pH-Regulated Strategy and Mechanism of Antibody Orientation on Magnetic Beads for Improving Capture Performance of Staphylococcus Species
Abstract
:Highlights
- An pH-regulated strategy can achieve antibody orientation on the surface of magnetic beads.
- The capture efficiency for Staphylococcus aureus of immunomagnetic beads prepared at pH 8.0 was improved.
- The antibody orientation mechanism was demonstrated using a quantum dots labeled antigen, antigen-binding fragment (Fab) accessibility assay and lysine mimicking.
Abstract
1. Introduction
2. Methods/Experimental Section
2.1. Reagents, Materials, and Apparatus
2.2. Preparation of IMBs
2.2.1. Activation of Carboxyl MBs
2.2.2. Antibody Immobilization on MBs
2.3. Characteristics of IMBs
2.3.1. Antibody Binding Quantification
2.3.2. Size Distribution and Dispersity Characteristics
2.3.3. Capture Efficiency
2.3.4. Measurement of the Maximum Binding Capacity
2.4. Mechanism of Antibody Orientation
2.4.1. Quantum Dots Labeling Antigen Assay
2.4.2. Fab Accessibility Assay
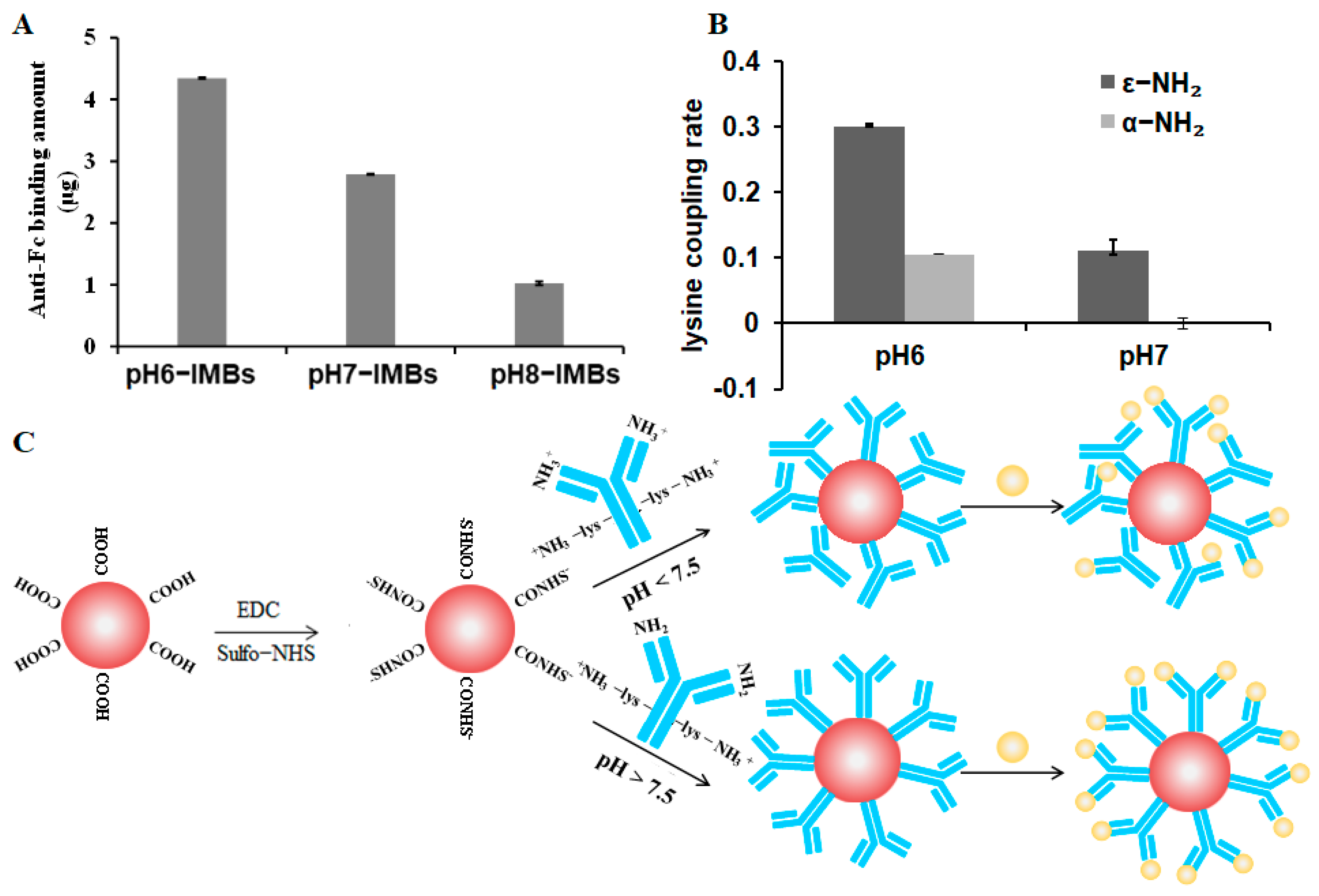
2.4.3. Crosslinking Ratio Analysis of ε-NH2-lys and α-NH2-lys on the Surface of the MBs
3. Results and Discussion
3.1. Characterization of IMBs Properties
3.2. Capture Efficiency of pH6-IMBs, pH7-IMBs and pH8-IMBs
3.3. Fluorescence Analysis of Antibody Orientation on MBs
3.4. Fab Accessibility Analysis on IMBs
3.5. Lysine Mimicking
4. Conclusions
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
Abbreviations
| IMBs | immunomagnetic beads |
| QDs | quantum dots |
| EDC | N-(3-dimethylaminopropyl)-N’-ethylcarbodiimide hydrochloride crystalline Sulfo-NHS: N-hydroxysulfosuccinimide sodium salt |
| BCA | bicinchoninic acid |
| BSA | bovine serum albumin |
| PCA | plate count agar |
| NB | nutrient broth |
| SEB | Staphylococcus enterotoxin B |
| BOC | tert-butyloxycarbonyl |
| CFU | colony-forming unit |
| Fab | antigen-binding fragment |
| Fc | fragment crystallizable |
References
- Nam, K.C.; Han, Y.S.; Lee, J.-M.; Kim, S.C.; Cho, G.; Park, B.J. Photo-Functionalized Magnetic Nanoparticles as a Nanocarrier of Photodynamic Anticancer Agent for Biomedical Theragnostics. Cancers 2020, 12, 571. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kyeong, S.; Kim, J.; Chang, H.; Lee, S.H.; Son, B.S.; Lee, J.H.; Rho, W.Y.; Pham, X.H.; Jun, B.H. Magnetic Nanoparticles. Adv. Exp. Med. Biol. 2021, 1309, 191–215. [Google Scholar] [PubMed]
