Microbiome-Metabolomics Insights into the Milk of Lactating Dairy Cows to Reveal the Health-Promoting Effects of Dietary Citrus Peel Extracts on the Mammary Metabolism
Abstract
:1. Introduction
2. Materials and Methods
2.1. Citrus Peel Extract Preparation
2.2. Animals, Experimental Design, and Treatments
2.3. Milk Sample Collection and Biochemical Parameters
2.4. Milk 16S rRNA Gene Sequencing and Bioinformatic Analysis
2.5. Metabolomics Analysis
2.6. Statistical Analyses
3. Results
3.1. Milk Biochemical Parameters
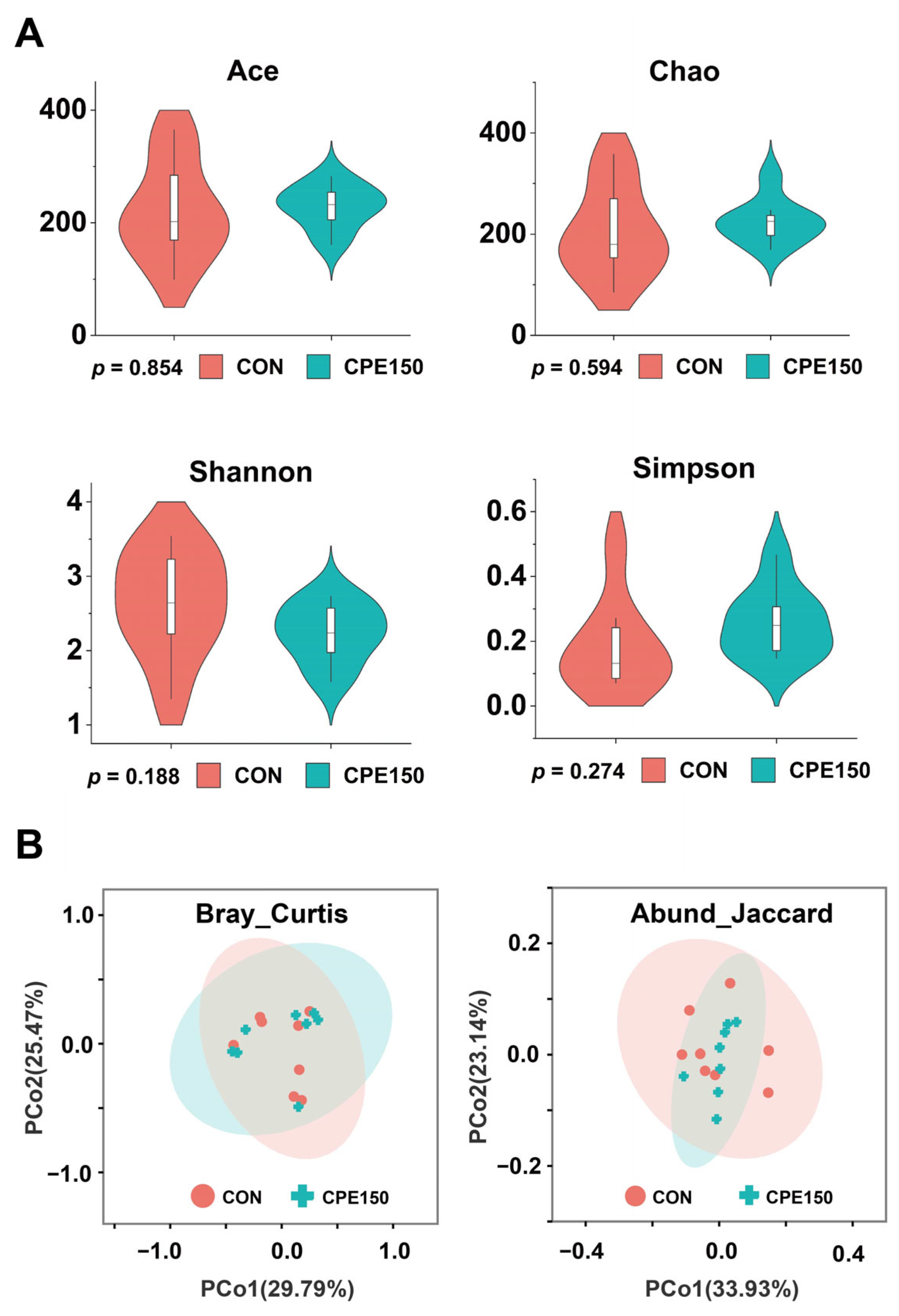
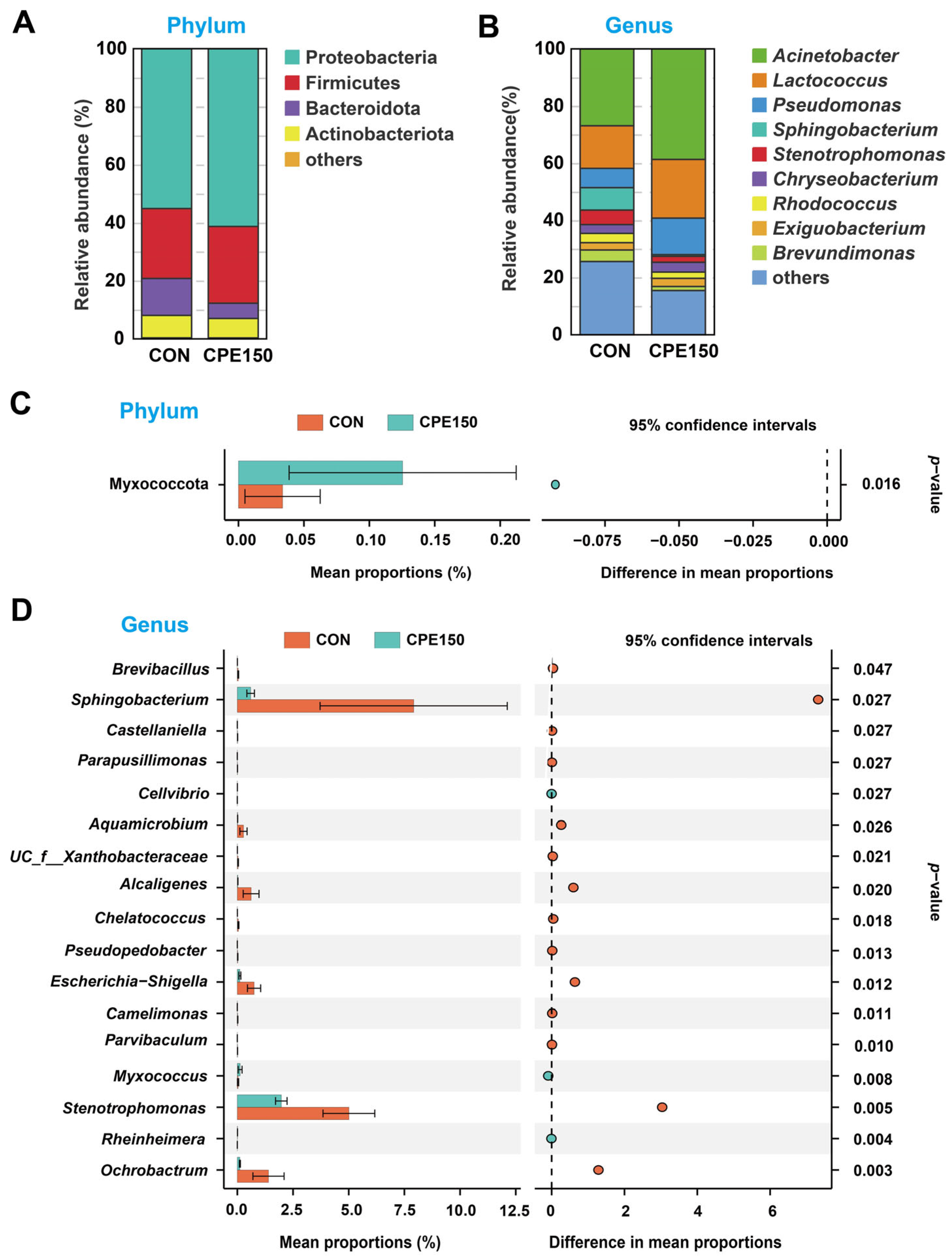
3.2. Milk Bacterial Community
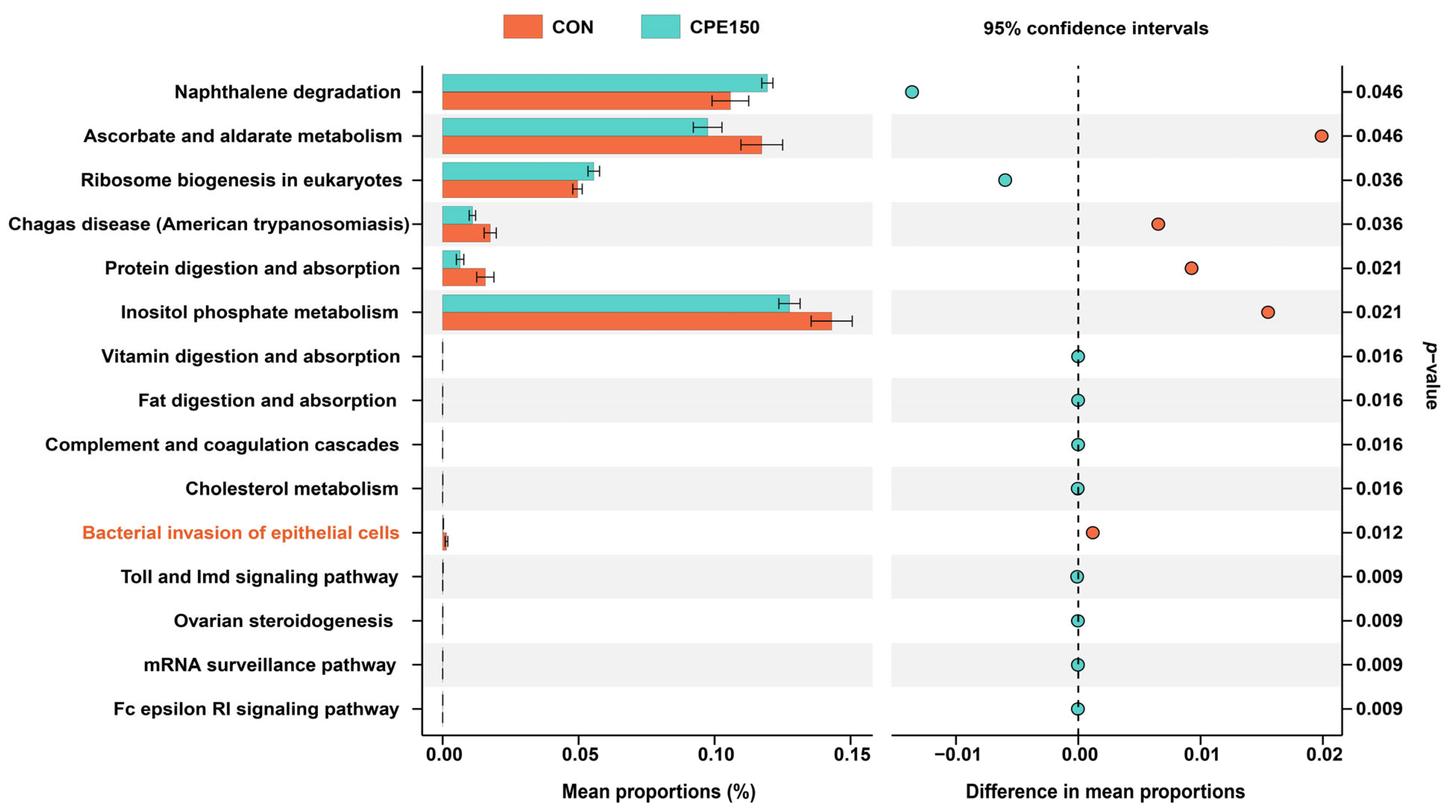
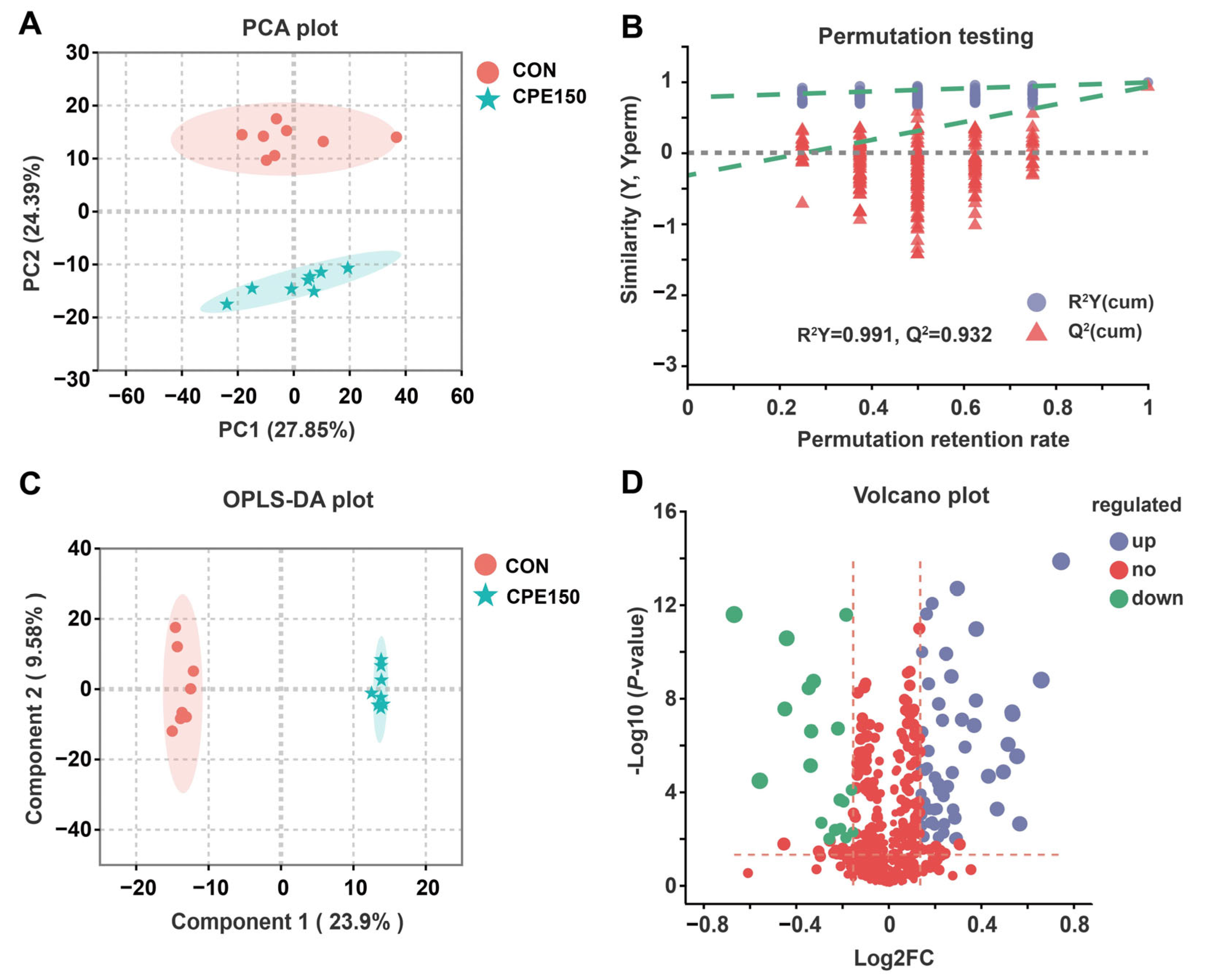
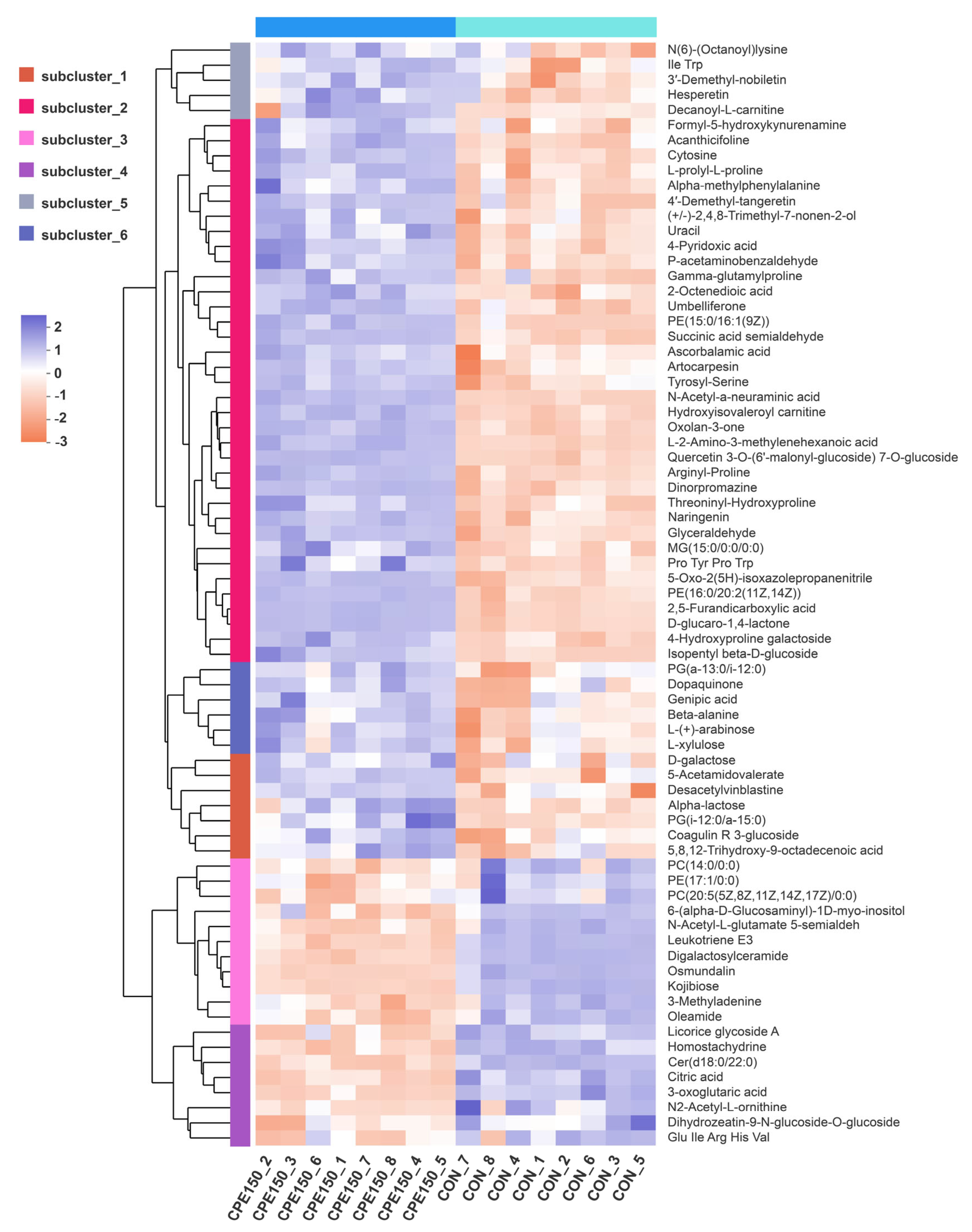
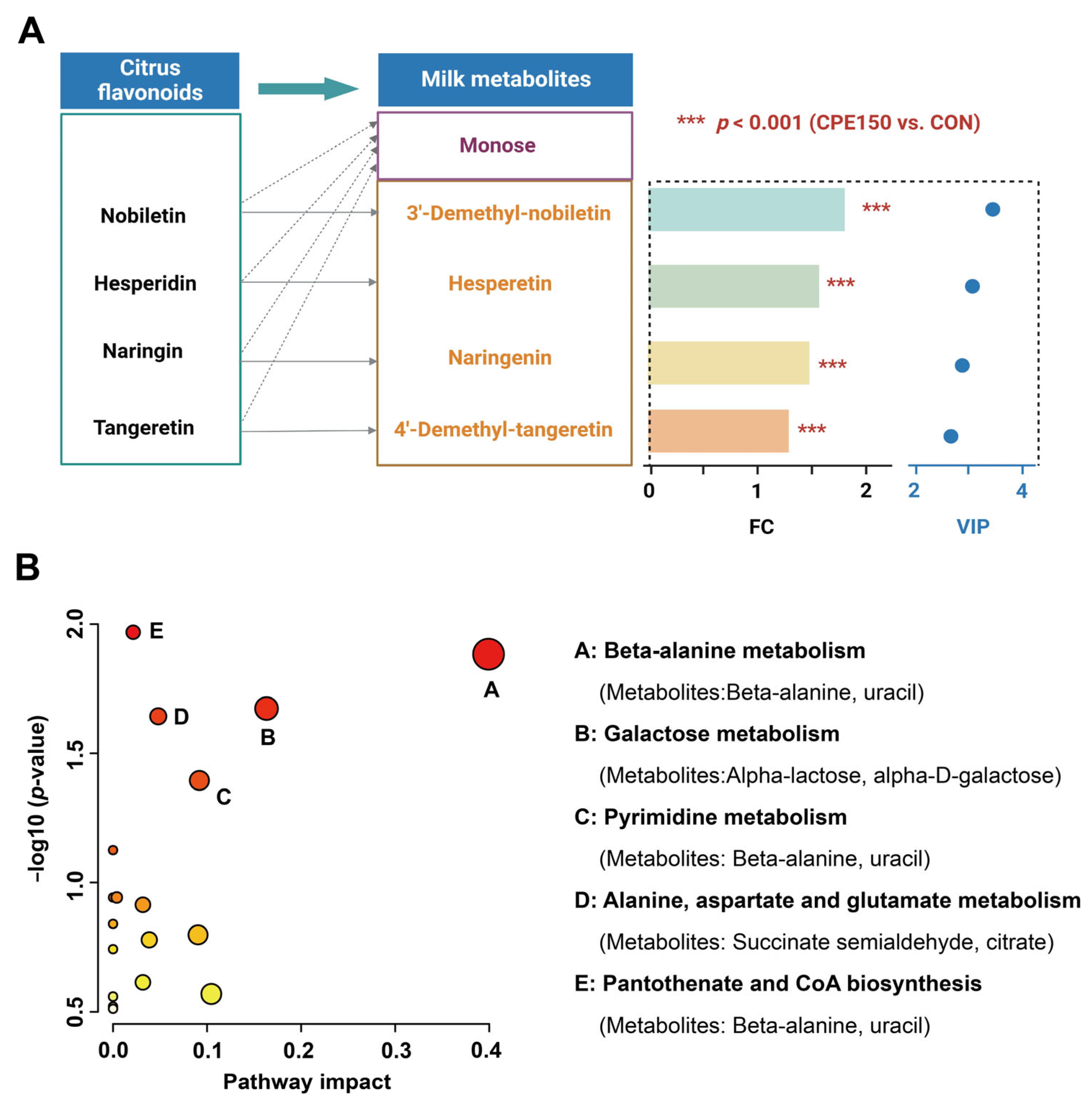
3.3. Milk Metabolomics Profiling
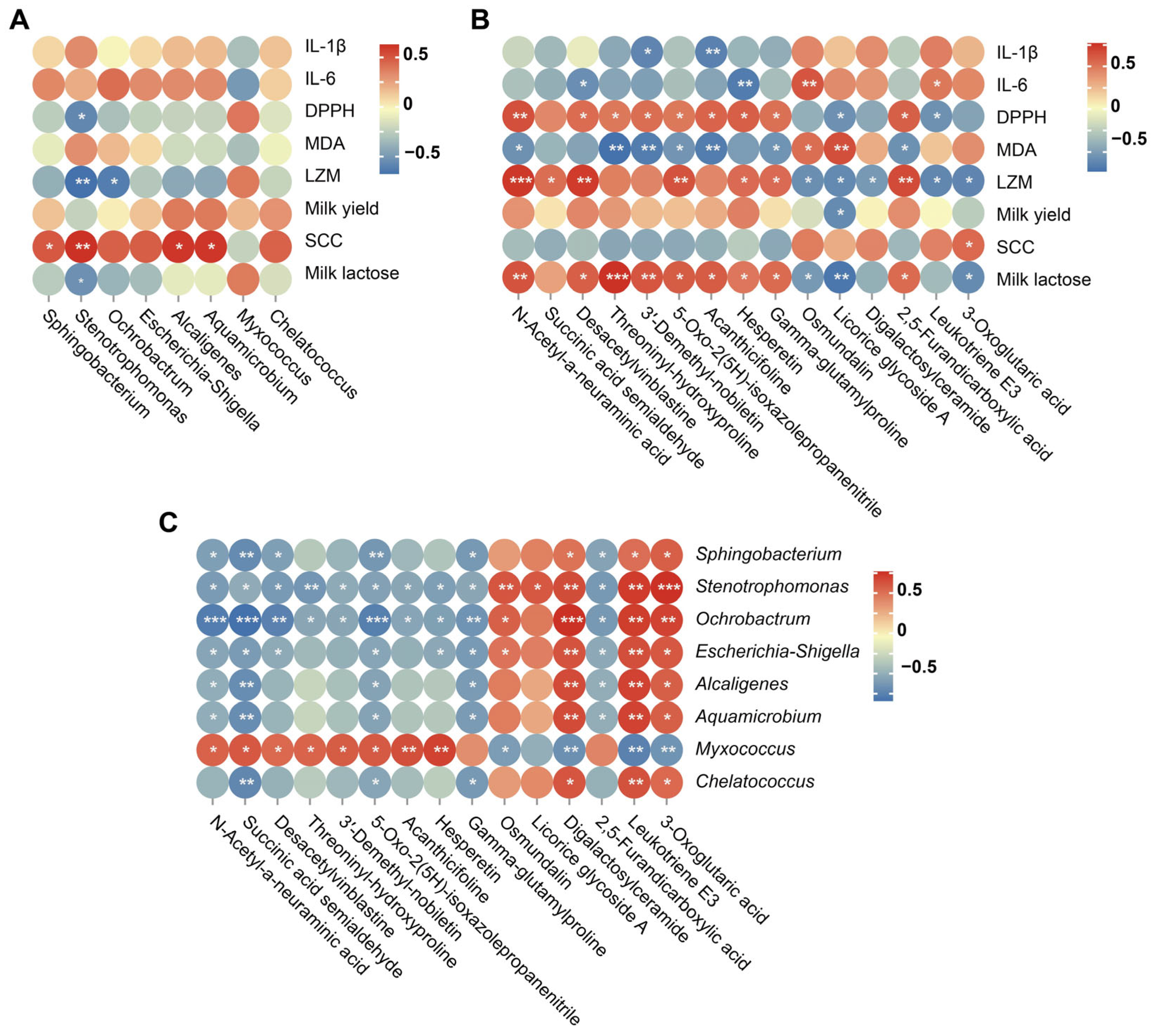
3.4. Correlation Analysis
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Dong, L.; Meng, L.; Liu, H.; Wu, H.; Hu, H.; Zheng, N.; Wang, J.; Schroyen, M. Effect of therapeutic administration of beta-lactam antibiotics on the bacterial community and antibiotic resistance patterns in milk. J. Dairy Sci. 2021, 104, 7018–7025. [Google Scholar] [CrossRef] [PubMed]
- Bruckmaier, R.M.; Gross, J.J. Lactational challenges in transition dairy cows. Anim. Prod. Sci. 2017, 57, 1471–1481. [Google Scholar] [CrossRef]
- Gross, J.J.; Bruckmaier, R.M. Invited review: Metabolic challenges and adaptation during different functional stages of the mammary gland in dairy cows: Perspectives for sustainable milk production. J. Dairy Sci. 2019, 102, 2828–2843. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, Y.; Nan, X.; Zhao, Y.; Jiang, L.; Wang, H.; Zhang, F.; Hua, D.; Liu, J.; Yao, J.; Yang, L.; et al. Consumption of supplementary inulin modulates milk microbiota and metabolites in dairy cows with subclinical mastitis. Appl. Environ. Microbiol. 2022, 88, e0205921. [Google Scholar] [CrossRef]
