A Droplet Digital PCR Based Approach for Identification and Quantification of Porcine and Chicken Derivatives in Beef
Abstract
1. Introduction
2. Materials and Methods
2.1. Samples Preparation
2.2. DNA Extraction
2.3. Specific Primers and Probes Synthesis
2.4. ddPCR Assay
2.5. Establishment of Quantitative Formula
2.6. Sensitivity, Repeatability and Reproducibility Analysis
2.7. Testing of Commercial Samples
3. Results and Discussion
3.1. Species-Specific Identification
3.2. Determination and Verification of Transfer Coefficient (K)
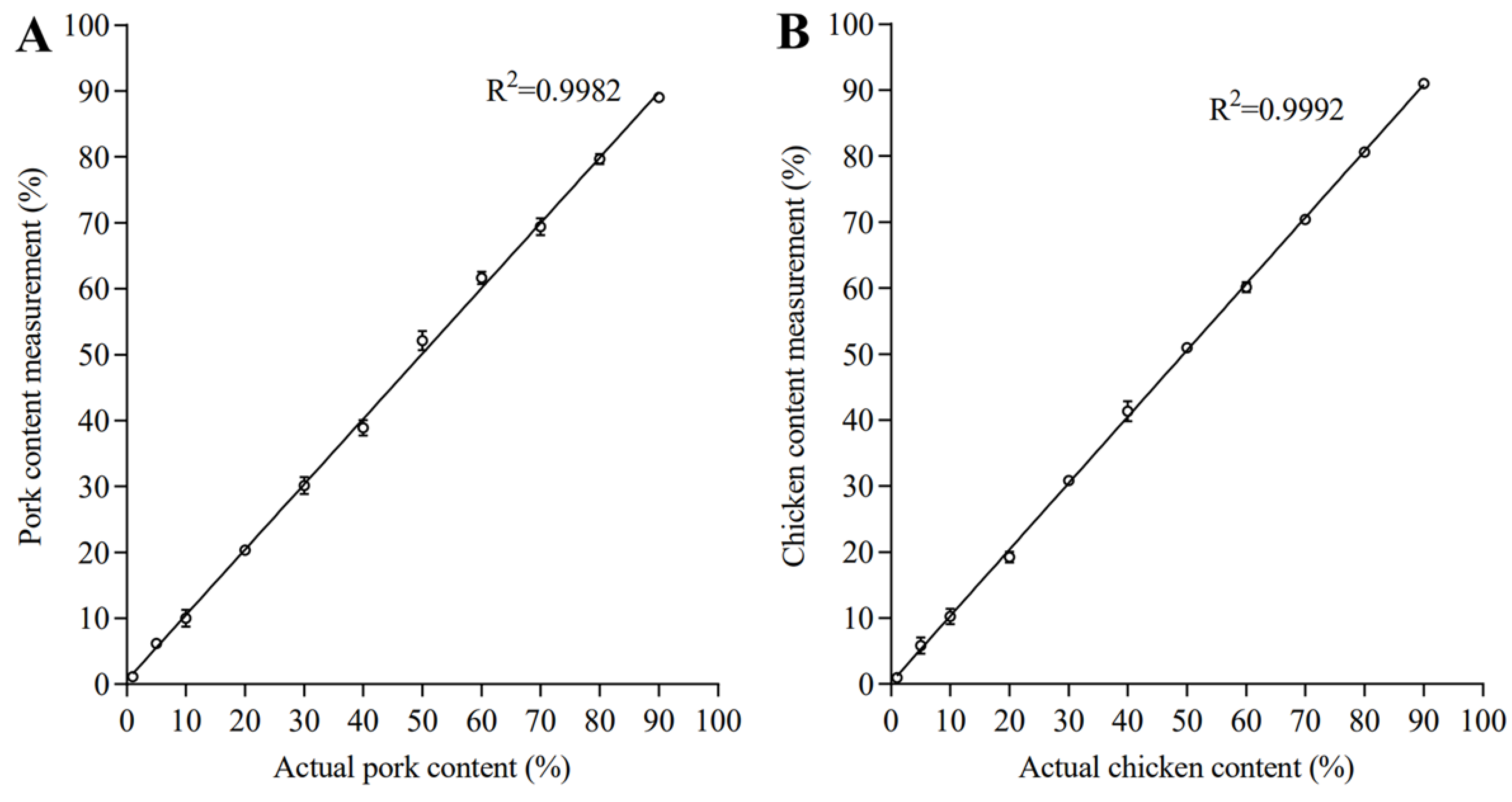
3.3. Linearity Range, LOD, LOQ of Quantification
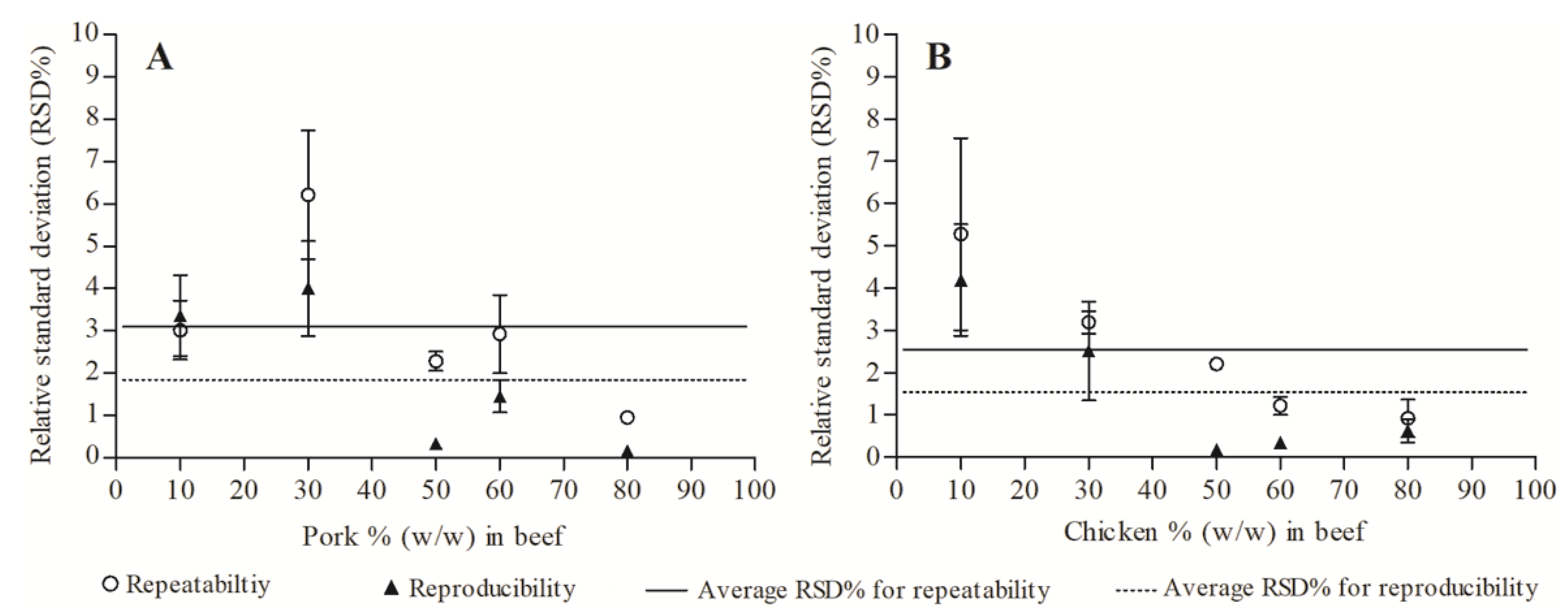
3.4. Repeatability and Reproducibility of Quantification
3.5. Effect of Thermal Treatment on Quantification
3.6. Test Commercial Products
4. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Lu, B.; Han, F.; Aheto, J.H.; Rashed, M.M.A.; Pan, Z. Artificial bionic taste sensors coupled with chemometrics for rapid detection of beef adulteration. Food Sci. Nutr. 2021, 9, 5220–5228. [Google Scholar] [CrossRef] [PubMed]
- Han, F.; Huang, X.; Aheto, J.H.; Zhang, D.; Feng, F. Detection of beef adulterated with pork using a low-cost electronic nose based on colorimetric sensors. Foods 2020, 9, 193. [Google Scholar] [CrossRef] [PubMed]
- Sarno, R.; Sabilla, S.I.; Wijaya, D.R.; Sunaryono, D.; Fatichah, C. Electronic nose dataset for pork adulteration in beef. Data Brief. 2020, 32, 106139. [Google Scholar] [CrossRef] [PubMed]
- Kamruzzaman, M.; Makino, Y.; Oshita, S.; Liu, S. Assessment of visible near-infrared hyperspectral imaging as a tool for detection of horsemeat adulteration in minced beef. Food Bioprocess Technol. 2015, 8, 1054–1062. [Google Scholar] [CrossRef]
- Alikord, M.; Momtaz, H.; keramat, J.; Kadivar, M.; Rad, A.H. Species identification and animal authentication in meat products: A review. J. Food Meas Charact. 2018, 12, 145–155. [Google Scholar] [CrossRef]