- Pastucha, M.; Farka, Z.; Lacina, K.; Mikušová, Z.; Skládal, P. Magnetic nanoparticles for smart electrochemical immunoassays: A review on recent developments. Mikrochim. Acta 2019, 186, 312. [Google Scholar] [CrossRef] [PubMed]
- Sharma, S.; Ram, P. Capturing of Magnetic Nanoparticles in a Fluidic Channel for Magnetic Drug Targeting. J. Nanosci. Nanotechnol. 2021, 21, 3588–3595. [Google Scholar] [CrossRef]
- Kim, J.-H.; Oh, S.-W. Pretreatment methods for nucleic acid-based rapid detection of pathogens in food: A review. Food Control 2021, 121, 107575. [Google Scholar] [CrossRef]
- Du, M.; Li, J.; Liu, Q.; Wang, Y.; Chen, E.; Kang, F.; Tu, C. Rapid detection of trace Salmonella in milk using an effective pretreatment combined with droplet digital polymerase chain reaction. Microbiol. Res. 2021, 251, 126838. [Google Scholar] [CrossRef]
- Du, M.; Li, J.; Zhao, R.; Yang, Y.; Wang, Y.; Ma, K.; Cheng, X.; Wan, Y.; Wu, X. Effective pre-treatment technique based on immune-magnetic separation for rapid detection of trace levels of Salmonella in milk. Food Control 2018, 91, 92–99. [Google Scholar] [CrossRef]
- Johari-Ahar, M.; Rashidi, M.R.; Barar, J.; Aghaie, M.; Mohammadnejad, D.; Ramazani, A.; Karami, P.; Coukos, G.; Omidi, Y. An ultra-sensitive impedimetric immunosensor for detection of the serum oncomarker CA-125 in ovarian cancer patients. Nanoscale 2015, 7, 3768–3779. [Google Scholar] [CrossRef]
- Wei, S.; Park, B.-J.; Seo, K.-H.; Oh, D.-H. Highly efficient and specific separation of Staphylococcus aureus from lettuce and milk using Dynabeads protein G conjugates. Food Sci. Biotechnol. 2016, 25, 1501–1505. [Google Scholar] [CrossRef]
- Luciani, M.; Di Febo, T.; Zilli, K.; Di Giannatale, E.; Armillotta, G.; Manna, L.; Minelli, F.; Tittarelli, M.; Caprioli, A. Rapid Detection and Isolation of Escherichia coli O104:H4 from Milk Using Monoclonal Antibody-coated Magnetic Beads. Front. Microbiol. 2016, 15, 942. [Google Scholar] [CrossRef]
- Kausaite-Minkstimiene, A.; Ramanaviciene, A.; Kirlyte, J.; Ramanavicius, A. Comparative Study of Random and Oriented Antibody Immobilization Techniques on the Binding Capacity of Immunosensor. Anal. Chem. 2010, 82, 6401–6408. [Google Scholar] [CrossRef] [PubMed]