- Zhan, J.; Shen, Y.; Li, X.; Zhang, H.; Niu, H.; Fang, L.; Xiong, B.; Tong, J.; Jiang, L. Microbiome and metabolic changes of milk in response to dietary supplementation with bamboo leaf extract in dairy cows. Front. Nutr. 2021, 8, 723446. [Google Scholar] [CrossRef]
- Olagaray, K.E.; Bradford, B.J. Plant flavonoids to improve productivity of ruminants—A review. Anim. Feed Sci. Technol. 2019, 251, 21–36. [Google Scholar] [CrossRef]
- Li, Y.Z.; Zhao, C.Y.; Lu, C.; Zhou, S.S.; Tian, G.F.; He, L.L.; Bao, Y.M.; Fauconnier, M.L.; Xiao, H.; Zheng, J.K. Simultaneous determination of 14 bioactive citrus flavonoids using thin-layer chromatography combined with surface enhanced Raman spectroscopy. Food Chem. 2021, 338, 128115. [Google Scholar] [CrossRef]
- Paniagua, M.; Crespo, J.; Aris, A.; Devant, M. Citrus aurantium flavonoid extract improves concentrate efficiency, animal behavior, and reduces rumen inflammation of Holstein bulls fed high-concentrate diets. Anim. Feed Sci. Technol. 2019, 258, 114304. [Google Scholar] [CrossRef]
- Eger, M.; Graz, M.; Riede, S.; Breves, G. Application of Mootral (TM) reduces methane production by altering the archaea community in the rumen simulation technique. Front. Microbiol. 2018, 9, 2094. [Google Scholar] [CrossRef]
- Simitzis, P.; Massouras, T.; Goliomytis, M.; Charismiadou, M.; Moschou, K.; Economou, C.; Papadedes, V.; Lepesioti, S.; Deligeorgis, S. The effects of hesperidin or naringin dietary supplementation on the milk properties of dairy ewes. J. Sci. Food Agric. 2019, 99, 6515–6521. [Google Scholar] [CrossRef]
