Metabolic Variations in Brown Rice Fertilised with Different Levels of Nitrogen
Abstract
:1. Introduction
2. Materials and Methods
2.1. Rice Samples
2.2. Sample Preparation and Extraction for Non-Targeted Metabolomics Analysis
2.3. Antioxidant Activity Assay
2.3.1. Determination of DPPH Radical Scavenging Activity
2.3.2. Determination of Hydroxyl Radical Scavenging Activity
2.4. KEGG Pathway-Enrichment Analysis
2.5. Data Analysis
3. Results
3.1. Non-Targeted Metabolomics Analysis in Brown Rice
3.2. Multivariate Analysis of Differentially Expressed Metabolites
3.3. Metabolomic Changes of Brown Rice with Different Nitrogen Amounts
3.4. KEGG Classification and Enrichment Analysis
3.5. Antioxidant Activity
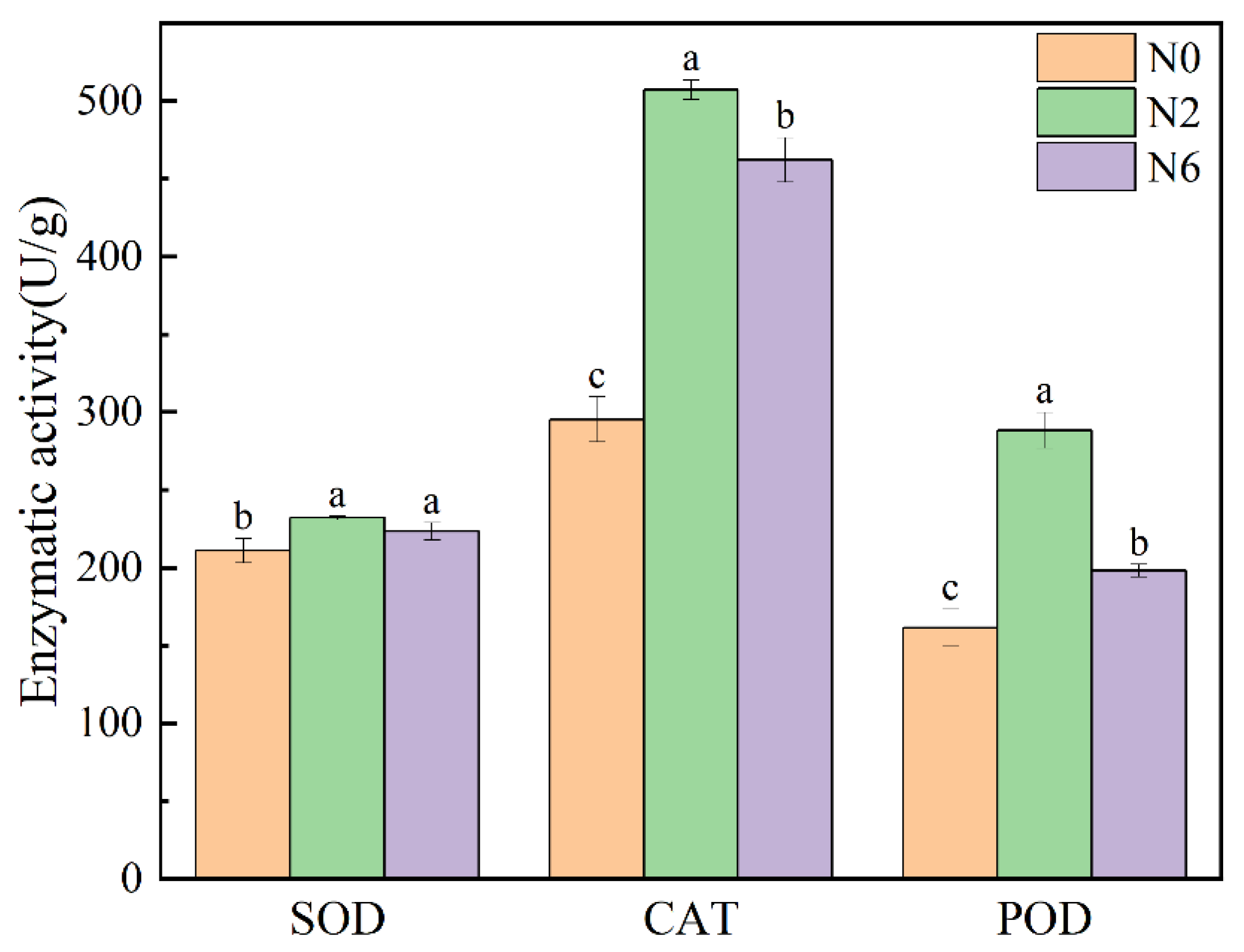
3.6. Enzymatic Activity
4. Discussion
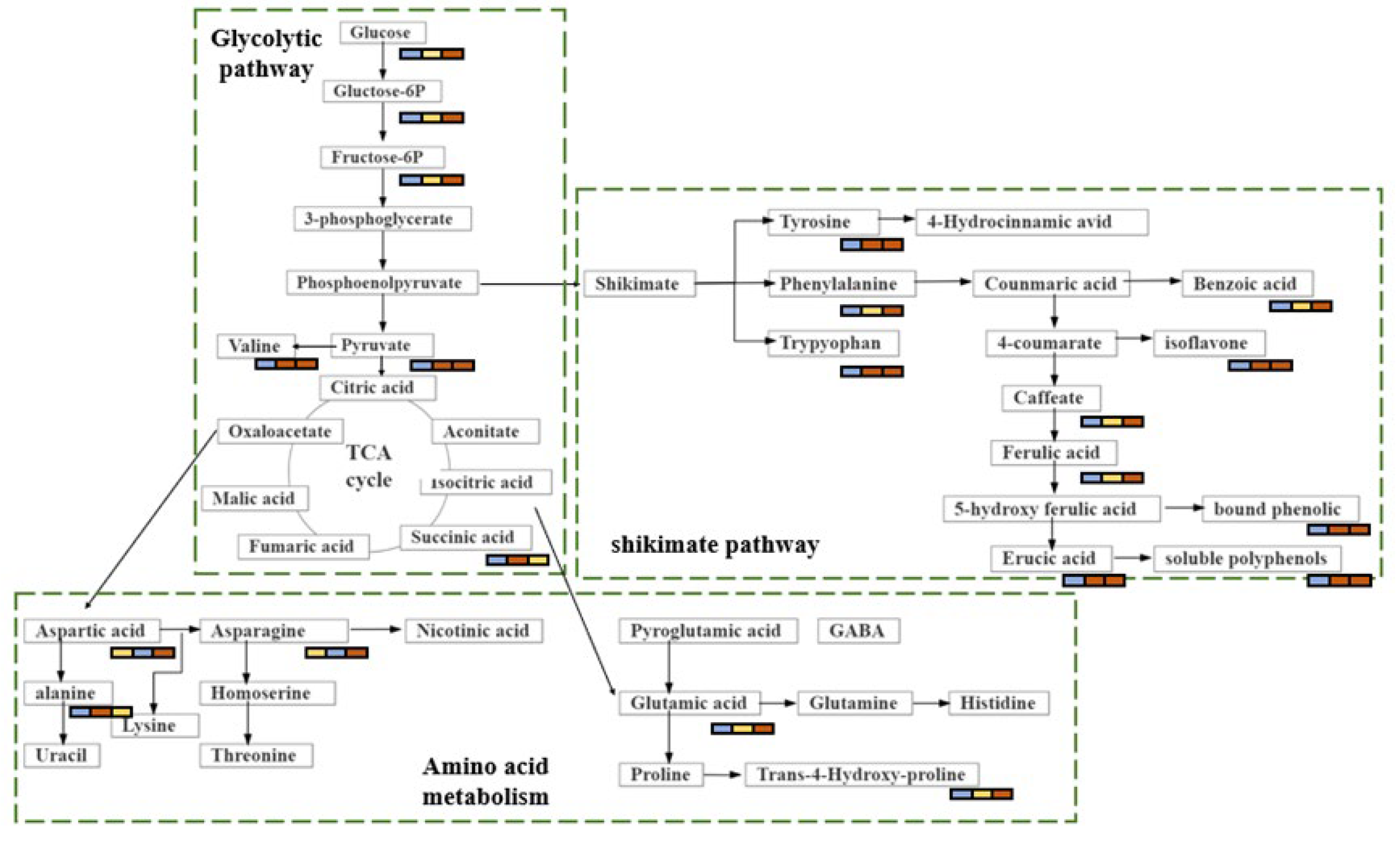
4.1. Carbohydrate Metabolism
4.2. Amino Acid Metabolism
4.3. Shikimate-Phenylpropanoid-Flavonoid Metabolism
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Liang, H.; Tao, D.; Zhang, Q.; Zhang, S.; Wang, J.; Liu, L.; Wu, Z.; Sun, W. Nitrogen fertilizer application rate impacts eating and cooking quality of rice after storage. PLoS ONE 2021, 16, e0253189. [Google Scholar] [CrossRef] [PubMed]
