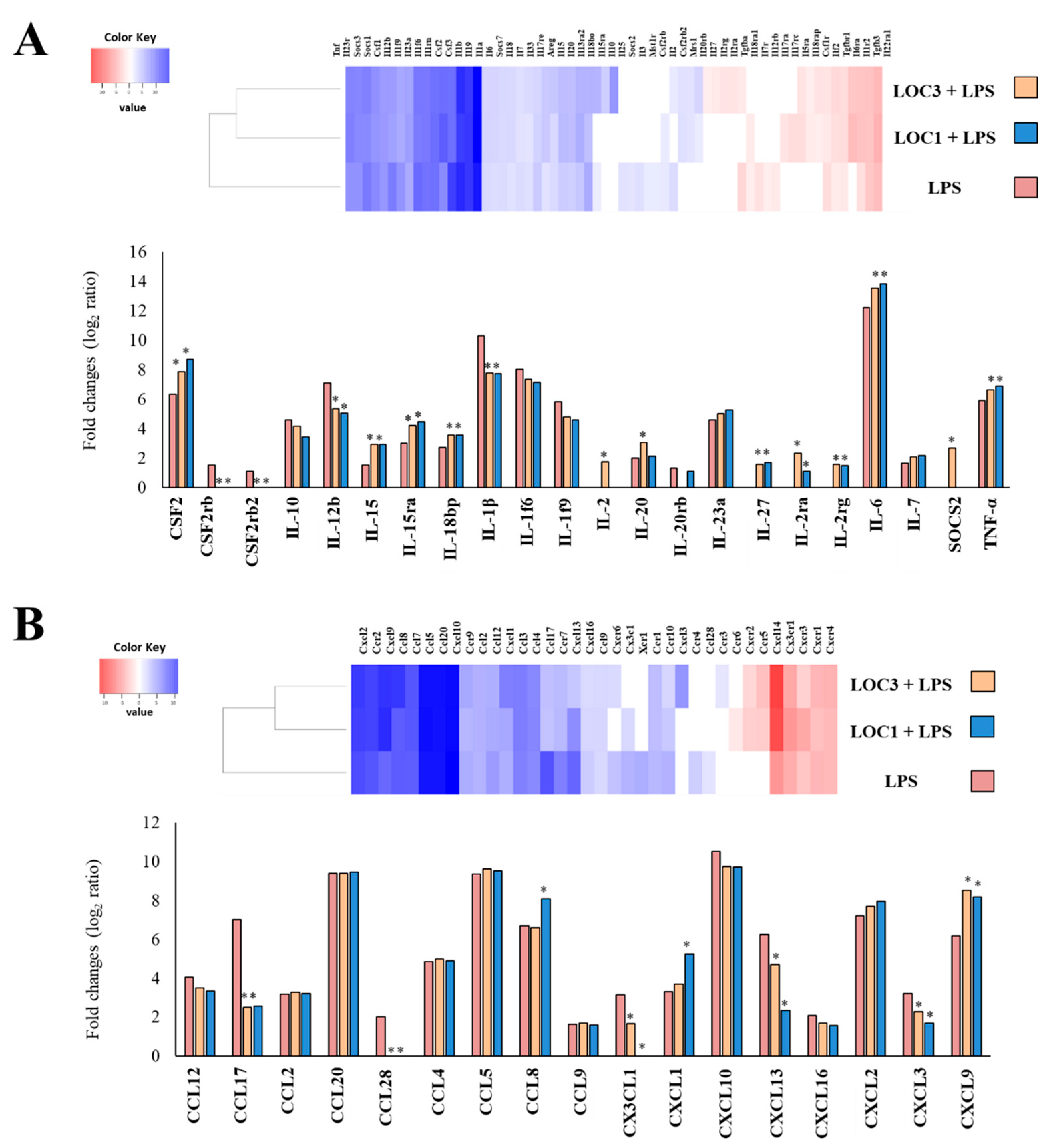
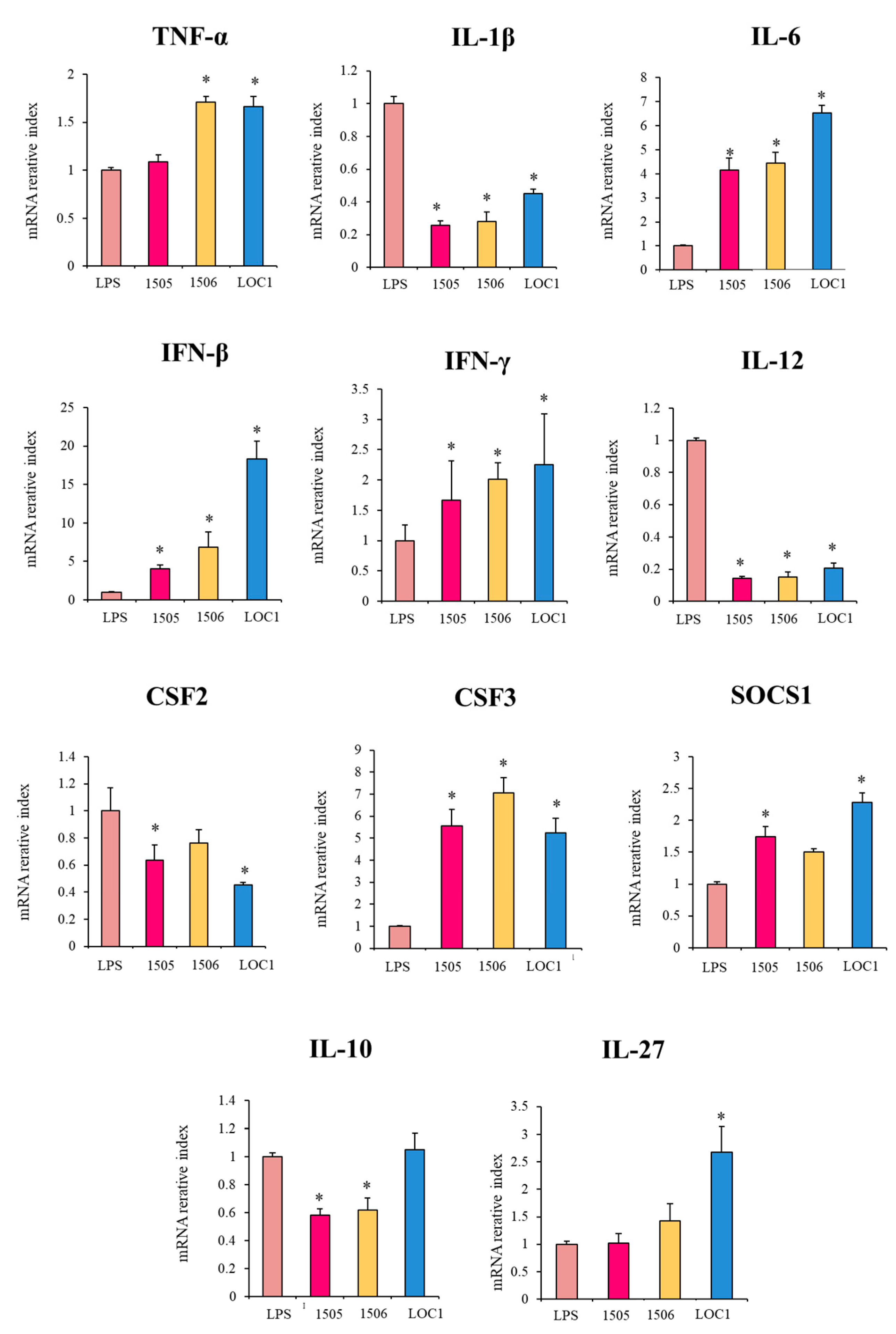
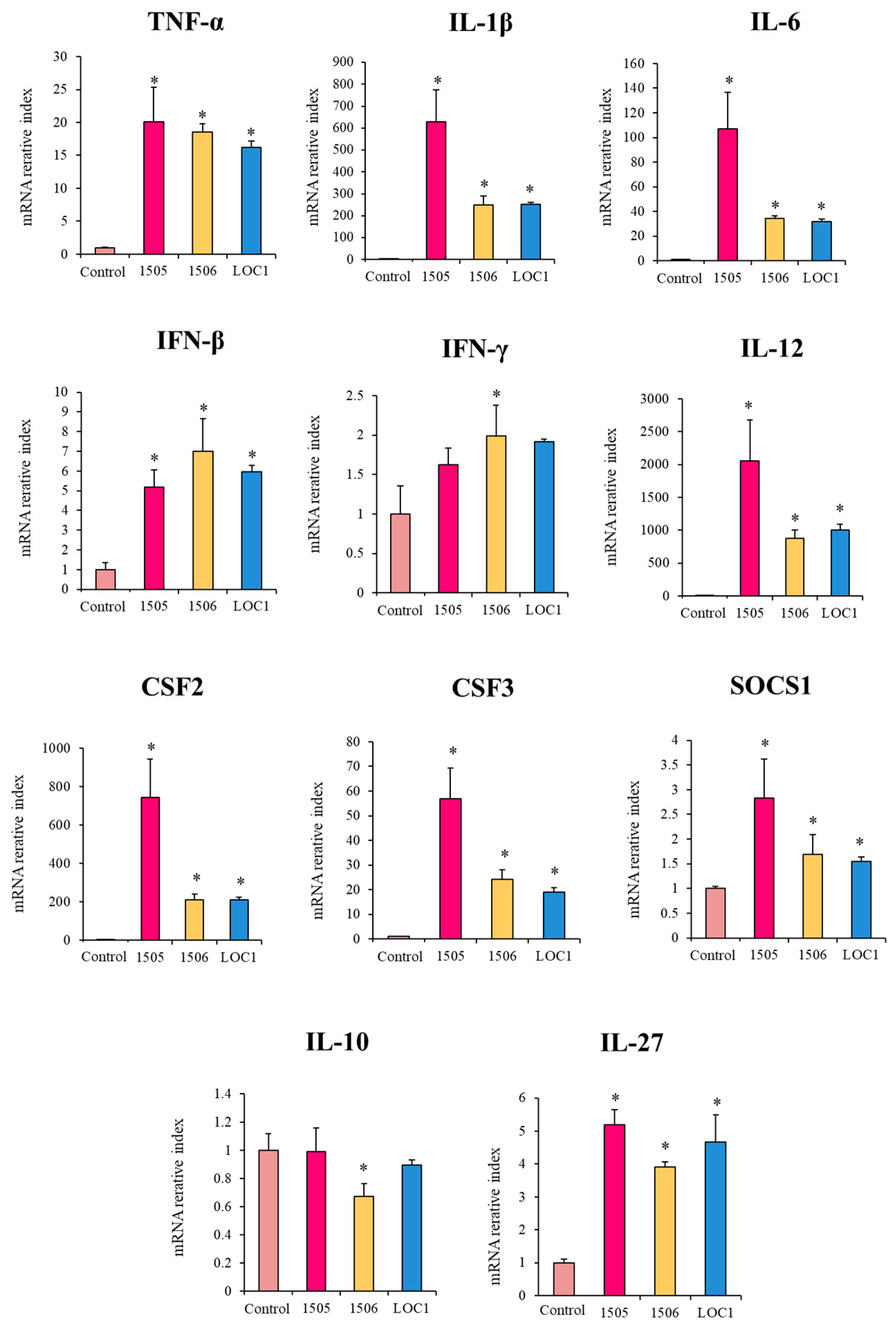
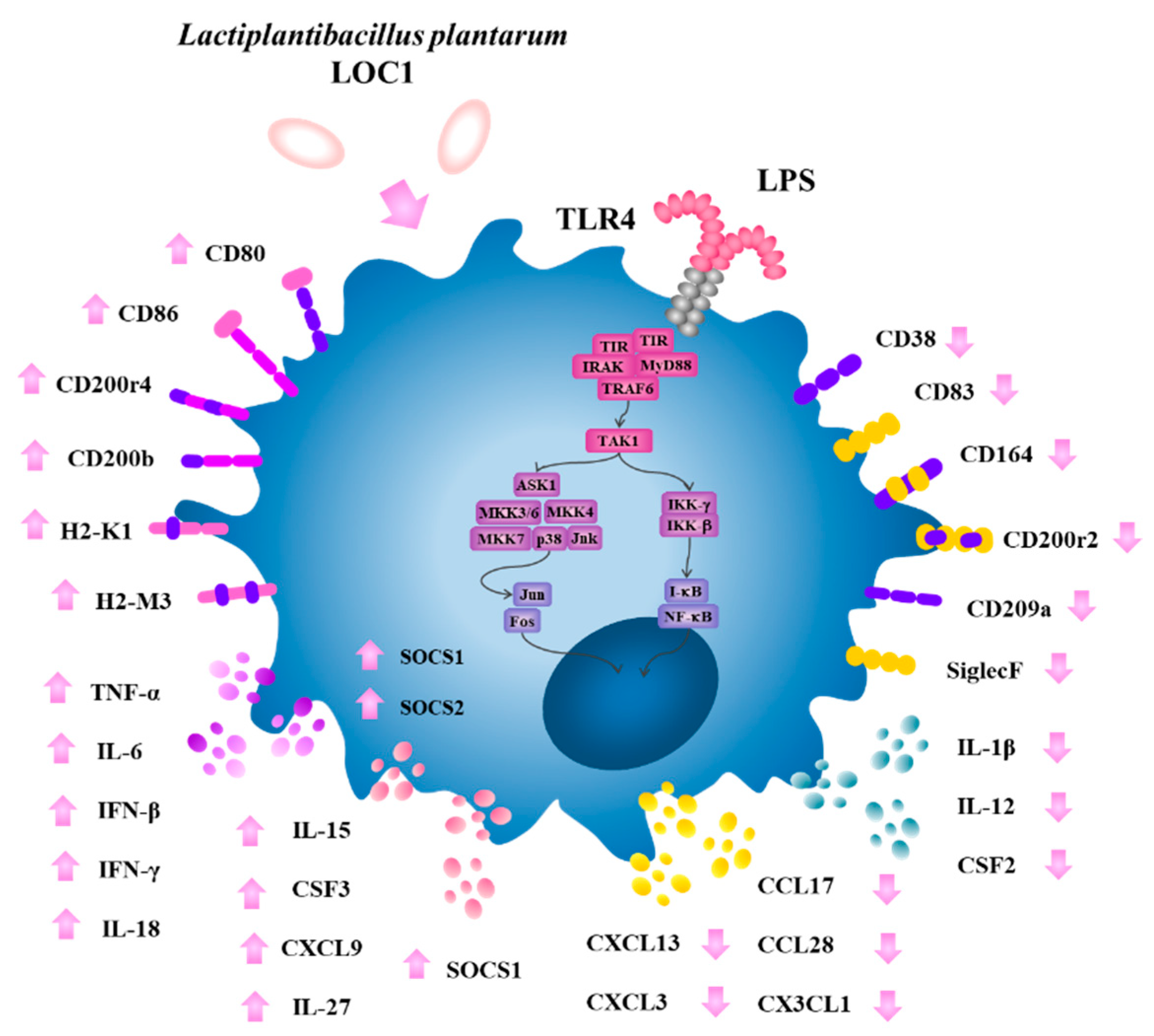
3. Results
3.1. L. plantarum Isolated from Fresh Tea Leaves Modulate the Immunotranscriptomic Response of Macrophages Triggered by TLR4 Activation
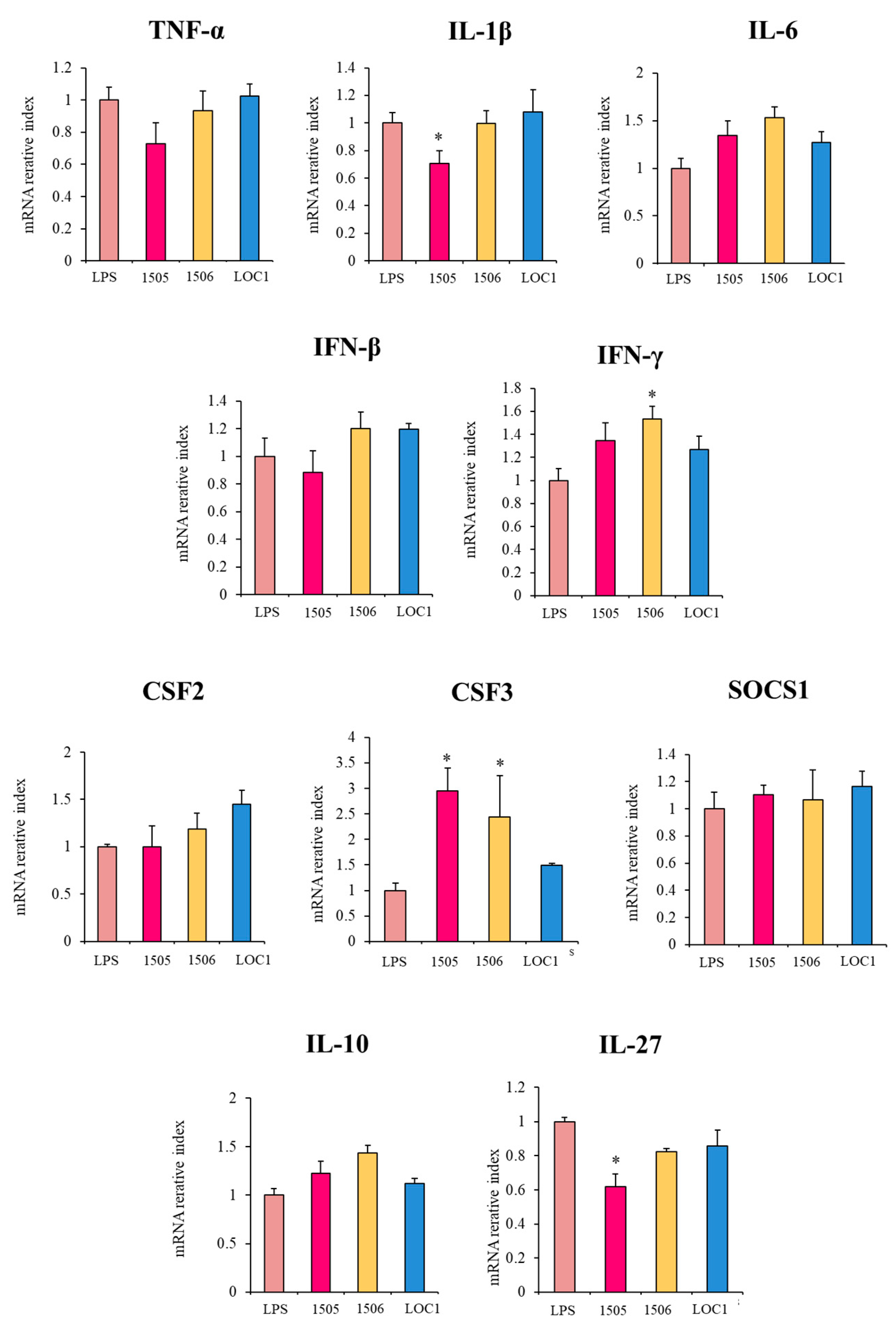
3.2. L. plantarum LOC1 Modulate the TLR4-Mediated Immune Response in Macrophages Similarly to Other Immunobiotic Strains
3.3. L. plantarum LOC1 Does Not Modulate the TLR4-Mediated Immune Response in Macrophages Indirectly Througth Intestinal Epithelial Cells
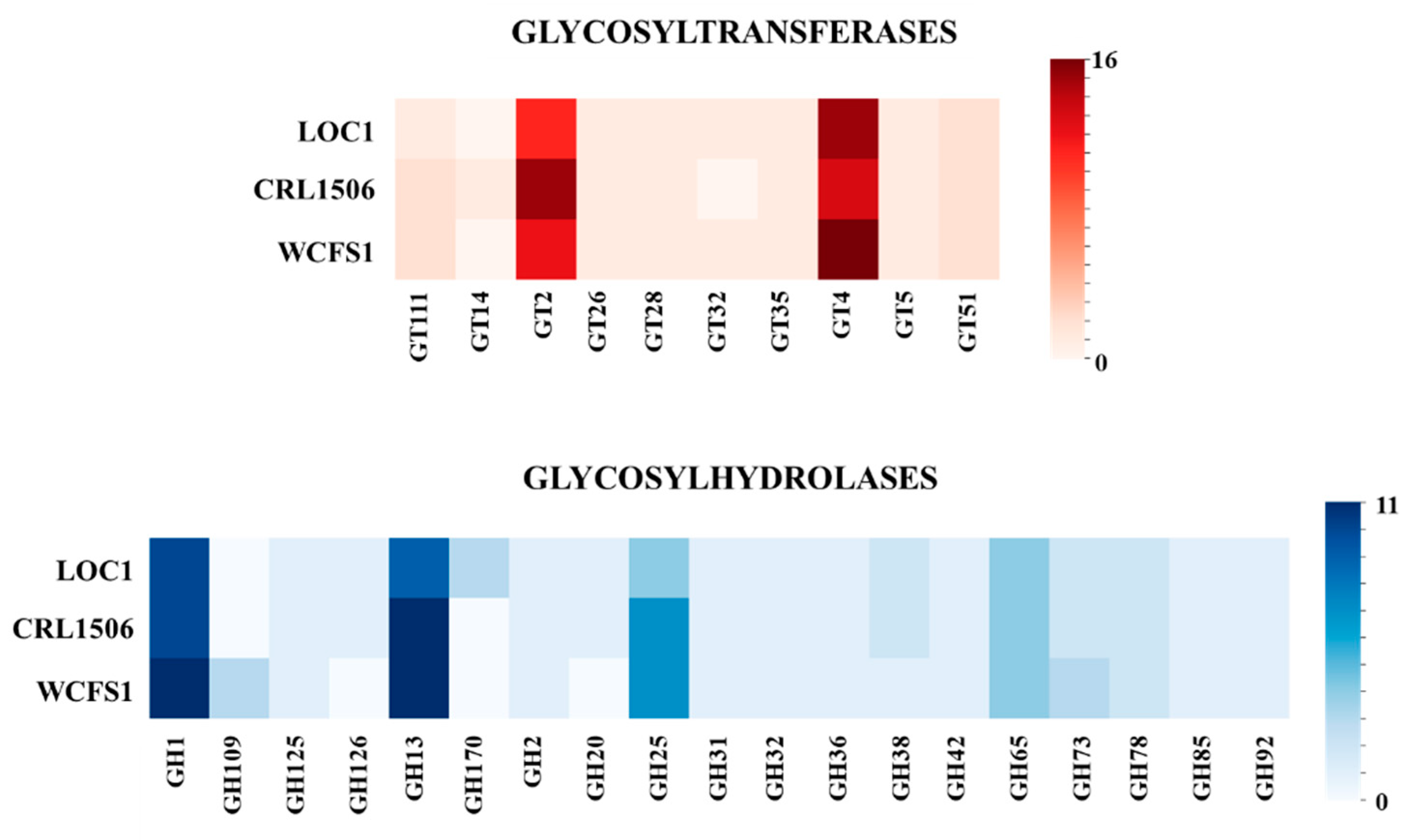
3.4. General Genomic Characteristics of L. plantarum LOC1
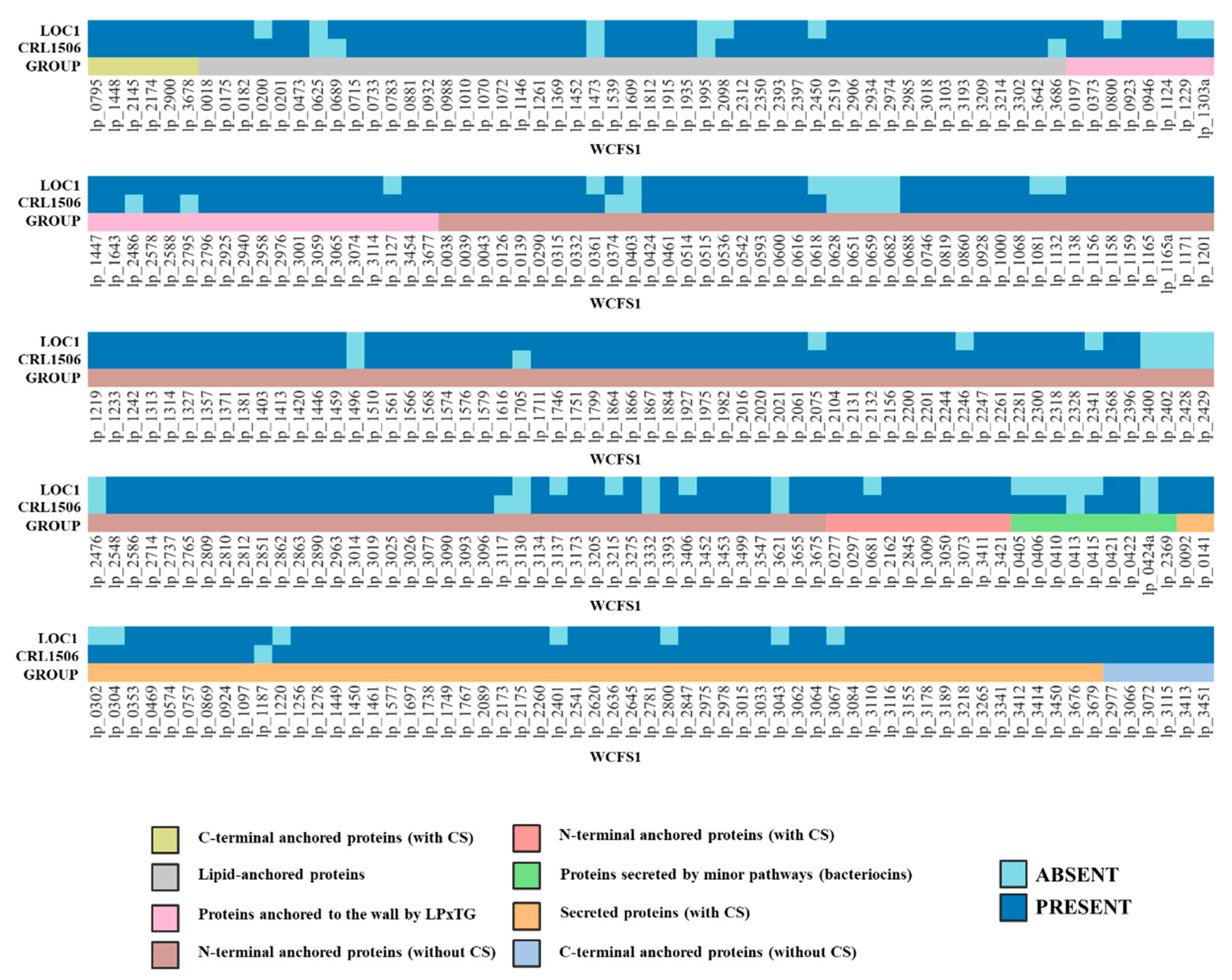
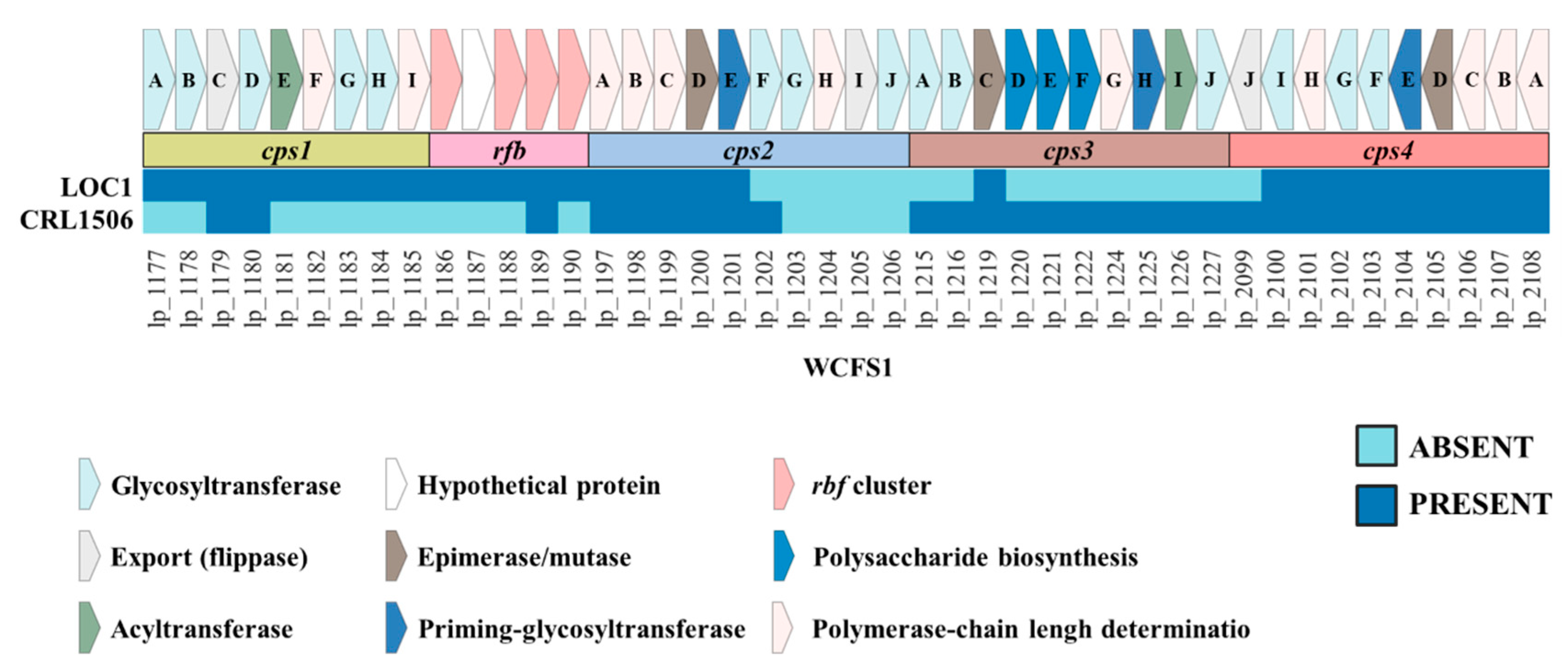
3.5. Study of the Genes Associated with the Expression of Surface Molecules in L. plantarum LOC1
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Duar, R.M.; Lin, X.B.; Zheng, J.; Martino, M.E.; Grenier, T.; Pérez-Muñoz, M.E.; Leulier, F.; Gänzle, M.; Walter, J. Lifestyles in transition: Evolution and natural history of the genus Lactobacillus. FEMS Microbiol. Rev. 2017, 41, S27–S48. [Google Scholar] [CrossRef]
- Yu, A.O.; Goldman, E.A.; Brooks, J.T.; Golomb, B.L.; Yim, I.S.; Gotcheva, V.; Angelov, A.; Kim, E.B.; Marco, M.L. Strain diversity of plant-associated Lactiplantibacillus plantarum. Microb. Biotechnol. 2021, 14, 1990–2008. [Google Scholar] [CrossRef]
- Salvetti, E.; Harris, H.M.B.; Felis, G.E.; O’Toole, P.W. Comparative Genomics of the Genus Lactobacillus Reveals Robust Phylogroups that Provide the Basis for Reclassification. Appl. Environ. Microbiol. 2018, 84, 00993-18. [Google Scholar] [CrossRef]
- Albarracin, L.; Kobayashi, H.; Iida, H.; Sato, N.; Nochi, T.; Aso, H.; Salva, S.; Alvarez, S.; Kitazawa, H.; Villena, J. Transcriptomic analysis of the innate antiviral immune response in porcine intestinal epithelial cells: Influence of immunobiotic lactobacilli. Front. Immunol. 2017, 8, 57. [Google Scholar] [CrossRef]
- Albarracin, L.; Garcia-Castillo, V.; Masumizu, Y.; Indo, Y.; Islam, M.A.M.A.M.A.; Suda, Y.; Garcia-Cancino, A.; Aso, H.; Takahashi, H.; Kitazawa, H.; et al. Efficient Selection of New Immunobiotic Strains with Antiviral Effects in Local and Distal Mucosal Sites by Using Porcine Intestinal Epitheliocytes. Front. Immunol. 2020, 11, 543. [Google Scholar] [CrossRef]
- Villena, J.; Chiba, E.; Vizoso-Pinto, M.G.; Tomosada, Y.; Takahashi, T.; Ishizuka, T.; Aso, H.; Salva, S.; Alvarez, S.; Kitazawa, H. Immunobiotic Lactobacillus rhamnosus strains differentially modulate antiviral immune response in porcine intestinal epithelial and antigen presenting cells. BMC Microbiol. 2014, 14, 126. [Google Scholar] [CrossRef]