- Lin, P.C.; Chen, S.H.; Wang, K.Y.; Chen, M.L.; Adak, A.K.; Hwu, J.R.; Chen, Y.J.; Lin, C.C. Fabrication of Oriented Antibody-Conjugated Magnetic Nanoprobes and Their Immunoaffinity Application. Anal. Chem. 2009, 81, 8774–8782. [Google Scholar] [CrossRef] [PubMed]
- Puertas, S.; Moros, M.; Fernández-Pacheco, R.; Ibarra, M.R.; Grazú, V.; De La Fuente, J.M. Designing novel nano-immunoassays: Antibody orientation versus sensitivity. J. Phys. D-Appl. Phys. 2010, 43, 474012. [Google Scholar] [CrossRef] [Green Version]
- Saha, B.; Songe, P.; Evers, T.H.; Prins, M.W.J. The influence of covalent immobilization conditions on antibody accessibility on nanoparticles. Analyst 2017, 142, 4247–4256. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Puertas, S.; Villa, M.D.G.; Mendoza, E.; Jiménez-Jorquera, C.; de la Fuente, J.M.; Fernández-Sánchez, C.; Grazú, V. Improving immunosensor performance through oriented immobilization of antibodies on carbon nanotube composite surfaces. Biosens. Bioelectron. 2013, 43, 274–280. [Google Scholar] [CrossRef]
- Song, H.Y.; Zhou, X.; Hobley, J.; Su, X. Comparative Study of Random and Oriented Antibody Immobilization as Measured by Dual Polarization Interferometry and Surface Plasnnon Resonance Spectroscopy. Langmuir 2012, 28, 997–1004. [Google Scholar] [CrossRef]
- Raghav, R.; Srivastava, S. Immobilization strategy for enhancing sensitivity of immunosensors: L-Asparagine-AuNPs as a promising alternative of EDC-NHS activated citrate-AuNPs for antibody immobilization. Biosens. Bioelectron. 2016, 78, 396–403. [Google Scholar] [CrossRef]
- Yan, Q.; Zheng, H.-N.; Jiang, C.; Li, K.; Xiao, S.-J. EDC/NHS activation mechanism of polymethacrylic acid: Anhydride versus NHS-ester. RSC Adv. 2015, 5, 69939–69947. [Google Scholar] [CrossRef]
- Duval, F.; van Beek, T.A.; Zuilhof, H. Key steps towards the oriented immobilization of antibodies using boronic acids. Analyst 2015, 140, 6467–6472. [Google Scholar] [CrossRef] [Green Version]
- Kumar, S.; Aaron, J.; Sokolov, K. Directional conjugation of antibodies to nanoparticles for synthesis of multiplexed optical contrast agents with both delivery and targeting moieties. Nat. Protoc. 2008, 3, 314–320. [Google Scholar] [CrossRef]