- Zhao, Y.; Yu, S.; Zhao, H.; Li, L.; Li, Y.; Tu, Y.; Jiang, L.; Zhao, G. Lipidomic profiling using GC and LC-MS/MS revealed the improved milk quality and lipid composition in dairy cows supplemented with citrus peel extract. Food Res. Int. 2022, 161, 111767. [Google Scholar] [CrossRef] [PubMed]
- Derakhshani, H.; Fehr, K.B.; Sepehri, S.; Francoz, D.; De Buck, J.; Barkema, H.W.; Plaizier, J.C.; Khafipour, E. Invited review: Microbiota of the bovine udder: Contributing factors and potential implications for udder health and mastitis susceptibility. J. Dairy Sci. 2018, 101, 10605–10625. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Oikonomou, G.; Bicalho, M.L.; Meira, E.; Rossi, R.E.; Foditsch, C.; Machado, V.S.; Teixeira, A.G.; Santisteban, C.; Schukken, Y.H.; Bicalho, R.C. Microbiota of cow’s milk; distinguishing healthy, sub-clinically and clinically diseased quarters. PLoS ONE 2014, 9, e85904. [Google Scholar] [CrossRef] [Green Version]
- Patel, S.H.; Vaidya, Y.H.; Patel, R.J.; Pandit, R.J.; Joshi, C.G.; Kunjadiya, A.P. Culture independent assessment of human milk microbial community in lactational mastitis. Sci. Rep. 2017, 7, 7804. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhu, D.; Kebede, B.; Chen, G.; McComb, K.; Frew, R. Changes in milk metabolome during the lactation of dairy cows based on 1H NMR and UHPLC–QToF/MS. Int. Dairy J. 2020, 111, 104836. [Google Scholar] [CrossRef]
- Zhu, D.; Kebede, B.; McComb, K.; Hayman, A.; Chen, G.; Frew, R. Milk biomarkers in relation to inherent and external factors based on metabolomics. Trends Food Sci. Tech. 2021, 109, 51–64. [Google Scholar] [CrossRef]