- Böhme, K.; Calo-Mata, P.; Barros-Velázquez, J.; Ortea, I. Review of recent DNA-based methods for main food-authentication topics. J. Agric. Food Chem. 2019, 67, 3854–3864. [Google Scholar] [CrossRef]
- Li, Y.C.; Liu, S.Y.; Meng, F.B.; Liu, D.Y.; Zhang, Y.; Wang, W.; Zhang, J.M. Comparative review and the recent progress in detection technologies of meat product adulteration. Compr. Rev. Food Sci. Food Saf. 2020, 19, 2256–2296. [Google Scholar] [CrossRef]
- Zia, Q.; Alawami, M.; Mokhtar, N.F.K.; Nhari, R.M.H.R.; Hanish, I. Current analytical methods for porcine identification in meat and meat products. Food Chem. 2020, 324, 126664. [Google Scholar] [CrossRef]
- Shi, Z.; Yin, B.; Yuquan, L.; Zhou, G.H.; Li, C.; Xu, X.L.; Luo, X.; Zhang, X.; Qi, J.; Voglmeir, J.; et al. N-Glycan profile as a tool in qualitative and quantitative analysis of meat adulteration. J. Agric. Food Chem. 2019, 67, 10543–10551. [Google Scholar] [CrossRef]
- Thienes, C.P.; Masiri, J.; Benoit, L.A.; Barrios-Lopez, B.; Samuel, S.A.; Krebs, R.A.; Cox, D.P.; Dobritsa, A.P.; Nadala, C.; Samadpour, M. Quantitative detection of beef contamination in cooked meat products by ELISA. J. AOAC Int. 2019, 102, 898–902. [Google Scholar] [CrossRef]
- Mandli, J.; El Fatimi, I.; Seddaoui, N.; Amine, A. Enzyme immunoassay (ELISA/immunosensor) for a sensitive detection of pork adulteration in meat. Food Chem. 2018, 255, 380–389. [Google Scholar] [CrossRef]
- Pan, X.D.; Chen, J.; Chen, Q.; Huang, B.F.; Han, J.L. Authentication of pork in meat mixtures using PRM mass spectrometry of myosin peptides. RSC Adv. 2018, 8, 11157–11162. [Google Scholar] [CrossRef]
- Stachniuk, A.; Sumara, A.; Montowska, M.; Fornal, E. Peptide markers for distinguishing guinea fowl meat from that of other species using liquid chromatography-mass spectrometry. Food Chem. 2021, 345, 128810. [Google Scholar] [CrossRef]
- Giaretta, N.; Di Giuseppe, A.M.; Lippert, M.; Parente, A.; Di Maro, A. Myoglobin as marker in meat adulteration: A UPLC method for determining the presence of pork meat in raw beef burger. Food Chem. 2013, 141, 1814–1820. [Google Scholar] [CrossRef]