- Kim, K.; Lee, G.; Thanh, H.D.; Kim, J.H.; Konkit, M.; Yoon, S.; Park, M.; Yang, S.; Park, E.; Kim, W. Exopolysaccharide from Lactobacillus plantarum LRCC5310 offers protection against rotavirus-induced diarrhea and regulates inflammatory response. J. Dairy Sci. 2018, 101, 5702–5712. [Google Scholar] [CrossRef]
- Kawashima, T.; Hayashi, K.; Kosaka, A.; Kawashima, M.; Igarashi, T.; Tsutsui, H.; Tsuji, N.M.; Nishimura, I.; Hayashi, T.; Obata, A. Lactobacillus plantarum strain YU from fermented foods activates Th1 and protective immune responses. Int. Immunopharmacol. 2011, 11, 2017–2024. [Google Scholar] [CrossRef]
- Kwon, M.; Lee, J.; Park, S.; Kwon, O.H.; Seo, J.; Roh, S. Exopolysaccharide Isolated from Lactobacillus plantarum L-14 Has Anti-Inflammatory Effects via the Toll-Like Receptor 4 Pathway in LPS-Induced RAW 264.7 Cells. Int. J. Mol. Sci. 2020, 21, 9283. [Google Scholar] [CrossRef]
- Yu, P.; Ke, C.; Guo, J.; Zhang, X.; Li, B. Lactobacillus plantarum L15 Alleviates Colitis by Inhibiting LPS-Mediated NF-κB Activation and Ameliorates DSS-Induced Gut Microbiota Dysbiosis. Front. Immunol. 2020, 11, 575173. [Google Scholar] [CrossRef]
- Dong, J.; Ping, L.; Zhang, K.; Tang, H.; Liu, J.; Liu, D.; Zhao, L.; Evivie, S.E.; Li, B.; Huo, G. Immunomodulatory effects of mixed Lactobacillus plantarum on lipopolysaccharide-induced intestinal injury in mice. Food Funct. 2022, 13, 4914–4929. [Google Scholar] [CrossRef] [PubMed]
- Mizuno, H.; Arce, L.; Tomotsune, K.; Albarracin, L.; Funabashi, R.; Vera, D.; Islam, M.A.M.A.; Vizoso-Pinto, M.G.M.G.; Takahashi, H.; Sasaki, Y.; et al. Lipoteichoic Acid Is Involved in the Ability of the Immunobiotic Strain Lactobacillus plantarum CRL1506 to Modulate the Intestinal Antiviral Innate Immunity Triggered by TLR3 Activation. Front. Immunol. 2020, 11, 571. [Google Scholar] [CrossRef] [PubMed]
- Tanasupawat, S.; Pakdeeto, A.; Thawai, C.; Yukphan, P.; Okada, S. Identification of lactic acid bacteria from fermented tea leaves (miang) in Thailand and proposals of Lactobacillus thailandensis sp. nov., Lactobacillus camelliae sp. nov., and Pediococcus siamensis sp. nov. J. Gen. Appl. Microbiol. 2007, 53, 7–15. [Google Scholar] [CrossRef]
- Klayraung, S.; Viernstein, H.; Sirithunyalug, J.; Okonogi, S. Probiotic Properties of Lactobacilli Isolated from Thai Traditional Food. Sci. Pharm. 2008, 76, 485–504. [Google Scholar] [CrossRef]