- Sundekilde, U.K.; Poulsen, N.A.; Larsen, L.B.; Bertram, H.C. Nuclear magnetic resonance metabonomics reveals strong association between milk metabolites and somatic cell count in bovine milk. J. Dairy Sci. 2013, 96, 290–299. [Google Scholar] [CrossRef] [Green Version]
- Jiang, J.P.; Yan, L.; Shi, Z.; Wang, L.X.; Shan, L.T.; Efferth, T. Hepatoprotective and anti-inflammatory effects of total flavonoids of Qu Zhi Ke (peel of Citrus changshan-huyou) on non-alcoholic fatty liver disease in rats via modulation of NF-kappa B and MAPKs. Phytomedicine 2019, 64, 153082. [Google Scholar] [CrossRef]
- Hou, K.; Tong, J.; Zhang, H.; Gao, S.; Guo, Y.; Niu, H.; Xiong, B.; Jiang, L. Microbiome and metabolic changes in milk in response to artemisinin supplementation in dairy cows. AMB Express 2020, 10, 154. [Google Scholar] [CrossRef]
- Zhang, Y.; He, L.; Chen, Y.; Xiong, B.; Wang, H.; Jiang, L. Effects of seabuckthorn flavonoids on performance, milk bioactive components and serum biochemical and antioxidant indexes of Holstein cows in mid-lactation. Chin. J. Anim. Nutr. 2022, 34, 3666–3675. [Google Scholar] [CrossRef]
- Wang, B.; Tu, Y.; Zhao, S.P.; Hao, Y.H.; Liu, J.X.; Liu, F.H.; Xiong, B.H.; Jiang, L.S. Effect of tea saponins on milk performance, milk fatty acids, and immune function in dairy cow. J. Dairy Sci. 2017, 100, 8043–8052. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Castro, I.; Alba, C.; Aparicio, M.; Arroyo, R.; Jiménez, L.; Fernández, L.; Arias, R.; Rodríguez, J.M. Metataxonomic and immunological analysis of milk from ewes with or without a history of mastitis. J. Dairy Sci. 2019, 102, 9298–9311. [Google Scholar] [CrossRef] [PubMed]
