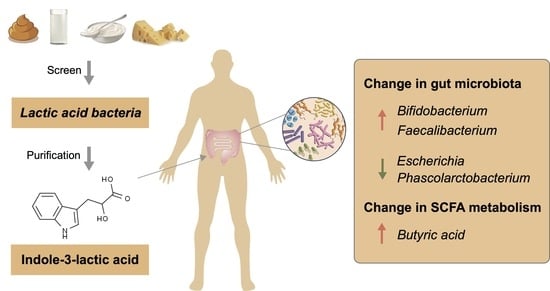
The Effect of Indole-3-Lactic Acid from Lactiplantibacillus plantarum ZJ316 on Human Intestinal Microbiota In Vitro
Abstract
1. Introduction
2. Materials and Methods
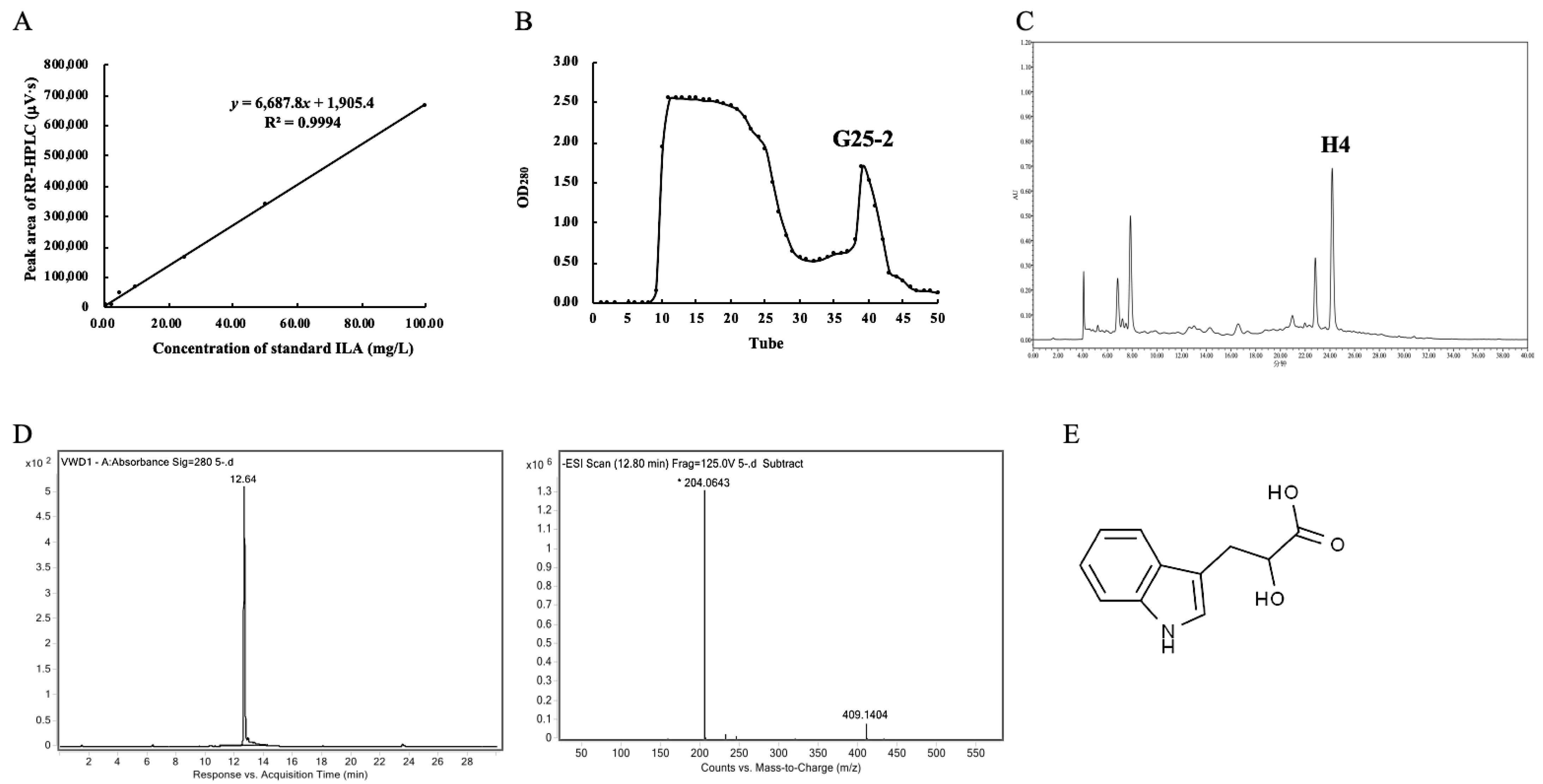
2.1. Determination of ILA Content
2.2. Purification of ILA from L. plantarum ZJ316
2.3. Identification of ILA
2.4. Antibacterial Activity of ILA
2.5. Intestinal Simulation Model In Vitro
2.6. 16S rRNA Sequencing
2.7. SCFA Concentrations by GC-MS
2.8. Statistical Analysis
3. Results
3.1. Screening of High ILA-Producing LAB Strains and ILA Purification
3.2. Antibacterial Activity and MICs of ILA
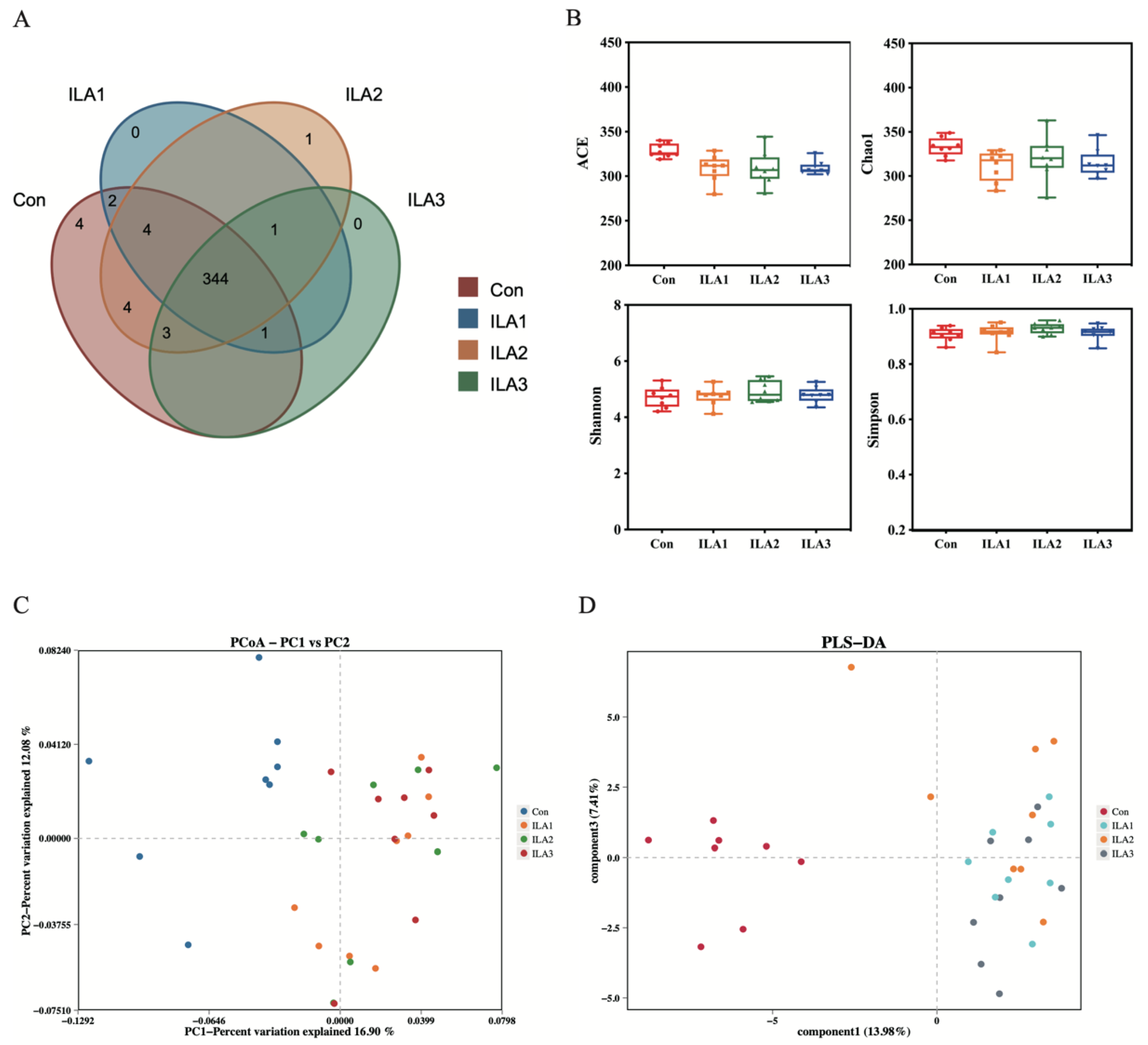
3.3. Diversity Analysis of Bacterial Communities
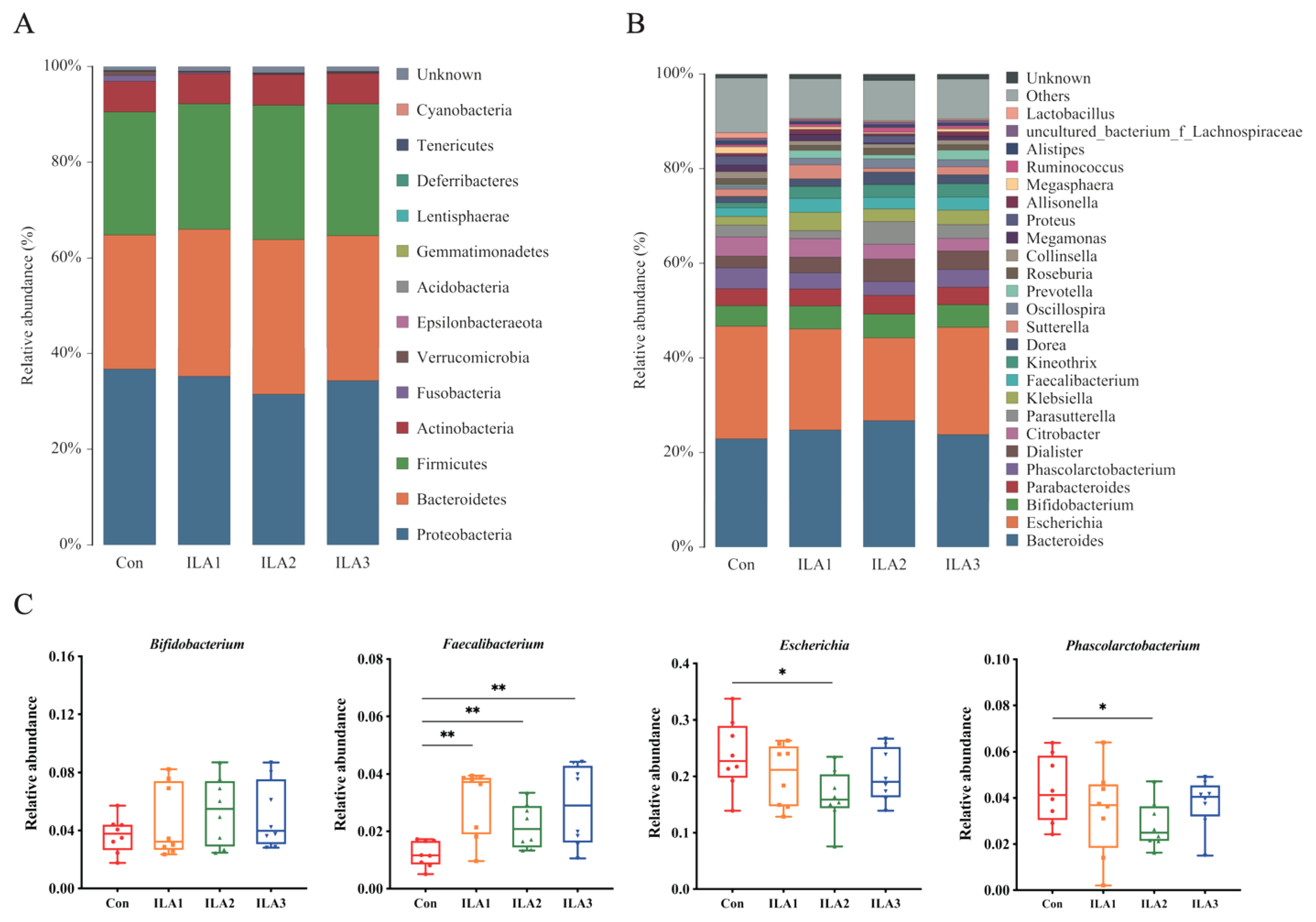
3.4. Relative Abundance of Bacterial Communities
3.5. Communities Difference by LEfSe Analysis
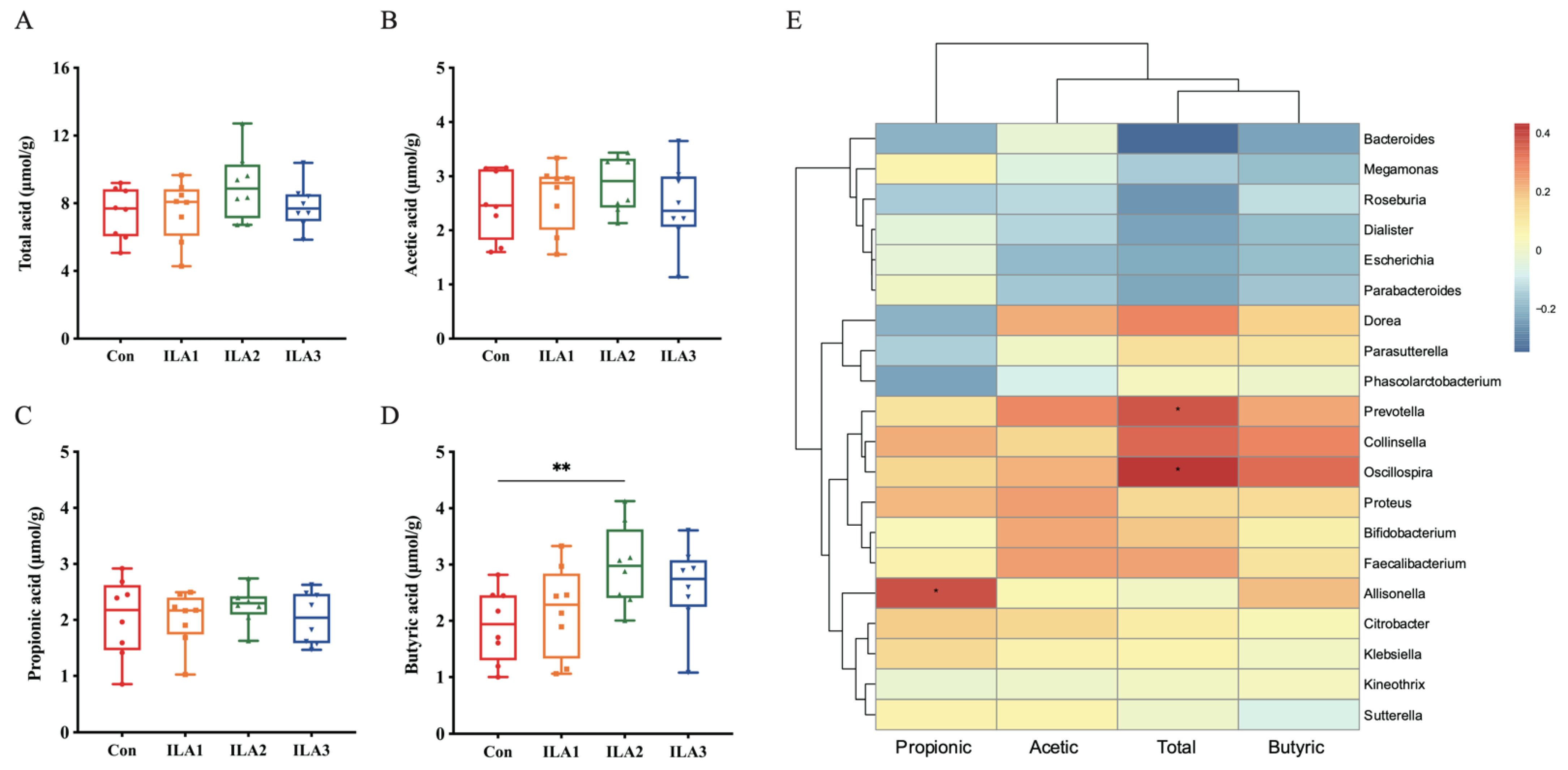
3.6. Correlation of SCFA Metabolism and Gut Microbiota
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Roager, H.M.; Licht, T.R. Microbial tryptophan catabolites in health and disease. Nat. Commun. 2018, 9, 3294. [Google Scholar] [CrossRef]
- Laursen, M.F.; Sakanaka, M.; von Burg, N.; Mörbe, U.; Andersen, D.; Moll, J.M.; Pekmez, C.T.; Rivollier, A.; Michaelsen, K.F.; Mølgaard, C.; et al. Bifidobacterium species associated with breastfeeding produce aromatic lactic acids in the infant gut. Nat. Microbiol. 2021, 6, 1367–1382. [Google Scholar] [CrossRef]
- Wilck, N.; Matus, M.G.; Kearney, S.M.; Olesen, S.W.; Forslund, K.; Bartolomaeus, H.; Haase, S.; Mahler, A.; Balogh, A.; Marko, L.; et al. Salt-responsive gut commensal modulates TH17 axis and disease. Nature 2017, 551, 585–589. [Google Scholar] [CrossRef]
- Nikolaus, S.; Schulte, B.; Al-Massad, N.; Thieme, F.; Schulte, D.M.; Bethge, J.; Rehman, A.; Tran, F.; Aden, K.; Häsler, R.; et al. Increased Tryptophan Metabolism Is Associated With Activity of Inflammatory Bowel Diseases. Gastroenterology 2017, 153, 1504–1516. [Google Scholar] [CrossRef]