- Horie, M.; Sato, H.; Tada, A.; Nakamura, S.; Sugino, S.; Tabei, Y.; Katoh, M.; Toyotome, T. Regional characteristics of Lactobacillus plantarum group strains isolated from two kinds of Japanese post-fermented teas, Ishizuchi-kurocha andAwa-bancha. Biosci. Microbiota Food Health 2019, 38, 11. [Google Scholar] [CrossRef] [PubMed]
- Gharaei-Fathabad, E.; Eslamifar, M. Isolation and applications of one strain of Lactobacillus paraplantarum from tea leaves (Camellia sinensis). Am. J. Food Technol. 2011, 6, 429–434. [Google Scholar] [CrossRef]
- Tsujikawa, Y.; Suzuki, M.; Sakane, I. Isolation, identification, and impact on intestinal barrier integrity of Lactiplantibacillus plantarum from fresh tea leaves (Camellia sinensis). Biosci. Microbiota Food Health 2021, 40, 186. [Google Scholar] [CrossRef]
- Kanaya, T.; Miyazawa, K.; Takakura, I.; Itani, W.; Watanabe, K.; Ohwada, S.; Kitazawa, H.; Rose, M.T.; McConochie, H.R.; Okano, H.; et al. Differentiation of a murine intestinal epithelial cell line (MIE) toward the M cell lineage. Am. J. Physiol. Gastrointest. Liver Physiol. 2008, 295, G273–G284. [Google Scholar] [CrossRef][Green Version]
- Kanehisa, M.; Sato, Y.; Morishima, K. BlastKOALA and GhostKOALA: KEGG Tools for Functional Characterization of Genome and Metagenome Sequences. J. Mol. Biol. 2016, 428, 726–731. [Google Scholar] [CrossRef]
- Zhang, H.; Yohe, T.; Huang, L.; Entwistle, S.; Wu, P.; Yang, Z.; Busk, P.K.; Xu, Y.; Yin, Y. dbCAN2: A meta server for automated carbohydrate-active enzyme annotation. Nucleic Acids Res. 2018, 46, W95–W101. [Google Scholar] [CrossRef]
- Johnson, M.; Zaretskaya, I.; Raytselis, Y.; Merezhuk, Y.; McGinnis, S.; Madden, T.L. NCBI BLAST: A better web interface. Nucleic Acids Res. 2008, 36, 5–9. [Google Scholar] [CrossRef] [PubMed]
- Villena, J.; Kitazawa, H. The Modulation of Mucosal Antiviral Immunity by Immunobiotics: Could They Offer Any Benefit in the SARS-CoV-2 Pandemic? Front. Physiol. 2020, 11, 699. [Google Scholar] [CrossRef] [PubMed]
- Villena, J.; Li, C.; Vizoso-Pinto, M.G.; Sacur, J.; Ren, L.; Kitazawa, H. Lactiplantibacillus plantarum as a Potential Adjuvant and Delivery System for the Development of SARS-CoV-2 Oral Vaccines. Microorganisms 2021, 9, 683. [Google Scholar] [CrossRef] [PubMed]
- Kayama, H.; Okumura, R.; Takeda, K. Interaction Between the Microbiota, Epithelia, and Immune Cells in the Intestine. Annu. Rev. Immunol. 2020, 38, 23–48. [Google Scholar] [CrossRef]
- Carpi, F.M.; Coman, M.M.; Silvi, S.; Picciolini, M.; Verdenelli, M.C.; Napolioni, V. Comprehensive pan-genome analysis of Lactiplantibacillus plantarum complete genomes. J. Appl. Microbiol. 2021, 132, 592–604. [Google Scholar] [CrossRef]