- Li, L.; Niu, H.; Zhan, J.; Tu, Y.; Jiang, L.; Zhao, Y. Matrine attenuates bovine mammary epithelial cells inflammatory responses induced by Streptococcus agalactiae through inhibiting NF-κB and MAPK signaling pathways. Int. Immunopharmacol. 2022, 112, 109206. [Google Scholar] [CrossRef] [PubMed]
- Cremonesi, P.; Ceccarani, C.; Curone, G.; Severgnini, M.; Pollera, C.; Bronzo, V.; Riva, F.; Addis, M.F.; Filipe, J.; Amadori, M.; et al. Milk microbiome diversity and bacterial group prevalence in a comparison between healthy Holstein Friesian and Rendena cows. PLoS ONE 2018, 13, e0205054. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gill, H.S.; Doull, F.; Rutherfurd, K.J.; Cross, M.L. Immunoregulatory peptides in bovine milk. Brit. J. Nutr. 2000, 84, S111–S117. [Google Scholar] [CrossRef]
- Newburg, D.S. Innate immunity and human milk. J. Nutr. 2005, 135, 1308–1312. [Google Scholar] [CrossRef] [Green Version]
- Oviedo-Boyso, J.; Valdez-Alarcon, J.J.; Cajero-Juarez, M.; Ochoa-Zarzosa, A.; Lopez-Meza, J.E.; Bravo-Patino, A.; Baizabal-Aguirre, V.M. Innate immune response of bovine mammary gland to pathogenic bacteria responsible for mastitis. J. Infect. 2007, 54, 399–409. [Google Scholar] [CrossRef]
- Burmanczuk, A.; Wojciechowska, B.; Gbylik-Sikorska, M.; Gajda, A.; Markiewicz, W.; Sosin, E.; Grabowski, T. The effect of hesperidin, chrysin, and naringenin on the number of somatic cell count in mastitis in dairy cows after multiple intramammary administration. Ann. Anim. Sci. 2022, 22, 155–172. [Google Scholar] [CrossRef]
- Boersma, B.J.; Patel, R.P.; Kirk, M.; Jackson, P.L.; Muccio, D.; Darley-Usmar, V.M.; Barnes, S. Chlorination and nitration of soy isoflavones. Arch. Biochem. Biophys. 1999, 368, 265–275. [Google Scholar] [CrossRef]