- Alexeev, E.E.; Lanis, J.M.; Kao, D.J.; Campbell, E.L.; Kelly, C.J.; Battista, K.D.; Gerich, M.E.; Jenkins, B.R.; Walk, S.T.; Kominsky, D.J.; et al. Microbiota-Derived Indole Metabolites Promote Human and Murine Intestinal Homeostasis through Regulation of Interleukin-10 Receptor. Am. J. Pathol. 2018, 188, 1183–1194. [Google Scholar] [CrossRef]
- Whitfield-Cargile, C.M.; Cohen, N.D.; Chapkin, R.S.; Weeks, B.R.; Davidson, L.A.; Goldsby, J.S.; Hunt, C.L.; Steinmeyer, S.H.; Menon, R.; Suchodolski, J.S.; et al. The microbiota-derived metabolite indole decreases mucosal inflammation and injury in a murine model of NSAID enteropathy. Gut Microbes 2016, 7, 246–261. [Google Scholar] [CrossRef]
- Yang, W.; Cong, Y. Gut microbiota-derived metabolites in the regulation of host immune responses and immune-related inflammatory diseases. Cell Mol. Immunol. 2021, 18, 866–877. [Google Scholar] [CrossRef]
- Venkatesh, M.; Mukherjee, S.; Wang, H.; Li, H.; Sun, K.; Benechet, A.P.; Qiu, Z.; Maher, L.; Redinbo, M.R.; Phillips, R.S.; et al. Symbiotic bacterial metabolites regulate gastrointestinal barrier function via the xenobiotic sensor PXR and Toll-like receptor 4. Immunity 2014, 41, 296–310. [Google Scholar] [CrossRef]
- Sakurai, T.; Odamaki, T.; Xiao, J.Z. Production of Indole-3-Lactic Acid by Bifidobacterium Strains Isolated fromHuman Infants. Microorganisms 2019, 7, 340. [Google Scholar] [CrossRef]
- Suzuki, Y.; Kosaka, M.; Shindo, K.; Kawasumi, T.; Kimoto-Nira, H.; Suzuki, C. Identification of antioxidants produced by Lactobacillus plantarum. Biosci. Biotechnol. Biochem. 2013, 77, 1299–1302. [Google Scholar] [CrossRef]
- Ménard, S.; Candalh, C.; Bambou, J.C.; Terpend, K.; Cerf-Bensussan, N.; Heyman, M. Lactic acid bacteria secrete metabolites retaining anti-inflammatory properties after intestinal transport. Gut 2004, 53, 821–828. [Google Scholar] [CrossRef]
- Dominguez-Bello, M.G.; Costello, E.K.; Contreras, M.; Magris, M.; Hidalgo, G.; Fierer, N.; Knight, R. Delivery mode shapes the acquisition and structure of the initial microbiota across multiple body habitats in newborns. Proc. Natl. Acad. Sci. USA 2010, 107, 11971–11975. [Google Scholar] [CrossRef]