- Prieto-Simon, B.; Saint, C.; Voelcker, N.H. Electrochemical Biosensors Featuring Oriented Antibody Immobilization via Electrografted and Self-Assembled Hydrazide Chemistry. Anal. Chem. 2014, 86, 1422–1429. [Google Scholar] [CrossRef] [PubMed]
- Seo, J.S.; Lee, S.; Poulter, C.D. Regioselective Covalent Immobilization of Recombinant Antibody-Binding Proteins A, G, and L for Construction of Antibody Arrays. J. Am. Chem. Soc. 2013, 135, 8973–8980. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Moon, J.; Byun, J.; Kim, H.; Jeong, J.; Lim, E.; Jung, J.; Cho, S.; Cho, W.K.; Kang, T. Surface-Independent and Oriented Immobilization of Antibody via One-Step Polydopamine/Protein G Coating: Application to Influenza Virus Immunoassay. Macromol. Biosci. 2019, 19, e1800486. [Google Scholar] [CrossRef] [PubMed]
- Kaittanis, C.; Santra, S.; Perez, J.M. Role of Nanoparticle Valency in the Nondestructive Magnetic-Relaxation-Mediated Detection and Magnetic Isolation of Cells in Complex Media. J. Am. Chem. Soc. 2009, 131, 12780–12791. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Granade, T.C.; Workman, S.; Wells, S.K.; Holder, A.N.; Owen, S.M.; Pau, C.-P. Rapid Detection and Differentiation of Antibodies to HIV-1 and HIV-2 Using Multivalent Antigens and Magnetic Immunochromatography Testing. Clin. Vaccine Immunol. 2010, 17, 1034–1039. [Google Scholar] [CrossRef] [Green Version]
- Baio, G.; Fabbi, M.; Salvi, S.; De Totero, D.; Truini, M.; Ferrini, S.; Neumaier, C.E. Two-Step In Vivo Tumor Targeting by Biotin-Conjugated Antibodies and Superparamagnetic Nanoparticles Assessed by Magnetic Resonance Imaging at 1.5 T. Mol. Imaging Biol. 2010, 12, 305–315. [Google Scholar] [CrossRef]
- Zhao, X.; Pan, F.; Garcia-Gancedo, L.; Flewitt, A.J.; Ashley, G.M.; Luo, J.; Lu, J.R. Interfacial recognition of human prostate-specific antigen by immobilized monoclonal antibody: Effects of solution conditions and surface chemistry. J. R. Soc. Interface 2012, 9, 2457–2467. [Google Scholar] [CrossRef]
- Wang, A.; Perera, Y.R.; Davidson, M.B.; Fitzkee, N.C. Electrostatic Interactions and Protein Competition Reveal a Dynamic Surface in Gold Nanoparticle-Protein Adsorption. J. Phys. Chem. C Nanomater. Interfaces 2016, 120, 24231–24239. [Google Scholar] [CrossRef] [Green Version]