- Khosravi, M.; Rouzbehan, Y.; Rezaei, M.; Rezaei, J. Total replacement of corn silage with sorghum silage improves milk fatty acid profile and antioxidant capacity of Holstein dairy cows. J. Dairy Sci. 2018, 101, 10953–10961. [Google Scholar] [CrossRef]
- Tian, X.Z.; Paengkoum, P.; Paengkoum, S.; Chumpawadee, S.; Ban, C.; Thongpea, S. Short communication: Purple corn (Zea mays L.) stover silage with abundant anthocyanins transferring anthocyanin composition to the milk and increasing antioxidant status of lactating dairy goats. J. Dairy Sci. 2019, 102, 413–418. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Teng, Z.W.; Yang, G.Q.; Wang, L.F.; Fu, T.; Lian, H.X.; Sun, Y.; Han, L.Q.; Zhang, L.Y.; Gao, T.Y. Effects of the circadian rhythm on milk composition in dairy cows: Does day milk differ from night milk? J. Dairy Sci. 2021, 104, 8301–8313. [Google Scholar] [CrossRef] [PubMed]
- Park, H.; Lee, M.; Kim, K.-T.; Park, E.; Paik, H.-D. Antioxidant and antigenotoxic effect of dairy products supplemented with red ginseng extract. J. Dairy Sci. 2018, 101, 8702–8710. [Google Scholar] [CrossRef] [Green Version]
- Zhang, R.Y.; Huo, W.J.; Zhu, W.Y.; Mao, S.Y. Characterization of bacterial community of raw milk from dairy cows during subacute ruminal acidosis challenge by high-throughput sequencing. J Sci. Food Agric. 2015, 95, 1072–1079. [Google Scholar] [CrossRef] [PubMed]
- Thiery, S.; Kaimer, C. The Predation Strategy of Myxococcus xanthus. Front. Microbiol. 2020, 11, 2. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Almeida, A.; Nayfach, S.; Boland, M.; Strozzi, F.; Beracochea, M.; Shi, Z.J.; Pollard, K.S.; Sakharova, E.; Parks, D.H.; Hugenholtz, P.; et al. A unified catalog of 204,938 reference genomes from the human gut microbiome. Nat. Biotechnol. 2021, 39, 105–114. [Google Scholar] [CrossRef] [PubMed]