- Zhen, S.; Zhou, J.; Deng, X.; Zhu, G.; Cao, H.; Wang, Z.; Yan, Y. Metabolite profiling of the response to high-nitrogen fertilizer during grain development of bread wheat (Triticum aestivum L.). J. Cereal Sci. 2016, 69, 85–94. [Google Scholar] [CrossRef]
- Yilan, X.U.; Aibin, F.U.; Liu, T. Effects of Different Nitrogen Application Patterns on Dry Matter Accumulation and Yield of Rice in Double Cropping Rice Field. Crop Res. 2019, 20, 77–83. [Google Scholar]
- Alves, G.H.; Ferreira, C.D.; Vivian, P.G.; Monks, J.L.F.; Elias, M.C.; Vanier, N.L.; de Oliveira, M. The revisited levels of free and bound phenolics in rice: Effects of the extraction procedure. Food Chem. 2016, 208, 116–123. [Google Scholar] [CrossRef] [PubMed]
- Kim, S.-H.; Yang, Y.-J.; Chung, I.-M. The Effect of Degree of Milling on the Nutraceutical Content in Ecofriendly and Conventional Rice (Oryza sativa L.). Foods 2020, 9, 1297. [Google Scholar] [CrossRef] [PubMed]
- Saleh, A.S.M.; Wang, P.; Wang, N.; Yang, L.; Xiao, Z. Brown Rice Versus White Rice: Nutritional Quality, Potential Health Benefits, Development of Food Products, and Preservation Technologies. Compr. Rev. Food Sci. Food Saf. 2019, 18, 1070–1096. [Google Scholar] [CrossRef] [Green Version]