- Li, Y.; Zhang, Y.; Li, H.; Zhao, W.; Guo, W.; Wang, S. Simultaneous determination of heat stable peptides for eight animal and plant species in meat products using UPLC-MS/MS method. Food Chem. 2018, 245, 125–131. [Google Scholar] [CrossRef]
- Kang, T.S.; Tanaka, T. Comparison of quantitative methods based on SYBR Green real-time qPCR to estimate pork meat adulteration in processed beef products. Food Chem. 2018, 269, 549–558. [Google Scholar] [CrossRef]
- Li, T.; Wang, J.; Wang, Z.; Qiao, L.; Liu, R.; Li, S.; Chen, A. Quantitative determination of mutton adulteration with single-copy nuclear genes by real-time PCR. Food Chem. 2021, 344, 128622. [Google Scholar] [CrossRef]
- Li, T.T.; Jalbani, Y.M.; Zhang, G.L.; Zhao, Z.Y.; Wang, Z.Y.; Zhao, X.Y.; Chen, A.L. Detection of goat meat adulteration by real-time PCR based on a reference primer. Food Chem. 2019, 277, 554–557. [Google Scholar] [CrossRef]
- Pinheiro, L.B.; Coleman, V.A.; Hindson, C.M.; Herrmann, J.; Hindson, B.J.; Bhat, S.; Emslie, K.R. Evaluation of a droplet digital polymerase chain reaction format for DNA copy number quantification. Anal Chem. 2012, 84, 1003–1101. [Google Scholar] [CrossRef]
- Kanagal-Shamanna, R. Digital PCR: Principles and applications. Methods Mol. Biol. 2016, 1392, 43–50. [Google Scholar]
- Ren, J.; Deng, T.; Huang, W.; Chen, Y.; Ge, Y.A. Digital PCR method for identifying and quantifying adulteration of meat species in raw and processed food. PLoS ONE 2017, 12, e0173567. [Google Scholar] [CrossRef]
- Deconinck, D.; Hostens, K.; Taverniers, I.; Volckaert, F.A.M.; Robbens, J.; Derycke, S. Identification and semi-quantification of Atlantic salmon in processed and mixed seafood products using Droplet Digital PCR (ddPCR). Food Chem. Toxicol. 2021, 154, 112329. [Google Scholar] [CrossRef]
- Ampaporn, K.; Phasuk, Y.; Duangjinda, M. Droplet digital polymerase chain reaction assay for identifying and quantifying pork products. Anim. Sci. J. 2021, 92, e13595. [Google Scholar] [CrossRef] [PubMed]