- Keefe, G. Update on Control of Staphylococcus aureus and Streptococcus agalactiae for management of mastitis. Vet. Clin. N. Am. Food Anim. Pract. 2012, 28, 203–216. [Google Scholar] [CrossRef] [PubMed]
- Kuehn, J.S.; Gorden, P.J.; Munro, D.; Rong, R.; Dong, Q.; Plummer, P.J.; Wang, C.; Phillips, G.J. Bacterial community profiling of milk samples as a means to understand culture-negative bovine clinical mastitis. PLoS ONE 2013, 8, e61959. [Google Scholar] [CrossRef] [Green Version]
- Yamamura, S.; Morita, Y.; Hasan, Q.; Yokoyama, K.; Tamiya, E. Keratin degradation: A cooperative action of two enzymes from Stenotrophomonas sp. Biochem. Bioph. Res. Commun. 2002, 294, 1138–1143. [Google Scholar] [CrossRef]
- Kromker, V.; Friedrich, J. Teat canal closure in non-lactating heifers and its association with udder health in the consecutive lactation. Vet. Microbiol. 2009, 134, 100–105. [Google Scholar] [CrossRef]
- Gowd, V.; Karim, N.; Shishir, M.R.I.; Xie, L.H.; Chen, W. Dietary polyphenols to combat the metabolic diseases via altering gut microbiota. Trends Food Sci. Technol. 2019, 93, 81–93. [Google Scholar] [CrossRef]
- Ghosh, S.; Sadowsky, M.J.; Roberts, M.C.; Gralnick, J.A.; LaPara, T.M. Sphingobacterium sp strain PM2-P1-29 harbours a functional tet(X) gene encoding for the degradation of tetracycline. J. Appl. Microbiol. 2009, 106, 1336–1342. [Google Scholar] [CrossRef] [PubMed]
- Rogier, E.W.; Frantz, A.L.; Bruno, M.E.C.; Wedlund, L.; Cohen, D.A.; Stromberg, A.J.; Kaetzel, C.S. Secretory antibodies in breast milk promote long-term intestinal homeostasis by regulating the gut microbiota and host gene expression. Proc. Natl. Acad. Sci. USA 2014, 111, 3074–3079. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ercolini, D.; Russo, F.; Ferrocino, I.; Villani, F. Molecular identification of mesophilic and psychrotrophic bacteria from raw cow’s milk. Food Microbiol. 2009, 26, 228–231. [Google Scholar] [CrossRef]