- Chen, M.; Huang, W.; Yin, Z.; Zhang, W.; Kong, Q.; Wu, S.; Li, W.; Bai, Z.; Fernie, A.R.; Huang, X.; et al. Environmentally-driven metabolite and lipid variations correspond to altered bioactivities of black wolfberry fruit. Food Chem. 2022, 372, 131342. [Google Scholar] [CrossRef]
- Fàbregas, N.; Fernie, A.R. The metabolic response to drought. J. Exp. Bot. 2019, 70, 1077–1085. [Google Scholar] [CrossRef] [Green Version]
- Ti, H.; Li, Q.; Zhang, R.; Zhang, M.; Deng, Y.; Wei, Z.; Chi, J.; Zhang, Y. Free and bound phenolic profiles and antioxidant activity of milled fractions of different indica rice varieties cultivated in southern China. Food Chem. 2014, 159, 166–174. [Google Scholar] [CrossRef]
- Qiu, Y.; Liu, Q.; Beta, T. Antioxidant properties of commercial wild rice and analysis of soluble and insoluble phenolic acids. Food Chem. 2010, 121, 140–147. [Google Scholar] [CrossRef]
- Liu, Y.; Kong, Z.; Liu, J.; Zhang, P.; Wang, Q.; Huan, X.; Li, L.; Qin, P. Non-targeted metabolomics of quinoa seed filling period based on liquid chromatography-mass spectrometry. Food Res. Int. 2020, 137, 109743. [Google Scholar] [CrossRef] [PubMed]
- Gong, E.S.; Luo, S.J.; Li, T.; Liu, C.M.; Zhang, G.W.; Chen, J.; Zeng, Z.C.; Liu, R.H. Phytochemical profiles and antioxidant activity of brown rice varieties. Food Chem. 2017, 227, 432–443. [Google Scholar] [CrossRef] [PubMed]