- Shehata, H.R.; Li, J.; Chen, S.; Redda, H.; Cheng, S.; Tabujara, N.; Li, H.; Warriner, K.; Hanner, R. Droplet digital polymerase chain reaction (ddPCR) assays integrated with an internal control for quantification of bovine, porcine, chicken and turkey species in food and feed. PLoS ONE 2017, 12, e0182872. [Google Scholar] [CrossRef] [PubMed]
- Cao, W.; Li, Y.; Chen, X.; Chang, Y.; Li, L.; Shi, L.; Bai, W.; Ye, L. Species identification and quantification of silver pomfret using the droplet digital PCR assay. Food Chem. 2020, 302, 125331. [Google Scholar] [CrossRef] [PubMed]
- Floren, C.; Wiedemann, I.; Brenig, B.; Schütz, E.; Beck, J. Species identification and quantification in meat and meat products using droplet digital PCR (ddPCR). Food Chem. 2015, 173, 1054–1058. [Google Scholar] [CrossRef]
- Wang, Q.; Cai, Y.; He, Y.; Yang, L.; Li, J.; Pan, L. Droplet digital PCR (ddPCR) method for the detection and quantification of goat and sheep derivatives in commercial meat products. Eur. Food Res. Technol. 2018, 244, 767–774. [Google Scholar] [CrossRef]
- Cai, Y.; He, Y.; Lv, R.; Chen, H.; Wang, Q.; Pan, L. Detection and quantification of beef and pork materials in meat products by duplex droplet digital PCR. PLoS ONE 2017, 12, e0181949. [Google Scholar] [CrossRef]
- Noh, E.S.; Park, Y.J.; Kim, E.M.; Park, J.Y.; Shim, K.B.; Choi, T.J.; Kim, K.H.; Kang, J.H. Quantitative analysis of Alaska pollock in seafood products by droplet digital PCR. Food Chem. 2019, 275, 638–643. [Google Scholar] [CrossRef]
- Yu, N.; Ren, J.; Huang, W.; Xing, R.; Deng, T.; Chen, Y. An effective analytical droplet digital PCR approach for identification and quantification of fur-bearing animal meat in raw and processed food. Food Chem. 2021, 355, 129525. [Google Scholar] [CrossRef]