- Lin, W.; Insley, T.; Tuttle, M.D.; Zhu, L.; Berthold, D.A.; Král, P.; Rienstra, C.M.; Murphy, C.J. Control of protein orientation on gold nanoparticles. J. Phys. Chem. C Nanomater. Interfaces 2015, 119, 21035–21043. [Google Scholar] [CrossRef] [Green Version]
- Siriwardana, K.; Wang, A.; Vangala, K.; Fitzkee, N.; Zhang, D. Probing the effects of cysteine residues on protein adsorption onto gold nanoparticles using wild-type and mutated GB3 proteins. Langmuir 2013, 29, 10990–10996. [Google Scholar] [CrossRef]
- Wiseman, M.E.; Frank, C.W. Antibody adsorption and orientation on hydrophobic surfaces. Langmuir 2012, 28, 1765–1774. [Google Scholar] [CrossRef] [PubMed]
- Ruiz, G.; Tripathi, K.; Okyem, S.; Driskell, J.D. pH Impacts the Orientation of Antibody Adsorbed onto Gold Nanoparticles. Bioconjugate Chem. 2019, 30, 1182–1191. [Google Scholar] [CrossRef] [PubMed]
- Zhou, J.; Tsao, H.-K.; Sheng, Y.-J.; Jiang, S. Monte Carlo simulations of antibody adsorption and orientation on charged surfaces. J. Chem. Phys. 2004, 121, 1050–1057. [Google Scholar] [CrossRef] [PubMed]
- Chen, S.; Liu, L.; Zhou, J.; Jiang, S. Controlling antibody orientation on charged self-assembled monolayers. Langmuir 2003, 19, 2859–2864. [Google Scholar] [CrossRef]
- Emaminejad, S.; Javanmard, M.; Gupta, C.; Chang, S.; Davis, R.W.; Howe, R.T. Tunable control of antibody immobilization using electric field. Proc. Natl. Acad. Sci. USA 2015, 112, 1995–1999. [Google Scholar] [CrossRef] [Green Version]
- Lou, D.; Ji, L.; Fan, L.; Ji, Y.; Gu, N.; Zhang, Y. Antibody-Oriented Strategy and Mechanism for the Preparation of Fluorescent Nanoprobes for Fast and Sensitive Immunodetection. Langmuir 2019, 35, 4860–4867. [Google Scholar] [CrossRef]
- Eriksson, A.E.; Baase, W.A.; Zhang, X.J.; Heinz, D.W.; Blaber, M.; Baldwin, E.P.; Matthews, B.W. Response of a Protein-Structure to Cavity-Creating Mutations and Its Relation to the Hydrophobic Effect. Science 1992, 255, 178–183. [Google Scholar] [CrossRef] [Green Version]
- Garcia-Seisdedos, H.; Ibarra-Molero, B.; Sanchez-Ruiz, J.M. How many ionizable groups can sit on a protein hydrophobic core? Proteins-Struct. Funct. Bioinform. 2012, 80, 1–7. [Google Scholar] [CrossRef]
- Xu, J.; Baase, W.A.; Baldwin, E.; Matthews, B.W. The response of T4 lysozyme to large-to-small substitutions within the core and its relation to the hydrophobic effect. Protein Sci. 1998, 7, 158–177. [Google Scholar] [CrossRef] [Green Version]