- Mbanjo, E.G.N.; Kretzschmar, T.; Jones, H.; Ereful, N.; Blanchard, C.; Boyd, L.A.; Sreenivasulu, N. The Genetic Basis and Nutritional Benefits of Pigmented Rice Grain. Front. Genet. 2020, 11, 229. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gao, Y.; Guo, X.; Liu, Y.; Zhang, M.; Zhang, R.; Abbasie, A.M.; You, L.; Li, T.; Liu, R.H. Comparative assessment of phytochemical profile, antioxidant capacity and anti-proliferative activity in different varieties of brown rice (Oryza sativa L.). LWT-Food Sci. Technol. 2018, 96, 19–25. [Google Scholar] [CrossRef]
- Lin, P.-Y.; Li, S.-C.; Lin, H.-P.; Shih, C.-K. Germinated brown rice combined with Lactobacillus acidophilus and Bifidobacterium animalis subsp. lactis inhibits colorectal carcinogenesis in rats. Food Sci. Nutr. 2019, 7, 216–224. [Google Scholar] [CrossRef] [Green Version]
- Quagliariello, V.; Iaffaioli, R.; Falcone, M.; Ferrari, G.; Pataro, G.; Donsì, F. Effect of pulsed electric fields—Assisted extraction on anti-inflammatory and cytotoxic activity of brown rice bioactive compounds. Food Res. Int. 2016, 87, 115–124. [Google Scholar] [CrossRef]
- Arbona, V.; Manzi, M.; De Ollas, C.; Gómez-Cadenas, A. Metabolomics as a Tool to Investigate Abiotic Stress Tolerance in Plants. Int. J. Mol. Sci. 2013, 14, 4885–4911. [Google Scholar] [CrossRef]