- Köppel, R.; Ganeshan, A.; van Velsen, F.; Weber, S.; Schmid, J.; Graf, C.; Hochegger, R. Digital duplex versus real-time pcr for the determination of meat proportions from sausages containing pork and beef. Eur. Food Res. Technol. 2019, 245, 151–157. [Google Scholar] [CrossRef]
- Köppel, R.; Ganeshan, A.; Weber, S.; Pietsch, K.; Graf, C.; Hochegger, R.; Griffiths, K.; Burkhardt, S. Duplex digital PCR for the determination of meat proportions of sausages containing meat from chicken, turkey, horse, cow, pig and sheep. Eur. Food Res. Technol. 2019, 245, 853–862. [Google Scholar] [CrossRef]
- Cai, Y.; Li, X.; Lv, R.; Yang, J.; Li, J.; He, Y.; Pan, L. Quantitative analysis of pork and chicken products by droplet digital PCR. Biomed Res. Int. 2014, 2014, 810209. [Google Scholar] [CrossRef] [PubMed]
- Köppel, R.; Ruf, J.; Zimmerli, F.; Rentsch, J. Multiplex real-time PCR for the detection and quantification of DNA from beef, pork, horse and sheep. Eur. Food Res. Technol. 2011, 232, 151–155. [Google Scholar] [CrossRef]
- Huggett, J.F.; Foy, C.A.; Benes, V.; Emslie, K.; Garson, J.A.; Haynes, R.; Hellemans, J.; Kubista, M.; Mueller, R.D.; Nolan, T.; et al. The digital MIQE guidelines: Minimum information for publication of quantitative digital PCR experiments. Clin. Chem. 2013, 59, 892–902. [Google Scholar] [CrossRef]
- Ha, J.; Kim, S.; Lee, J.; Lee, S.; Lee, H.; Choi, Y.; Oh, H.; Yoon, Y. Identification of pork adulteration in processed meat products using the developed mitochondrial DNA-based primers. Korean J. Food Sci. Anim. Resour. 2017, 37, 464–468. [Google Scholar] [CrossRef]
- Cho, A.R.; Dong, H.J.; Cho, S. Meat Species Identification using loop-mediated isothermal amplification assay targeting species-specific mitochondrial DNA. Korean J. Food Sci. Anim. Resour. 2014, 34, 799–807. [Google Scholar] [CrossRef]
- Ladenburger, E.M.; Dehmer, M.; Grünberg, R.; Waiblinger, H.U.; Stoll, D.; Bergemann, J. Highly sensitive matrix-independent quantification of major food allergens peanut and soy by competitive real-time PCR targeting mitochondrial DNA. J. AOAC Int. 2018, 101, 170–184. [Google Scholar] [CrossRef]