- Burgel, P.H.; Marina, C.L.; Saavedra, P.H.V.; Albuquerque, P.; de Oliveira, S.A.M.; Veloso Janior, P.H.H.; de Castro, R.A.; Heyman, H.M.; Coelho, C.; Cordero, R.J.B.; et al. Cryptococcus neoformans Secretes Small Molecules That Inhibit IL-1beta Inflammasome-Dependent Secretion. Mediat. Inflamm. 2020, 2020, 3412763. [Google Scholar] [CrossRef]
- Xia, J.; Jiang, S.; Lv, L.; Wu, W.; Wang, Q.; Xu, Q.; Ye, J.; Fang, D.; Li, Y.; Wu, J.; et al. Modulation of the immune response and metabolism in germ-free rats colonized by the probiotic Lactobacillus salivarius LI01. Appl. Microbiol. Biotechnol. 2021, 105, 1629–1645. [Google Scholar] [CrossRef]
- Walker, W.A.; Meng, D. Breast Milk and Microbiota in the Premature Gut: A Method of Preventing Necrotizing Enterocolitis. Nestle Nutr. Inst. Workshop Ser. 2020, 94, 103–112. [Google Scholar] [CrossRef]
- Lei, C.; Mu, J.; Teng, Y.; He, L.; Xu, F.; Zhang, X.; Sundaram, K.; Kumar, A.; Sriwastva, M.K.; Lawrenz, M.B.; et al. Lemon Exosome-like Nanoparticles-Manipulated Probiotics Protect Mice from C. d iff Infection. iScience 2020, 23, 101571. [Google Scholar] [CrossRef]
- Tremaroli, V.; Bäckhed, F. Functional interactions between the gut microbiota and host metabolism. Nature 2012, 489, 242–249. [Google Scholar] [CrossRef]
- Caruso, R.; Lo, B.C.; Núñez, G. Host–microbiota interactions in inflammatory bowel disease. Nat. Rev. Immunol. 2020, 20, 411–426. [Google Scholar] [CrossRef]
- Zhu, Y.; Zhou, Q.; Li, P.; Gu, Q. Purification, characterization, and mode of action of Paracin 54, a novel bacteriocin against Staphylococci. Appl. Microbiol. Biotechnol. 2021, 105, 6735–6748. [Google Scholar] [CrossRef]
- Li, P.; Zhou, Q.; Gu, Q. Complete genome sequence of Lactobacillus plantarum LZ227, a potential probiotic strain producing B-group vitamins. J. Biotechnol. 2016, 234, 66–70. [Google Scholar] [CrossRef]
- Li, X.; Gu, Q.; Lou, X.; Zhang, X.; Song, D.; Shen, L.; Zhao, Y. Complete genome sequence of the probiotic Lactobacillus plantarum strain ZJ316. Genome Announc. 2013, 1, e0009413. [Google Scholar] [CrossRef]