- Shen, T.; Xiong, Q.; Zhong, L.; Shi, X.; Cao, C.; He, H.; Chen, X. Analysis of main metabolisms during nitrogen deficiency and compensation in rice. Acta Physiol. Plant. 2019, 41, 68. [Google Scholar] [CrossRef]
- Glaubitz, U.; Erban, A.; Kopka, J.; Hincha, D.K.; Zuther, E. Metabolite Profiling Reveals Sensitivity-Dependent Metabolic Shifts in Rice (Oryza sativa L.) Cultivars under High Night Temperature Stress. Procedia Environ. Sci. 2015, 29, 72. [Google Scholar] [CrossRef] [Green Version]
- Desta, K.T.; Hur, O.S.; Lee, S.; Yoon, H.; Shin, M.-J.; Yi, J.; Lee, Y.; Ro, N.Y.; Wang, X.; Choi, Y.-M. Origin and seed coat color differently affect the concentrations of metabolites and antioxidant activities in soybean (Glycine max (L.) Merrill) seeds. Food Chem. 2022, 381, 132249. [Google Scholar] [CrossRef]
- Dare, A.P.; Günther; Grey, A.; Guo, G.; Demarais, N.; Cordiner, S.; McGhie, T.; Boldingh, H.; Hunt, M.; Deng, C.; et al. Resolving the developmental distribution patterns of polyphenols and related primary metabolites in bilberry (Vaccinium myrtillus) fruit. Food Chem. 2022, 374, 131703. [Google Scholar] [CrossRef] [PubMed]
- Campos, R.I. Studies of Wound-Induced Phenylpropanoid Metabolism in Fresh-Cut Romaine Lettuce and Its Modification by Heat Shock; University of California: Davis, CA, USA, 2001. [Google Scholar]