- Deprez, L.; Corbisier, P.; Kortekaas, A.M.; Mazoua, S.; Beaz Hidalgo, R.; Trapmann, S.; Emons, H. Validation of a digital PCR method for quantification of DNA copy number concentrations by using a certified reference material. Biomol. Detect. Quantif. 2016, 9, 29–39. [Google Scholar] [CrossRef]
- Codex Committee on Methods of Analysis and Sampling. Guidelines on Performance Criteria and Validation of Methods for Detection, Identification and Quantification of Specific DNA Sequences and Specific Proteins in Foods; Codex Alimentarius-FAO: Rome, Italy, 2010. [Google Scholar]
- Hird, H.; Chisholm, J.; Sanchez, A.; Hernandez, M.; Goodier, R.; Schneede, K.; Boltz, C.; Popping, B. Effect of heat and pressure processing on DNA fragmentation and implications for the detection of meat using a real-time polymerase chain reaction. Food Addit. Contam. 2006, 23, 645–650. [Google Scholar] [CrossRef]
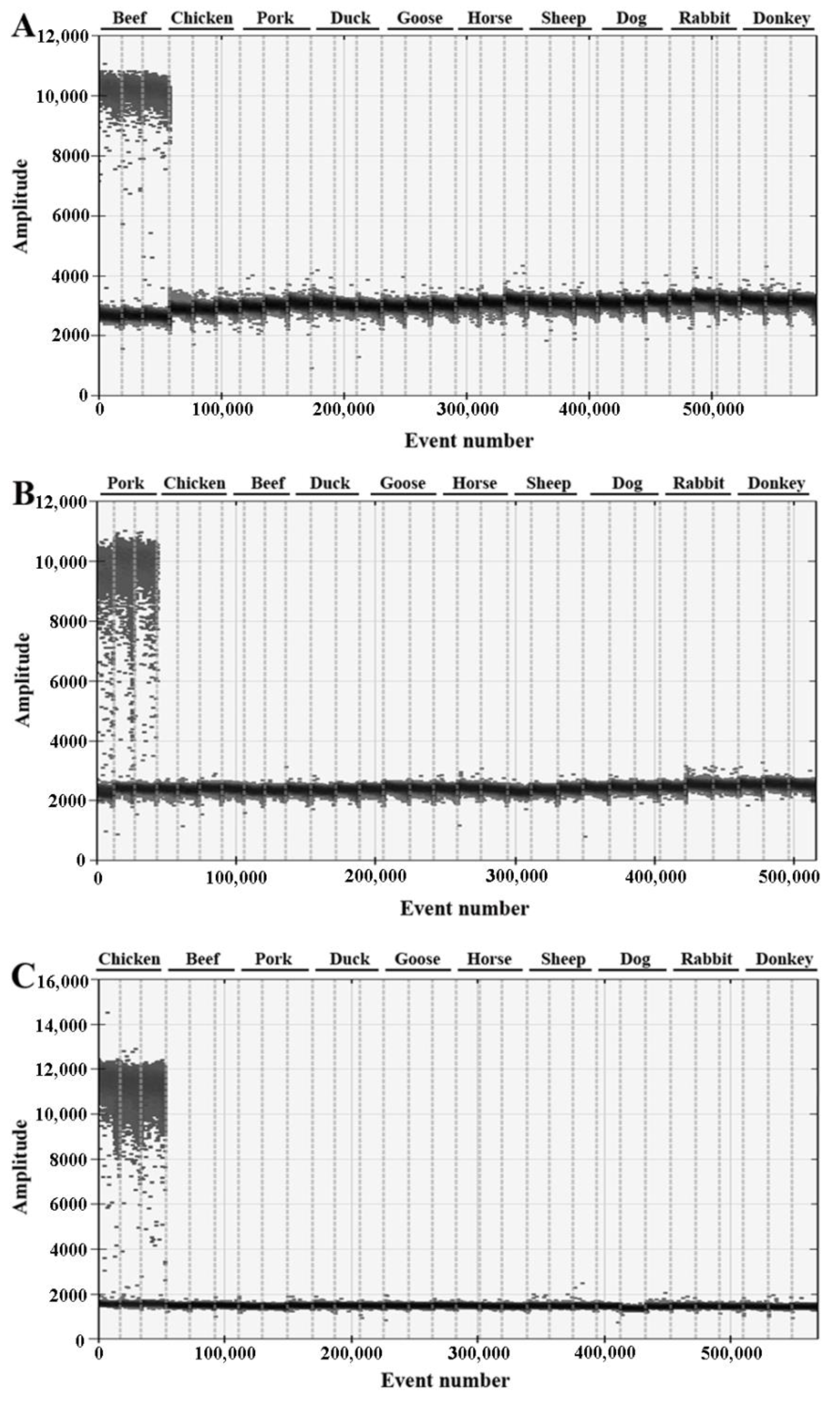
| Primer/Probe | Sequence (5′-3′) | Target Gene | Length |
|---|---|---|---|
| Beef-F | GTAGGTGCACAGTACGTTCTGAAG | ACTB [34] | 96 bp |
| Beef-R | GGCCAGACTGGGCACATG | ||
| Beef-P | HEX-CGGCACACTCGGCTGTGTTCCTTGC-BHQ1 | ||
| Pork F | GGAGTGTGTATCCCGTAGGTG | ACTB [34] | 103 bp |
| Pork R | CTGGGGACATGCAGAGAGTG | ||
| Pork-P | FAM-TCTGACGTGACTCCCCGACCTGG-BHQ1 | ||
| Chicken-F | GGCTGCAAGTCACCGTGGTA | TGFB3 [33] | 129 bp |
| Chicken-F | CCGCTAGCCAGAAGCTCAGC | ||
| Chicken-P | FAM-CAGGAGCCACGTGAGCAGCACAG-BHQ1 |
| Mass Proportion | Six Parallel Tests of Pork or Chicken (Copies/μL) | Six Parallel Tests of Beef (copies/μL) | K Value | K Mean (RSD) | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Pork | ||||||||||||||
| 10% | 29.7 | 31 | 30.3 | 31 | 32.2 | 31.7 | 332 | 350 | 345 | 344 | 355 | 361 | 1.25 | 1.19 (5.79%) |
| 30% | 80.3 | 71.1 | 76.7 | 77.8 | 81.2 | 87 | 201 | 190 | 189 | 205 | 223 | 233 | 1.12 | |
| 50% | 109 | 121 | 123 | 127 | 130 | 140 | 122 | 130 | 136 | 143 | 141 | 158 | 1.11 | |