- Wan, C.; Qian, W.W.; Liu, W.; Pi, X.; Tang, M.T.; Wang, X.L.; Gu, Q.; Li, P.; Zhou, T. Exopolysaccharide from Lactobacillus rhamnosus ZFM231 alleviates DSS-induced colitis in mice by regulating gut microbiota. J. Sci. Food Agric. 2022. [Google Scholar] [CrossRef]
- Zhou, Q.; Gu, R.; Li, P.; Lu, Y.; Chen, L.; Gu, Q. Anti-Salmonella mode of action of natural L-phenyl lactic acid purified from Lactobacillus plantarum ZJ316. Appl. Microbiol. Biotechnol. 2020, 104, 5283–5292. [Google Scholar] [CrossRef]
- Tong, Z.; Zhang, Y.; Ling, J.; Ma, J.; Huang, L.; Zhang, L. An in vitro study on the effects of nisin on the antibacterial activities of 18 antibiotics against Enterococcus faecalis. PLoS ONE 2014, 9, e89209. [Google Scholar] [CrossRef]
- Li, P.; Yao, X.; Zhou, Q.; Meng, X.; Zhou, T.; Gu, Q. Citrus Peel Flavonoid Extracts: Health-Beneficial Bioactivities and Regulation of Intestinal Microecology in vitro. Front. Nutr. 2022, 9, 888745. [Google Scholar] [CrossRef]
- Tidjani Alou, M.; Naud, S.; Khelaifia, S.; Bonnet, M.; Lagier, J.C.; Raoult, D. State of the Art in the Culture of the Human Microbiota: New Interests and Strategies. Clin. Microbiol. Rev. 2020, 34, e00129-19. [Google Scholar] [CrossRef]
- Lin, R.; Liu, W.; Piao, M.; Zhu, H. A review of the relationship between the gut microbiota and amino acid metabolism. Amino Acids 2017, 49, 2083–2090. [Google Scholar] [CrossRef]
- Huang, W.; Cho, K.Y.; Meng, D.; Walker, W.A. The impact of indole-3-lactic acid on immature intestinal innate immunity and development: A transcriptomic analysis. Sci. Rep. 2021, 11, 8088. [Google Scholar] [CrossRef]
- Meng, D.; Sommella, E.; Salviati, E.; Campiglia, P.; Ganguli, K.; Djebali, K.; Zhu, W.; Walker, W.A. Indole-3-lactic acid, a metabolite of tryptophan, secreted by Bifidobacterium longum subspecies infantis is anti-inflammatory in the immature intestine. Pediatr. Res. 2020, 88, 209–217. [Google Scholar] [CrossRef]