- Liu, Y.; Liu, J.; Liu, M.; Liu, Y.; Strappe, P.; Sun, H.; Zhou, Z. Comparative non-targeted metabolomic analysis reveals insights into the mechanism of rice yellowing. Food Chem. 2020, 308, 125621. [Google Scholar] [CrossRef] [PubMed]
- Adom, K.K.; Sorrells, A.M.E.; Liu, R.H. Phytochemicals and Antioxidant Activity of Milled Fractions of Different Wheat Varieties. J. Agric. Food Chem. 2005, 53, 2297–2306. [Google Scholar] [CrossRef] [PubMed]
- De Guzman, M.K.; Parween, S.; Butardo, V.M.; Alhambra, C.M.; Anacleto, R.; Seiler, C.; Bird, A.R.; Chow, C.-P.; Sreenivasulu, N. Investigating glycemic potential of rice by unraveling compositional variations in mature grain and starch mobilization patterns during seed germination. Sci. Rep. 2017, 7, 5854. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, H.; Shao, Y.; Bao, J.; Beta, T. Phenolic compounds and antioxidant properties of breeding lines between the white and black rice. Food Chem. 2015, 172, 630–639. [Google Scholar] [CrossRef] [PubMed]
- Miller, S.B.; Heuberger, A.L.; Broeckling, C.D.; Jahn, C.E. Non-Targeted Metabolomics Reveals Sorghum Rhizosphere-Associated Exudates are Influenced by the Belowground Interaction of Substrate and Sorghum Genotype. Int. J. Mol. Sci. 2019, 20, 431. [Google Scholar] [CrossRef] [Green Version]