- Mao, B.; Yin, R.; Li, X.; Cui, S.; Zhang, H.; Zhao, J.; Chen, W. Comparative Genomic Analysis of Lactiplantibacillus plantarum Isolated from Different Niches. Genes 2021, 12, 241. [Google Scholar] [CrossRef]
- Fernández Ramírez, M.D.; Nierop Groot, M.N.; Smid, E.J.; Hols, P.; Kleerebezem, M.; Abee, T. Role of cell surface composition and lysis in static biofilm formation by Lactobacillus plantarum WCFS1. Int. J. Food Microbiol. 2018, 271, 15–23. [Google Scholar] [CrossRef]
- Albarracin, L.; Tonetti, F.R.; Fukuyama, K.; Suda, Y.; Zhou, B.; Baillo, A.A.; Fadda, S.; Saavedra, L.; Kurata, S.; Hebert, E.M.; et al. Genomic Characterization of Lactiplantibacillus plantarum Strains Possessing Differential Antiviral Immunomodulatory Activities. Bacteria 2022, 1, 136–160. [Google Scholar] [CrossRef]
- Boekhorst, J.; Wels, M.; Kleerebezem, M.; Siezen, R.J. The predicted secretome of Lactobacillus plantarum WCFS1 sheds light on interactions with its environment. Microbiology 2006, 152, 3175–3183. [Google Scholar] [CrossRef]
- Jia, F.-F.; Zheng, H.-Q.; Sun, S.-R.; Pang, X.-H.; Liang, Y.; Shang, J.-C.; Zhu, Z.-T.; Meng, X.-C. Role of luxS in Stress Tolerance and Adhesion Ability in Lactobacillus plantarum KLDS1.0391. Biomed Res. Int. 2018, 2018, 4506829. [Google Scholar] [CrossRef]
- Wang, G.; Zhang, M.; Zhao, J.; Xia, Y.; Lai, P.F.H.; Ai, L. A surface protein from Lactobacillus plantarumincreases the adhesion of lactobacillus strains to human epithelial cells. Front. Microbiol. 2018, 9, 2858. [Google Scholar] [CrossRef] [PubMed]
- Holst, B.; Glenting, J.; Holmstrøm, K.; Israelsen, H.; Vrang, A.; Antonsson, M.; Ahrné, S.; Madsen, S.M. Molecular Switch Controlling Expression of the Mannose-Specific Adhesin, Msa, in Lactobacillus plantarum. Appl. Environ. Microbiol. 2019, 85, e02954-18. [Google Scholar] [CrossRef] [PubMed]
- Huang, T.; Peng, Z.; Hu, M.; Xiao, Y.; Liu, Z.-g.; Guan, Q.-q.; Xie, M.-y.; Xiong, T. Interactions between Lactobacillus plantarum NCU116 and its environments based on extracellular proteins and polysaccharides prediction by comparative analysis. Genomics 2020, 112, 3579–3587. [Google Scholar] [CrossRef] [PubMed]
- Remus, D.M.; van Kranenburg, R.; van Swam, I.I.; Taverne, N.; Bongers, R.S.; Wels, M.; Wells, J.M.; Bron, P.A.; Kleerebezem, M. Impact of 4 Lactobacillus plantarum capsular polysaccharide clusters on surface glycan composition and host cell signaling. Microb. Cell Fact. 2012, 11, 149. [Google Scholar] [CrossRef]
- Péant, B.; LaPointe, G.; Gilbert, C.; Atlan, D.; Ward, P.; Roy, D. Comparative analysis of the exopolysaccharide biosynthesis gene clusters from four strains of Lactobacillus rhamnosus. Microbiology 2005, 151, 1839–1851. [Google Scholar] [CrossRef]