- Guimaraes, A.; Venancio, A.; Abrunhosa, L. Antifungal effect of organic acids from lactic acid bacteria on Penicillium nordicum. Food Addit. Contam. Part A Chem. Anal. Control. Expo. Risk Assess. 2018, 35, 1803–1818. [Google Scholar] [CrossRef]
- Lee, J.H.; Lee, J. Indole as an intercellular signal in microbial communities. FEMS Microbiol. Rev. 2010, 34, 426–444. [Google Scholar] [CrossRef]
- Honoré, A.H.; Aunsbjerg, S.D.; Ebrahimi, P.; Thorsen, M.; Benfeldt, C.; Knøchel, S.; Skov, T. Metabolic footprinting for investigation of antifungal properties of Lactobacillus paracasei. Anal. Bioanal. Chem. 2016, 408, 83–96. [Google Scholar] [CrossRef]
- Narayanan, T.K.; Rao, G.R. Beta-indoleethanol and beta-indolelactic acid production by Candida species: Their antibacterial and autoantibiotic action. Antimicrob. Agents Chemother. 1976, 9, 375–380. [Google Scholar] [CrossRef]
- Yang, W.H.; Heithoff, D.M.; Aziz, P.V.; Sperandio, M.; Nizet, V.; Mahan, M.J.; Marth, J.D. Recurrent infection progressively disables host protection against intestinal inflammation. Science 2017, 358, eaao5610. [Google Scholar] [CrossRef]
- Sivan, A.; Corrales, L.; Hubert, N.; Williams, J.B.; Aquino-Michaels, K.; Earley, Z.M.; Benyamin, F.W.; Lei, Y.M.; Jabri, B.; Alegre, M.L.; et al. Commensal Bifidobacterium promotes antitumor immunity and facilitates anti-PD-L1 efficacy. Science 2015, 350, 1084–1089. [Google Scholar] [CrossRef]
- Collinson, S.; Deans, A.; Padua-Zamora, A.; Gregorio, G.V.; Li, C.; Dans, L.F.; Allen, S.J. Probiotics for treating acute infectious diarrhoea. Cochrane Database Syst. Rev. 2020, 12, Cd003048. [Google Scholar] [CrossRef]
- Enomoto, T.; Sowa, M.; Nishimori, K.; Shimazu, S.; Yoshida, A.; Yamada, K.; Furukawa, F.; Nakagawa, T.; Yanagisawa, N.; Iwabuchi, N.; et al. Effects of bifidobacterial supplementation to pregnant women and infants in the prevention of allergy development in infants and on fecal microbiota. Allergol. Int. 2014, 63, 575–585. [Google Scholar] [CrossRef]