| 70% | 120 | 120 | 121 | 122 | 123 | 134 | 62.8 | 64.5 | 64.6 | 65.3 | 64.2 | 71.1 | 1.24 | |
| 90% | 137 | 137 | 140 | 142 | 143 | 164 | 18.2 | 18.5 | 19.4 | 19.7 | 20.3 | 22 | 1.23 | |
| Chicken | ||||||||||||||
| 10% | 230 | 247 | 255 | 225 | 221 | 226 | 756 | 792 | 711 | 736 | 746 | 831 | 0.36 | 0.38 (6.32%) |
| 30% | 281 | 282 | 272 | 286 | 221 | 247 | 266 | 209 | 170 | 265 | 274 | 258 | 0.39 | |
| 50% | 375 | 392 | 377 | 401 | 384 | 394 | 130.1 | 146 | 121 | 144 | 142 | 129 | 0.35 | |
| 70% | 535 | 582 | 583 | 593 | 601 | 600 | 100 | 106 | 102 | 105 | 105 | 99 | 0.41 | |
| 90% | 581 | 591 | 596 | 632 | 638 | 591 | 23.8 | 22.5 | 24.8 | 24.3 | 29 | 28 | 0.38 | |
| Actual Value | Pork | Chicken | ||
|---|---|---|---|---|
| Measured Value (%) | Deviation | Measured Value (%) | Deviation | |
| 20% | 20.44 ± 0.30 | 2.18% | 18.60 ± 0.38 | −6.99% |
| 40% | 38.68 ± 0.49 | −3.29% | 41.49 ± 0.64 | 3.73% |
| 60% | 62.39 ± 0.28 | 3.98% | 60.08 ± 0.51 | 0.13% |
| 80% | 79.62 ± 0.48 | −0.48% | 80.15 ± 0.53 | −0.19% |
| Actual Value | Pork | Chicken | ||||
|---|---|---|---|---|---|---|
| Measured Value (%) | Deviation | Detection Rate | Measured Value (%) | Deviation | Detection Rate | |
| 0.01% | 0 | 100% | 0% | 0 | −100% | 0% |
| 0.05% | 0.01 ± 0.01 | 80.89% | 33.3% | 0 | −100% | 0% |
| 0.1% | 0.22 ± 0.21 | 115.72% | 100% | 0.17 ± 0.07 | −70.88% | 100% |
| 0.2% | 0.32 ± 0.02 | 58.28 % | 100% | 0.32 ± 0.13 | −57.89% | 100% |
| 0.5% | 0.68 ± 0.13 | 36.12% | 100% | 0.33 ± 0.13 | 33.99% | 100% |
| 0.8% | 0.58 ± 0.12 | −26.96% | 100% | 0.59 ± 0.15 | −25.89% | 100% |
| 1% | 1.13 ± 0.02 | 12.68 % | 100% | 0.98 ± 0.04 | −2.39 % | 100% |
| 5% | 5.66 ± 0.33 | 13.26% | 100% | 4.57 ± 0.16 | −8.52% | 100% |
| 10% | 9.68 ± 0.06 | −3.17% | 100% | 10.91 ± 0.01 | 9.13% | 100% |
| Actual Value | Treatment Time (min) | Pork | Chicken | ||
|---|---|---|---|---|---|
| Measured Value (%) | Deviation | Measured Value (%) | Deviation | ||
| 10% | 5 | 9.50 ± 0.36 | −4.97% | 9.22 ± 0.31 | −7.77% |