- Lee, I.C.; van Swam, I.I.; Boeren, S.; Vervoort, J.; Meijerink, M.; Taverne, N.; Starrenburg, M.; Bron, P.A.; Kleerebezem, M. Lipoproteins Contribute to the Anti-inflammatory Capacity of Lactobacillus plantarum WCFS1. Front. Microbiol. 2020, 11, 1822. [Google Scholar] [CrossRef]
- Grangette, C.; Nutten, S.; Palumbo, E.; Morath, S.; Hermann, C.; Dewulf, J.; Pot, B.; Hartung, T.; Hols, P.; Mercenier, A. Enhanced antiinflammatory capacity of a Lactobacillus plantarum mutant synthesizing modified teichoic acids. Proc. Natl. Acad. Sci. USA 2005, 102, 10321–10326. [Google Scholar] [CrossRef]
- Hirose, Y.; Murosaki, S.; Fujiki, T.; Yamamoto, Y.; Yoshikai, Y.; Yamashita, M. Lipoteichoic acids on Lactobacillus plantarum cell surfaces correlate with induction of interleukin-12p40 production. Microbiol. Immunol. 2010, 54, 143–151. [Google Scholar] [CrossRef]
- Bron, P.A.; Tomita, S.; van Swam, I.I.; Remus, D.M.; Meijerink, M.; Wels, M.; Okada, S.; Wells, J.M.; Kleerebezem, M. Lactobacillus plantarum possesses the capability for wall teichoic acid backbone alditol switching. Microb. Cell Fact. 2012, 11, 123. [Google Scholar] [CrossRef]
- Kota, R.S.; Rutledge, J.C.; Gohil, K.; Kumar, A.; Enelow, R.I.; Ramana, C.V. Regulation of gene expression in RAW 264.7 macrophage cell line by interferon-gamma. Biochem. Biophys. Res. Commun. 2006, 342, 1137–1146. [Google Scholar] [CrossRef]
- Muller, P.A.; Matheis, F.; Mucida, D. Gut macrophages: Key players in intestinal immunity and tissue physiology. Curr. Opin. Immunol. 2020, 62, 54–61. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Liu, H.; Zhao, J. Macrophage Polarization Induced by Probiotic Bacteria: A Concise Review. Probiotics Antimicrob. Proteins 2020, 12, 798–808. [Google Scholar] [CrossRef] [PubMed]
- Pradhan, B.; Guha, D.; Ray, P.; Das, D.; Aich, P. Comparative Analysis of the Effects of Two Probiotic Bacterial Strains on Metabolism and Innate Immunity in the RAW 264.7 Murine Macrophage Cell Line. Probiotics Antimicrob. Proteins 2016, 8, 73–84. [Google Scholar] [CrossRef] [PubMed]
- Jorjão, A.L.; De Oliveira, F.E.; Leão, M.V.P.; Carvalho, C.A.T.; Jorge, A.O.C.; De Oliveira, L.D. Live and Heat-Killed Lactobacillus rhamnosus ATCC 7469 May Induce Modulatory Cytokines Profiles on Macrophages RAW 264.7. Sci. World J. 2015, 2015, 716749. [Google Scholar] [CrossRef]
- Maldonado Galdeano, C.; De Moreno De Leblanc, A.; Vinderola, G.; Bibas Bonet, M.E.; Perdigón, G. Proposed model: Mechanisms of immunomodulation induced by probiotic bacteria. Clin. Vaccine Immunol. 2007, 14, 485–492. [Google Scholar] [CrossRef]