- Saez-Lara, M.J.; Gomez-Llorente, C.; Plaza-Diaz, J.; Gil, A. The role of probiotic lactic acid bacteria and bifidobacteria in the prevention and treatment of inflammatory bowel disease and other related diseases: A systematic review of randomized human clinical trials. Biomed. Res. Int. 2015, 2015, 505878. [Google Scholar] [CrossRef]
- Ehrlich, A.M.; Pacheco, A.R.; Henrick, B.M.; Taft, D.; Xu, G.; Huda, M.N.; Mishchuk, D.; Goodson, M.L.; Slupsky, C.; Barile, D.; et al. Indole-3-lactic acid associated with Bifidobacterium-dominated microbiota significantly decreases inflammation in intestinal epithelial cells. BMC Microbiol. 2020, 20, 357. [Google Scholar] [CrossRef]
- Falony, G.; Joossens, M.; Vieira-Silva, S.; Wang, J.; Darzi, Y.; Faust, K.; Kurilshikov, A.; Bonder, M.J.; Valles-Colomer, M.; Vandeputte, D.; et al. Population-level analysis of gut microbiome variation. Science 2016, 352, 560–564. [Google Scholar] [CrossRef]
- Lopez-Siles, M.; Duncan, S.H.; Garcia-Gil, L.J.; Martinez-Medina, M. Faecalibacterium prausnitzii: From microbiology to diagnostics and prognostics. ISME J. 2017, 11, 841–852. [Google Scholar] [CrossRef] [PubMed]
- O’Toole, P.W.; Marchesi, J.R.; Hill, C. Next-generation probiotics: The spectrum from probiotics to live biotherapeutics. Nat. Microbiol. 2017, 2, 17057. [Google Scholar] [CrossRef] [PubMed]
- Bao, R.; Hesser, L.A.; He, Z.; Zhou, X.; Nadeau, K.C.; Nagler, C.R. Fecal microbiome and metabolome differ in healthy and food-allergic twins. J. Clin. Investig. 2021, 131, e141935. [Google Scholar] [CrossRef] [PubMed]
- Chen, H.; Fink, G.R. Feedback control of morphogenesis in fungi by aromatic alcohols. Genes Dev. 2006, 20, 1150–1161. [Google Scholar] [CrossRef] [PubMed]
- Elleuch, L.; Shaaban, M.; Smaoui, S.; Mellouli, L.; Karray-Rebai, I.; Fourati-Ben Fguira, L.; Shaaban, K.A.; Laatsch, H. Bioactive secondary metabolites from a new terrestrial Streptomyces sp. TN262. Appl. Biochem. Biotechnol. 2010, 162, 579–593. [Google Scholar] [CrossRef] [PubMed]
- Sivaprakasam, S.; Prasad, P.D.; Singh, N. Benefits of short-chain fatty acids and their receptors in inflammation and carcinogenesis. Pharmacol. Ther. 2016, 164, 144–151. [Google Scholar] [CrossRef]