- Rao, S.V.; Anderson, K.W.; Bachas, L.G. Oriented Immobilization of Proteins. Protein Sci. 1998, 128, 127–143. [Google Scholar] [CrossRef]
- Fuentes, M.; Mateo, C.; Guisán, J.; Fernández-Lafuente, R. Preparation of Inert Magnetic Nanoparticles for the Directed Immobilization of Antibodies. Biosens. Bioelectron. 2005, 20, 1380–1387. [Google Scholar] [CrossRef] [PubMed]
- Sun, J.; He, M.; Liu, X.; Gu, N. Optimizing colloidal dispersity of magnetic nanoparticles based on magnetic separation with magnetic nanowires array. Appl. Phys. A-Mater. Sci. Process. 2015, 118, 569–577. [Google Scholar] [CrossRef]
- Zhao, Y.; Ye, M.; Chao, Q.; Jia, N.; Ge, Y.; Shen, H. Simultaneous Detection of Multifood-Borne Pathogenic Bacteria Based on Functionalized Quantum Dots Coupled with Immunomagnetic Separation in Food Samples. J. Agric. Food Chem. 2009, 57, 517–524. [Google Scholar] [CrossRef] [PubMed]


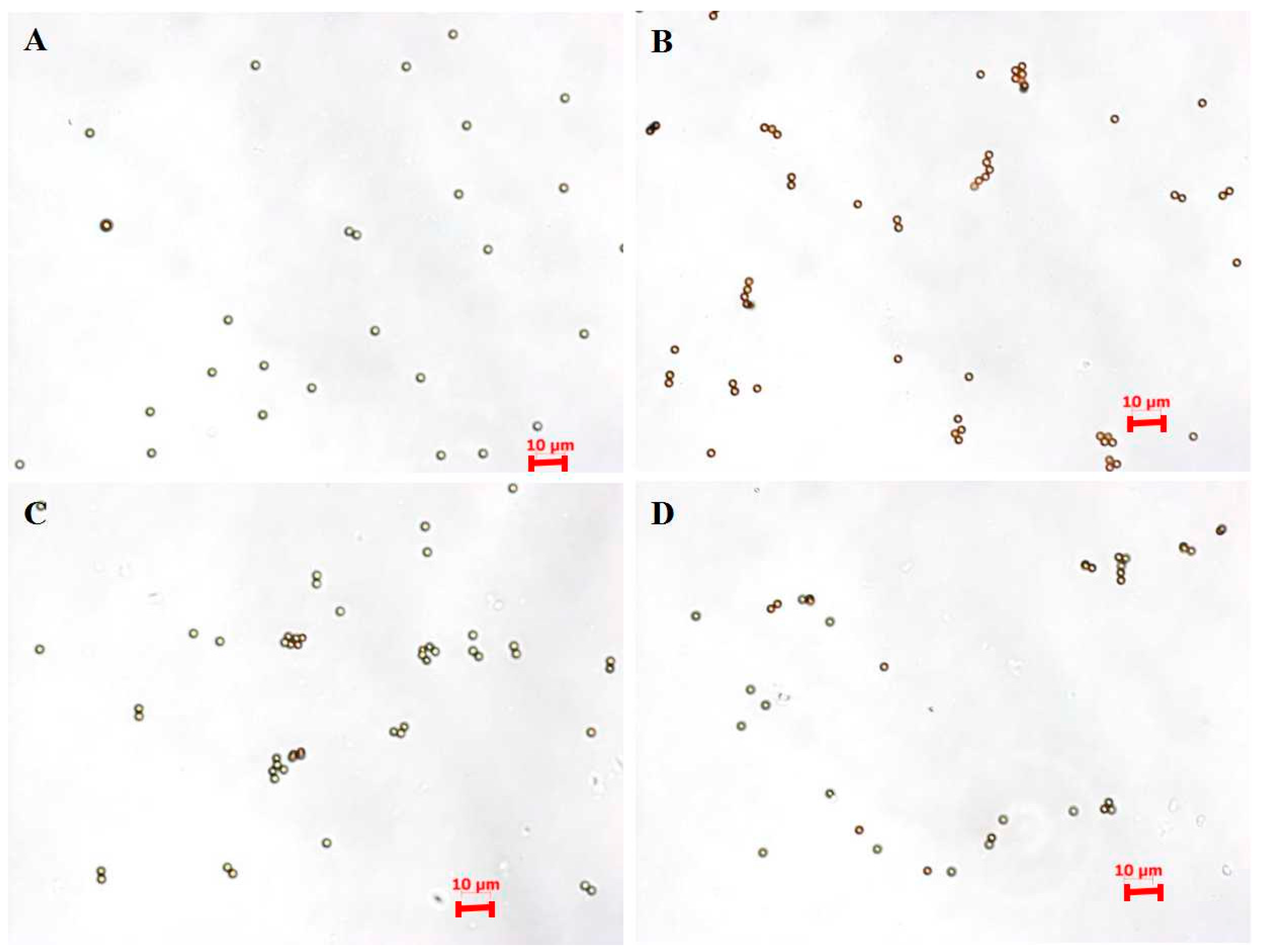


| Sample | Zeta Potential (mV) |
|---|---|
| aMBs-pH6 | −47.73 |
| aMBs-pH7 | −41.87 |
| aMBs-pH8 | −57.07 |
| pH6-IMBs | −15.33 |
| pH7-IMBs | −17.23 |
| pH8-IMBs | −15.83 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kang, F.; Yang, Y.; Li, J.; Chen, E.; Hong, T.; Zhao, L.; Du, M. pH-Regulated Strategy and Mechanism of Antibody Orientation on Magnetic Beads for Improving Capture Performance of Staphylococcus Species. Foods 2022, 11, 3599. https://doi.org/10.3390/foods11223599
Kang F, Yang Y, Li J, Chen E, Hong T, Zhao L, Du M. pH-Regulated Strategy and Mechanism of Antibody Orientation on Magnetic Beads for Improving Capture Performance of Staphylococcus Species. Foods. 2022; 11(22):3599. https://doi.org/10.3390/foods11223599
Chicago/Turabian StyleKang, Fuying, Yin Yang, Jingwen Li, Erning Chen, Tian Hong, Lulu Zhao, and Meihong Du. 2022. "pH-Regulated Strategy and Mechanism of Antibody Orientation on Magnetic Beads for Improving Capture Performance of Staphylococcus Species" Foods 11, no. 22: 3599. https://doi.org/10.3390/foods11223599
APA StyleKang, F., Yang, Y., Li, J., Chen, E., Hong, T., Zhao, L., & Du, M. (2022). pH-Regulated Strategy and Mechanism of Antibody Orientation on Magnetic Beads for Improving Capture Performance of Staphylococcus Species. Foods, 11(22), 3599. https://doi.org/10.3390/foods11223599