- Salva, S.; Villena, J.; Alvarez, S. Immunomodulatory activity of Lactobacillus rhamnosus strains isolated from goat milk: Impact on intestinal and respiratory infections. Int. J. Food Microbiol. 2010, 141, 82–89. [Google Scholar] [CrossRef]
- Kojima, N.; Kojima, S.; Hosokawa, S.; Oda, Y.; Zenke, D.; Toura, Y.; Onohara, E.; Yokota, S.I.; Nagaoka, M.; Kuroda, Y. Wall teichoic acid-dependent phagocytosis of intact cell walls of Lactiplantibacillus plantarum elicits IL-12 secretion from macrophages. Front. Microbiol. 2022, 13, 986396. [Google Scholar] [CrossRef]
- Villena, J.; Salva, S.; Agüero, G.; Alvarez, S. Immunomodulatory and protective effect of probiotic Lactobacillus casei against Candida albicans infection in malnourished mice. Microbiol. Immunol. 2011, 55, 434–445. [Google Scholar] [CrossRef]
- Wormald, S.; Hilton, D.J. Inhibitors of cytokine signal transduction. J. Biol. Chem. 2004, 279, 821–824. [Google Scholar] [CrossRef]
- De Oca, M.M.; De Labastida Rivera, F.; Winterford, C.; Frame, T.C.M.; Ng, S.S.; Amante, F.H.; Edwards, C.L.; Bukali, L.; Wang, Y.; Uzonna, J.E.; et al. IL-27 signalling regulates glycolysis in Th1 cells to limit immunopathology during infection. PLoS Pathog. 2020, 16, e1008994. [Google Scholar] [CrossRef]
- de Bus, I.; van Krimpen, S.; Hooiveld, G.J.; Boekschoten, M.V.; Poland, M.; Witkamp, R.F.; Albada, B.; Balvers, M.G.J. Immunomodulating effects of 13- and 16-hydroxylated docosahexaenoyl ethanolamide in LPS stimulated RAW264.7 macrophages. Biochim. Biophys. Acta Mol. Cell Biol. Lipids 2021, 1866, 158908. [Google Scholar] [CrossRef] [PubMed]
- Kopczynski, M.; Rumienczyk, I.; Kulecka, M.; Statkiewicz, M.; Pysniak, K.; Sandowska-Markiewicz, Z.; Wojcik-Trechcinska, U.; Goryca, K.; Pyziak, K.; Majewska, E.; et al. Selective Extracellular Signal-Regulated Kinase 1/2 (ERK1/2) Inhibition by the SCH772984 Compound Attenuates In Vitro and In Vivo Inflammatory Responses and Prolongs Survival in Murine Sepsis Models. Int. J. Mol. Sci. 2021, 22, 10204. [Google Scholar] [CrossRef] [PubMed]
- Takano, M.; Nishimura, H.; Kimura, Y.; Mokuno, Y.; Washizu, J.; Itohara, S.; Nimura, Y.; Yoshikai, Y. Protective roles of gamma delta T cells and interleukin-15 in Escherichia coli infection in mice. Infect. Immun. 1998, 66, 3270–3278. [Google Scholar] [CrossRef]
- Elia, G.; Guglielmi, G. CXCL9 chemokine in ulcerative colitis. Clin. Ter. 2018, 169, E235–E241. [Google Scholar] [CrossRef]
- Tarique, A.A.; Logan, J.; Thomas, E.; Holt, P.G.; Sly, P.D.; Fantino, E. Phenotypic, functional, and plasticity features of classical and alternatively activated human macrophages. Am. J. Respir. Cell Mol. Biol. 2015, 53, 676–688. [Google Scholar] [CrossRef] [PubMed]