- Dalile, B.; Van Oudenhove, L.; Vervliet, B.; Verbeke, K. The role of short-chain fatty acids in microbiota-gut-brain communication. Nat. Rev. Gastroenterol. Hepatol. 2019, 16, 461–478. [Google Scholar] [CrossRef]
- Stoeva, M.K.; Garcia-So, J.; Justice, N.; Myers, J.; Tyagi, S.; Nemchek, M.; McMurdie, P.J.; Kolterman, O.; Eid, J. Butyrate-producing human gut symbiont, Clostridium butyricum, and its role in health and disease. Gut Microbes 2021, 13, 1907272. [Google Scholar] [CrossRef]

| Lactic Acid Bacteria | Sources | Culture Condition | Retention Time (min) | ILA Content (mg/L) |
|---|---|---|---|---|
| Lactiplantibacillus plantarum ZFM9 | Healthy newborn feces | 37 °C, MRS | 22.720 | 25.08 ± 1.13 |
| Lactiplantibacillus plantarum ZJ316 | 37 °C, MRS | 22.765 | 43.14 ± 1.02 | |
| Lactiplantibacillus plantarum ZFM55 | 37 °C, MRS | 22.734 | 30.89 ± 0.57 | |
| Lactiplantibacillus plantarum LZ206 | 37 °C, MRS | 22.699 | 22.31 ± 0.81 | |
| Lacticaseibacillus paracasei ZFM54 | 37 °C, MRS | 22.743 | 14.58 ± 0.25 | |
| Lactiplantibacillus plantarum LZ227 | Fresh milk | 37 °C, MRS | 22.697 | 12.55 ± 0.24 |
| Lactilactobacillus sakei ZFM225 | 37 °C, MRS | 22.700 | 6.39 ± 0.75 | |
| Lactilactobacillus sakei ZFM220 | 37 °C, MRS | 22.727 | 12.92 ± 0.76 | |
| Lacticaseibacillus rhamnosus ZFM231 | 37 °C, MRS | / | / | |
| Lacticaseibacillus rhamnosus ZFM202 | 37 °C, MRS | / | / | |
| Lactiplantibacillus plantarum ZFM806 | Cheese | 37 °C, MRS | 22.719 | 36.35 ± 1.37 |
| Leuconostoc mesenteroides ZFM802 | 37 °C, MRS | 22.712 | 7.50 ± 0.02 | |
| Limosilactobacillus fermentum ZFM001 | Yogurt | 37 °C, MRS | 22.685 | 6.56 ± 0.35 |
| Lacticaseibacillus rhamnosus GG | Commercial strain | 37 °C, MRS | / | / |
| Indicator Bacteria | Sources | Culture Condition | Inhibition Zone Diameter (mm) | MIC (mg/mL) | |
|---|---|---|---|---|---|
| G− | Salmonella paratyphi-B CMCC 50094 | CMCC | 37 °C, LB | 11.24 ± 0.35 | 1.6 |
| Salmonella paratyphi-A CMCC 50093 | CMCC | 37 °C, LB | 12.89 ± 0.28 | 0.8 | |
| Salmonella enterica subsp. arizonae CMCC(B)47001 | CMCC | 37 °C, LB | 11.36 ± 0.26 | 1.6 | |