| 10 | 8.96 ± 0.33 | −10.36% | 9.00 ± 0.40 | −10.03% | |
| 20 | 8.39 ± 0.10 | −16.10% | 8.49 ± 0.04 | −15.10% | |
| 50% | 5 | 51.63 ± 0.62 | 3.26% | 51.87 ± 0.48 | 3.75% |
| 10 | 53.48 ± 0.77 | 6.97% | 52.79 ± 0.99 | 5.59% | |
| 20 | 53.99 ± 0.17 | 7.99% | 42.77 ± 5.26 | −14.46% | |
| Sample | Pork % (w/w) | Chicken % (w/w) | ||
|---|---|---|---|---|
| Declared Value | Measured Value | Declared Value | Measured Value | |
| Beef roll | 0 | 0 | 0 | 0 |
| Beef ball_1 | 0 | 0 | 0 | 0 |
| Beef ball_2 | 19.00% | 31.67% | 0 | 0 |
| Beef kebabs_1 | 0 | 0 | 0 | 0 |
| Beef kebabs_2 | 0 | 0 | 0 | 0 |
| Beefsteak | 0 | 0 | 0 | 0 |
| Minced beef | 0 | 0 | 0 | 65.90 |
| Beef stick | 0 | 0 | 0 | 0 |
| Beef slice | 0 | 0 | 0 | 0 |
| Veal sausage | 0 | 0 | 0 | 0 |
| Beef sausage | 0 | 0 | 49.40% | 50.25% |
| Canned beef | 0 | 0 | 0 | 0 |
| Dried beef jerky | LC | 17.47% | 0 | 0 |
| Dried beef floss | 0 | 0 | 0 | 0 |
| Cured beef | 0 | 0 | 0 | 0 |
| Bresaola | 0 | 0 | 0 | 0 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Xu, H.; Ma, X.; Ye, Z.; Yu, X.; Liu, G.; Wang, Z. A Droplet Digital PCR Based Approach for Identification and Quantification of Porcine and Chicken Derivatives in Beef. Foods 2022, 11, 3265. https://doi.org/10.3390/foods11203265
Xu H, Ma X, Ye Z, Yu X, Liu G, Wang Z. A Droplet Digital PCR Based Approach for Identification and Quantification of Porcine and Chicken Derivatives in Beef. Foods. 2022; 11(20):3265. https://doi.org/10.3390/foods11203265
Chicago/Turabian StyleXu, Huili, Xiaoyu Ma, Zihong Ye, Xiaoping Yu, Guangfu Liu, and Zhengliang Wang. 2022. "A Droplet Digital PCR Based Approach for Identification and Quantification of Porcine and Chicken Derivatives in Beef" Foods 11, no. 20: 3265. https://doi.org/10.3390/foods11203265
APA StyleXu, H., Ma, X., Ye, Z., Yu, X., Liu, G., & Wang, Z. (2022). A Droplet Digital PCR Based Approach for Identification and Quantification of Porcine and Chicken Derivatives in Beef. Foods, 11(20), 3265. https://doi.org/10.3390/foods11203265