- Zhao, C.-N.; Zhang, J.-J.; Li, Y.; Meng, X.; Li, H.-B. Microwave-Assisted Extraction of Phenolic Compounds from Melastoma sanguineum Fruit: Optimization and Identification. Molecules 2018, 23, 2498. [Google Scholar] [CrossRef] [Green Version]
- Krasensky, J.; Jonak, C. Drought, salt, and temperature stress-induced metabolic rearrangements and regulatory networks. J. Exp. Bot. 2012, 63, 1593–1608. [Google Scholar] [CrossRef] [Green Version]
- Byeon, Y.S.; Hong, Y.-S.; Kwak, H.S.; Lim, S.-T.; Kim, S.S. Metabolite profile and antioxidant potential of wheat (Triticum aestivum L.) during malting. Food Chem. 2022, 384, 132443. [Google Scholar] [CrossRef]
- Chung, R.-S.; Chen, C.-C.; Ng, L.-T. Nitrogen fertilization affects the growth performance, betaine and polysaccharide concentrations of Lycium barbarum. Ind. Crops Prod. 2010, 32, 650–655. [Google Scholar] [CrossRef]
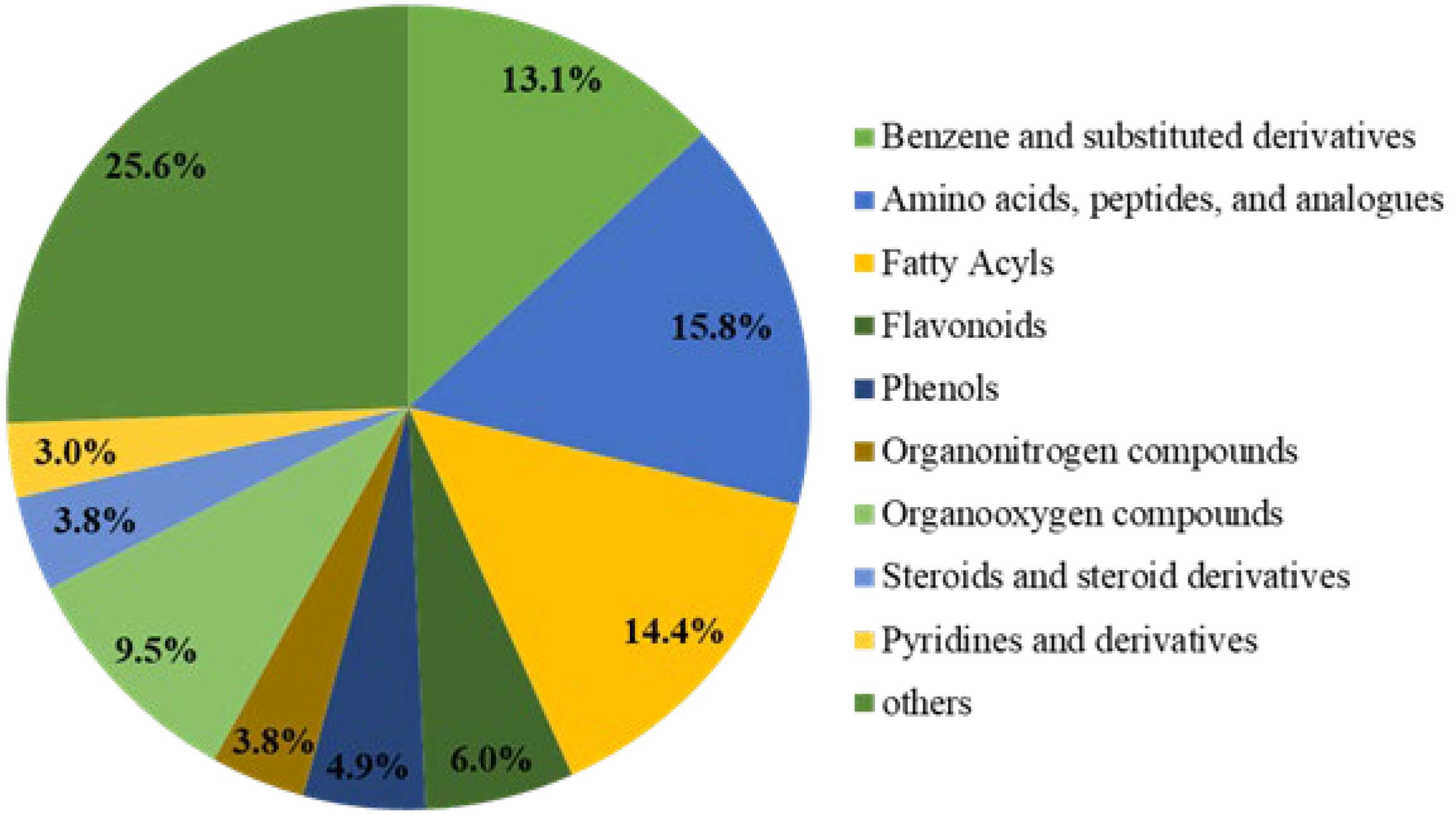
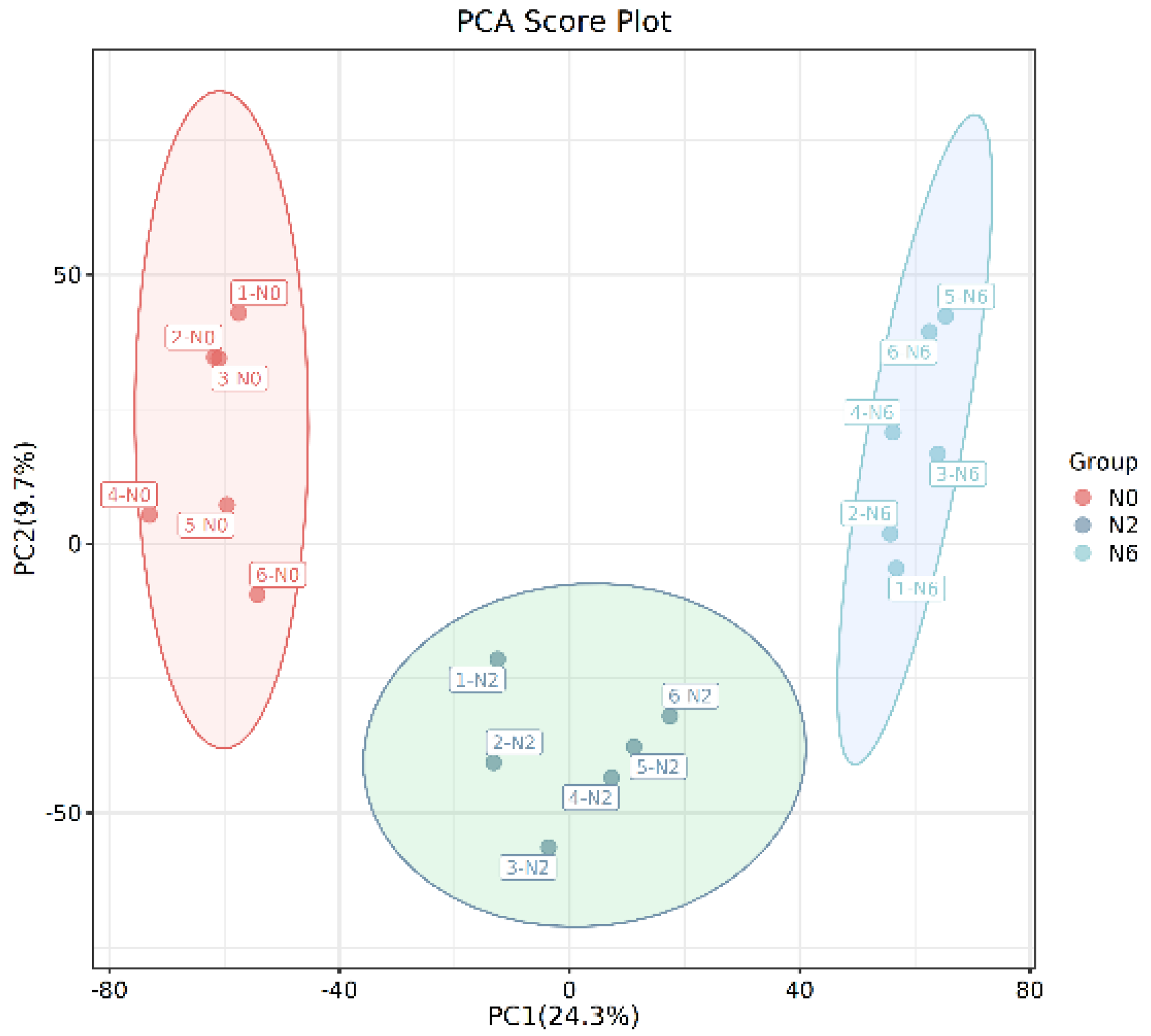

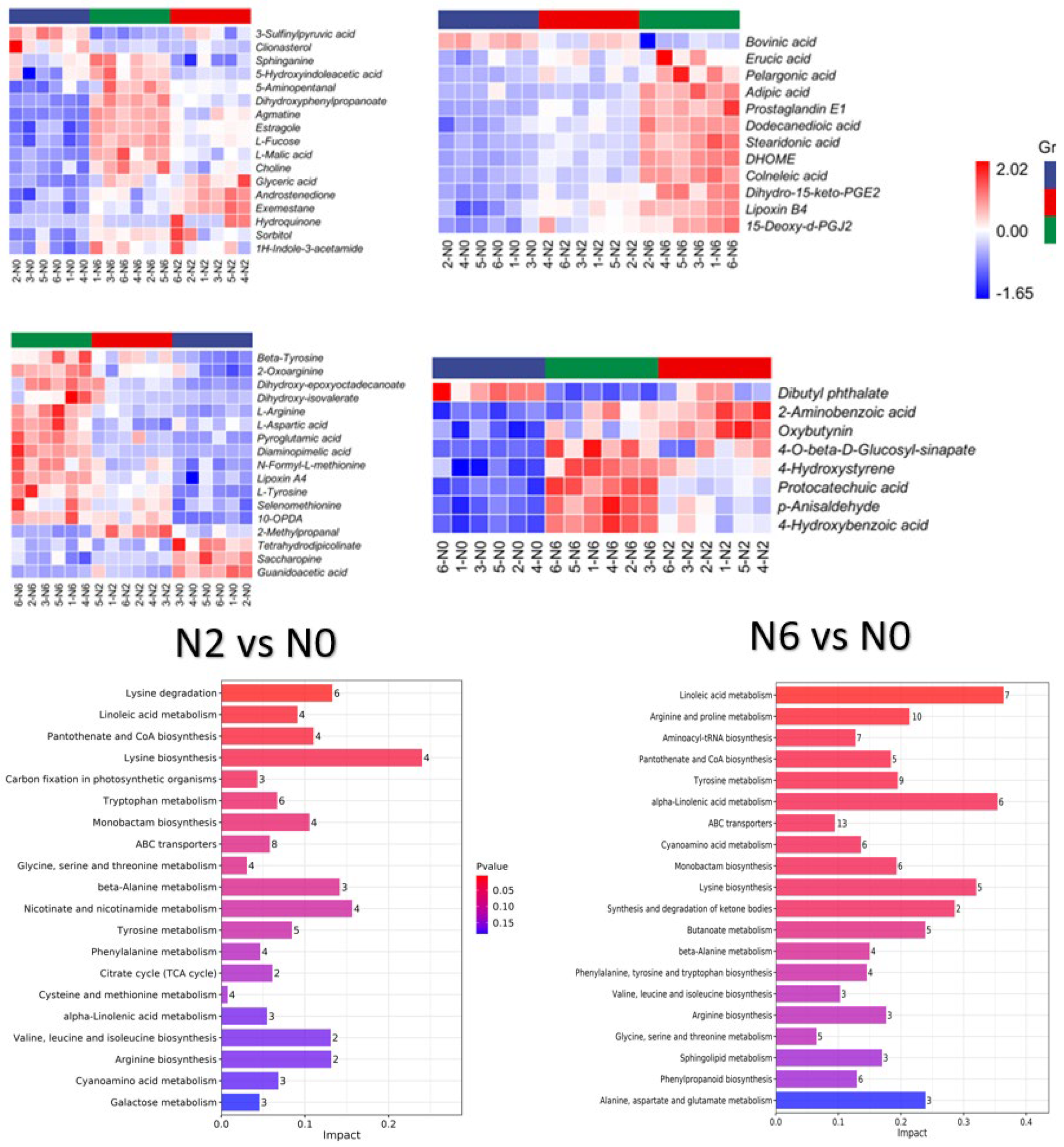
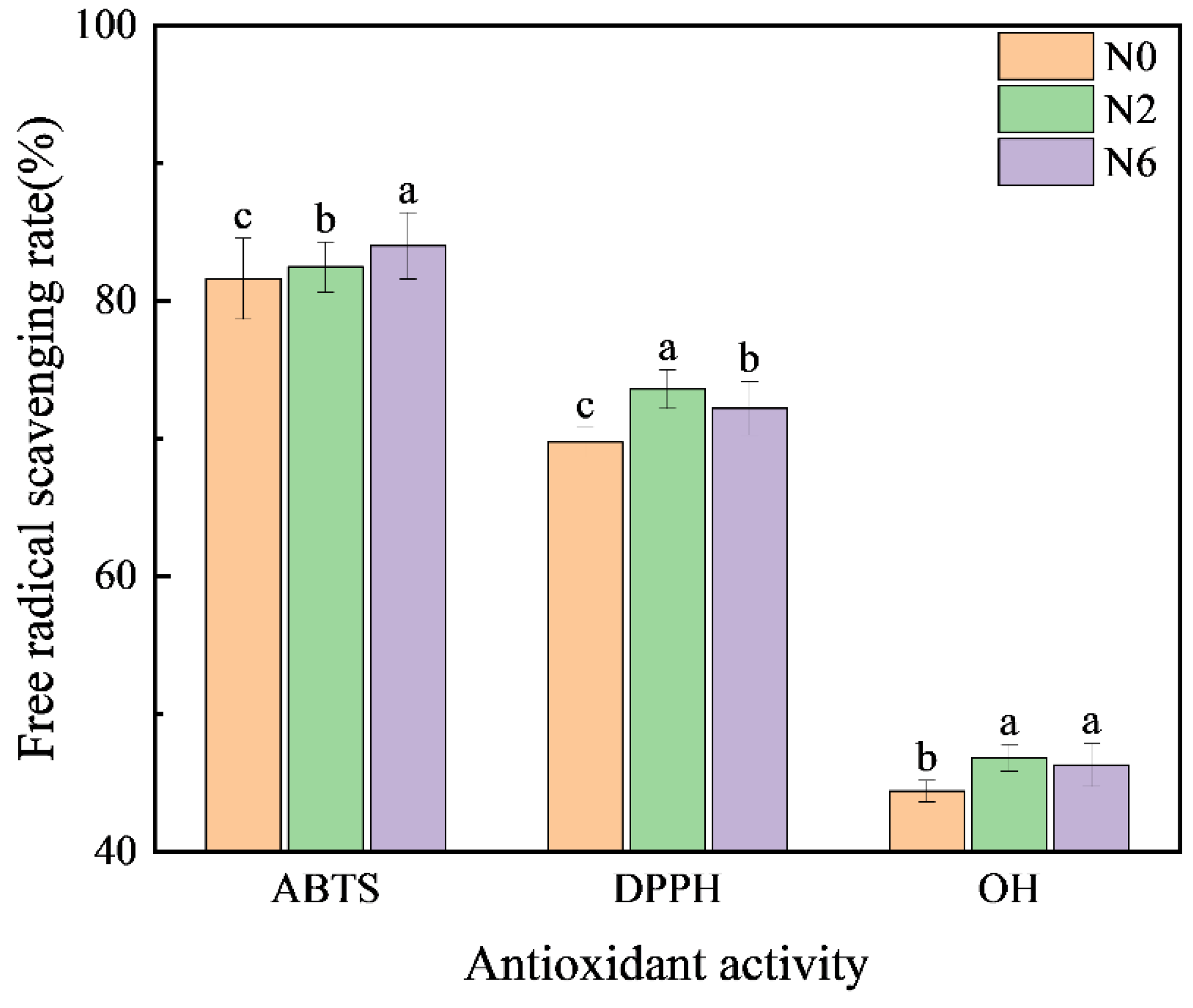
| Soil pH | Organic Matter (mg/kg) | Total Nitrogen (g/kg) | Available Nutrient (mg/kg) | Bulk Density (g/cm3) | ||
|---|---|---|---|---|---|---|