- Noh, H.J.; Park, J.M.; Kwon, Y.J.; Kim, K.; Park, S.Y.; Kim, I.; Lim, J.H.; Kim, B.K.; Kim, B.Y. Immunostimulatory Effect of Heat-Killed Probiotics on RAW264.7 Macrophages. J. Microbiol. Biotechnol. 2022, 32, 638–644. [Google Scholar] [CrossRef] [PubMed]
- Khanna, S.; Bishnoi, M.; Kondepudi, K.K.; Shukla, G. Isolation, characterization and anti-inflammatory mechanism of probiotics in lipopolysaccharide-stimulated RAW 264.7 macrophages. World J. Microbiol. Biotechnol. 2020, 36, 1323–1327. [Google Scholar] [CrossRef]
- Qi, S.R.; Cui, Y.J.; Liu, J.X.; Luo, X.; Wang, H.F. Lactobacillus rhamnosus GG components, SLP, gDNA and CpG, exert protective effects on mouse macrophages upon lipopolysaccharide challenge. Lett. Appl. Microbiol. 2020, 70, 118–127. [Google Scholar] [CrossRef]
- Kern, K.; Pierre, S.; Schreiber, Y.; Angioni, C.; Thomas, D.; Ferreirós, N.; Geisslinger, G.; Scholich, K. CD200 selectively upregulates prostaglandin E 2 and D 2 synthesis in LPS-treated bone marrow-derived macrophages. Prostaglandins Other Lipid Mediat. 2017, 133, 53–59. [Google Scholar] [CrossRef]
- Amici, S.A.; Young, N.A.; Narvaez-Miranda, J.; Jablonski, K.A.; Arcos, J.; Rosas, L.; Papenfuss, T.L.; Torrelles, J.B.; Jarjour, W.N.; Guerau-de-Arellano, M. CD38 is robustly induced in human macrophages and monocytes in inflammatory conditions. Front. Immunol. 2018, 9, 1593. [Google Scholar] [CrossRef]
- Zhou, X.; Hong, T.; Yu, Q.; Nie, S.; Gong, D.; Xiong, T.; Xie, M. Exopolysaccharides from Lactobacillus plantarum NCU116 induce c-Jun dependent Fas/Fasl-mediated apoptosis via TLR2 in mouse intestinal epithelial cancer cells. Sci. Rep. 2017, 7, 14247. [Google Scholar] [CrossRef] [PubMed]
- Deo, D.; Davray, D.; Kulkarni, R. A Diverse Repertoire of Exopolysaccharide Biosynthesis Gene Clusters in Lactobacillus Revealed by Comparative Analysis in 106 Sequenced Genomes. Microorganisms 2019, 7, 444. [Google Scholar] [CrossRef] [PubMed]
- Garcia-Gonzalez, N.; Bottacini, F.; van Sinderen, D.; Gahan, C.G.M.; Corsetti, A. Comparative Genomics of Lactiplantibacillus plantarum: Insights into Probiotic Markers in Strains Isolated from the Human Gastrointestinal Tract and Fermented Foods. Front. Microbiol. 2022, 13, 1353. [Google Scholar] [CrossRef] [PubMed]
- Pretzer, G.; Snel, J.; Molenaar, D.; Wiersma, A.; Bron, P.A.; Lambert, J.; De Vos, W.M.; Van Der Meer, R.; Smits, M.A.; Kleerebezem, M. Biodiversity-based identification and functional characterization of the mannose-specific adhesin of Lactobacillus plantarum. J. Bacteoriology 2005, 187, 6128–6136. [Google Scholar] [CrossRef] [PubMed]