| Salmonella enterica subsp. enterica ATCC 14028 | ATCC | 37 °C, LB | 12.35 ± 0.44 | 1.6 | |
| Salmonella choleraesuis ATCC 13312 | ATCC | 37 °C, LB | 11.51 ± 0.34 | 1.6 | |
| Salmonella typhimurium CMCC 50015 | CMCC | 37 °C, LB | 12.28 ± 0.40 | 1.6 | |
| Escherichia coli DH5α | CMCC | 37 °C, LB | 12.50 ± 0.17 | 1.6 | |
| Pseudomonas aeruginosa ATCC 47085 | ATCC | 37 °C, LB | 10.66 ± 0.08 | 3.2 | |
| G+ | Staphylococcus aureus D48 | Gift from Eefjan Breukink, Utrecht University, The Netherlands | 37 °C, LB | 11.93 ± 0.47 | 0.8 |
| Staphylococcus warneri | 37 °C, LB | 20.25 ± 0.50 | 0.4 | ||
| Staphylococcus carnosus pCA 44 | 37 °C, LB | 12.93 ± 0.40 | 1.6 | ||
| Staphylococcus carnosus pot 20 | 37 °C, LB | 14.73 ± 0.53 | 0.8 | ||
| Staphylococcus simulans | 37 °C, LB | 15.13 ± 1.15 | 0.4 | ||
| Micrococcus luteus CICC 10209 | CICC | 30 °C, TSB | 19.06 ± 0.76 | 0.4 | |
| Listeria monocytogenes ATCC 19111 | ATCC | 37 °C, BHI | 10.95 ± 0.07 | 3.2 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhou, Q.; Xie, Z.; Wu, D.; Liu, L.; Shi, Y.; Li, P.; Gu, Q. The Effect of Indole-3-Lactic Acid from Lactiplantibacillus plantarum ZJ316 on Human Intestinal Microbiota In Vitro. Foods 2022, 11, 3302. https://doi.org/10.3390/foods11203302
Zhou Q, Xie Z, Wu D, Liu L, Shi Y, Li P, Gu Q. The Effect of Indole-3-Lactic Acid from Lactiplantibacillus plantarum ZJ316 on Human Intestinal Microbiota In Vitro. Foods. 2022; 11(20):3302. https://doi.org/10.3390/foods11203302
Chicago/Turabian StyleZhou, Qingqing, Zuorui Xie, Danli Wu, Lingli Liu, Yongqing Shi, Ping Li, and Qing Gu. 2022. "The Effect of Indole-3-Lactic Acid from Lactiplantibacillus plantarum ZJ316 on Human Intestinal Microbiota In Vitro" Foods 11, no. 20: 3302. https://doi.org/10.3390/foods11203302
APA StyleZhou, Q., Xie, Z., Wu, D., Liu, L., Shi, Y., Li, P., & Gu, Q. (2022). The Effect of Indole-3-Lactic Acid from Lactiplantibacillus plantarum ZJ316 on Human Intestinal Microbiota In Vitro. Foods, 11(20), 3302. https://doi.org/10.3390/foods11203302