| N (Nitrogen) | P (Phosphorus) | K (Potassium) | ||||
| 8.2 | 22.57 | 1.42 | 105.24 | 21.61 | 164.22 | 1.39 |
| Multivariate Analysis | Group | Positive Ion Model | Negative Ion Model | ||||
|---|---|---|---|---|---|---|---|
| R2X | R2Y | Q2 | R2X | R2Y | Q2 | ||
| PLS-DA | N2 vs. N0 | 0.305 | 0.994 | 0.757 | 0.389 | 0.999 | 0.816 |
| N6 vs. N0 | 0.471 | 0.999 | 0.854 | 0.467 | 0.997 | 0.831 | |
| N6 vs. N2 | 0.412 | 1.0 | 0.896 | 0.415 | 0.998 | 0.812 | |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ma, Y.; Zhang, S.; Wu, Z.; Sun, W. Metabolic Variations in Brown Rice Fertilised with Different Levels of Nitrogen. Foods 2022, 11, 3539. https://doi.org/10.3390/foods11213539
Ma Y, Zhang S, Wu Z, Sun W. Metabolic Variations in Brown Rice Fertilised with Different Levels of Nitrogen. Foods. 2022; 11(21):3539. https://doi.org/10.3390/foods11213539
Chicago/Turabian StyleMa, Yichao, Shuang Zhang, Zhaoxia Wu, and Wentao Sun. 2022. "Metabolic Variations in Brown Rice Fertilised with Different Levels of Nitrogen" Foods 11, no. 21: 3539. https://doi.org/10.3390/foods11213539
APA StyleMa, Y., Zhang, S., Wu, Z., & Sun, W. (2022). Metabolic Variations in Brown Rice Fertilised with Different Levels of Nitrogen. Foods, 11(21), 3539. https://doi.org/10.3390/foods11213539