- Smelt, M.J.; de Haan, B.J.; Bron, P.A.; van Swam, I.; Meijerink, M.; Wells, J.M.; Kleerebezem, M.; Faas, M.M.; de Vos, P. The Impact of Lactobacillus plantarum WCFS1 Teichoic Acid D-Alanylation on the Generation of Effector and Regulatory T-cells in Healthy Mice. PLoS ONE 2013, 8, e63099. [Google Scholar] [CrossRef]
- Molenaar, D.; Bringel, F.; Schuren, F.H.; De Vos, W.M.; Siezen, R.J.; Kleerebezem, M. Exploring Lactobacillus plantarum Genome Diversity by Using Microarrays. J. Bacteriol. 2005, 187, 6119. [Google Scholar] [CrossRef]
- Siezen, R.J.; Tzeneva, V.A.; Castioni, A.; Wels, M.; Phan, H.T.K.; Rademaker, J.L.W.; Starrenburg, M.J.C.; Kleerebezem, M.; van Hylckama Vlieg, J.E.T. Phenotypic and genomic diversity of Lactobacillus plantarum strains isolated from various environmental niches. Environ. Microbiol. 2010, 12, 758–773. [Google Scholar] [CrossRef]




| Lactoplantibacillus plantarum | LOC1 | CRL1506 | WCFS1 |
|---|---|---|---|
| Host | Camellia sinensis | Capra aegagrus hircus | Homo sapiens |
| Origen | Fresh tea leaves | Milk | Saliva |
| Genome size (pb) | 3,138,505 | 3,228,096 | 3,348,624 |
| G+C content (%) | 44.7 | 44.5 | 44.4 |
| Genes | 3078 | 3051 | 3154 |
| Coding sequences (total) | 2907 | 2967 | 3062 |
| Protein coding sequences | 2834 | 2918 | 3015 |
| ARNr (5s, 16s, 23s) | 3 (1, 1, 1) | 13 (6, 4, 3) | 16 (6, 5, 5) |
| ARNt | 57 | 67 | 72 |
| Access number | BOUN00000000 | LNCP00000000 | AL935263.2 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Suzuki, M.; Albarracin, L.; Tsujikawa, Y.; Fukuyama, K.; Sakane, I.; Villena, J.; Kitazawa, H. Lactiplantibacillus plantarum LOC1 Isolated from Fresh Tea Leaves Modulates Macrophage Response to TLR4 Activation. Foods 2022, 11, 3257. https://doi.org/10.3390/foods11203257
Suzuki M, Albarracin L, Tsujikawa Y, Fukuyama K, Sakane I, Villena J, Kitazawa H. Lactiplantibacillus plantarum LOC1 Isolated from Fresh Tea Leaves Modulates Macrophage Response to TLR4 Activation. Foods. 2022; 11(20):3257. https://doi.org/10.3390/foods11203257
Chicago/Turabian StyleSuzuki, Masahiko, Leonardo Albarracin, Yuji Tsujikawa, Kohtaro Fukuyama, Iwao Sakane, Julio Villena, and Haruki Kitazawa. 2022. "Lactiplantibacillus plantarum LOC1 Isolated from Fresh Tea Leaves Modulates Macrophage Response to TLR4 Activation" Foods 11, no. 20: 3257. https://doi.org/10.3390/foods11203257
APA StyleSuzuki, M., Albarracin, L., Tsujikawa, Y., Fukuyama, K., Sakane, I., Villena, J., & Kitazawa, H. (2022). Lactiplantibacillus plantarum LOC1 Isolated from Fresh Tea Leaves Modulates Macrophage Response to TLR4 Activation. Foods, 11(20), 3257. https://doi.org/10.3390/foods11203257