- Hantsis-Zacharov, E.; Halpern, M. Culturable psychrotrophic bacterial communities in raw milk and their proteolytic and lipolytic traits. Appl. Environ. Microbiol. 2007, 73, 7162–7168. [Google Scholar] [CrossRef] [Green Version]
- Gladine, C.; Rock, E.; Morand, C.; Bauchart, D.; Durand, D. Bioavailability and antioxidant capacity of plant extracts rich in polyphenols, given as a single acute dose, in sheep made highly susceptible to lipoperoxidation. Brit. J. Nutr. 2007, 98, 691–701. [Google Scholar] [CrossRef] [Green Version]
- Griffith, O.W. β-amino acids: Mammalian metabolism and utility as α-amino acid analogues. Annu. Rev. Biochem. 1986, 55, 855–878. [Google Scholar] [CrossRef]
- Smith, A.E.; Stout, J.R.; Kendall, K.L.; Fukuda, D.H.; Cramer, J.T. Exercise-induced oxidative stress: The effects of β-alanine supplementation in women. Amino Acids 2012, 43, 77–90. [Google Scholar] [CrossRef]
- Lee, J.S.; Kim, J.H.; Bae, J.S.; Kim, J.Y.; Park, T.J.; Pasaje, C.F.; Park, B.L.; Cheong, H.S.; Park, J.S.; Uh, S.T.; et al. Association analysis of UBE3C polymorphisms in Korean aspirin-intolerant asthmatic patients. Ann. Allerg. Asthma Immunol. 2010, 105, 307–312. [Google Scholar] [CrossRef]
- Brown, E.M.; Ke, X.B.; Hitchcock, D.; Jeanfavre, S.; Avila-Pacheco, J.; Nakata, T.; Arthur, T.D.; Fornelos, N.; Heim, C.; Franzosa, E.A.; et al. Bacteroides-derived sphingolipids are critical for maintaining intestinal homeostasis and symbiosis. Cell Host Microbe 2019, 25, 668–680. [Google Scholar] [CrossRef]
- Gaggini, M.; Ndreu, R.; Michelucci, E.; Rocchiccioli, S.; Vassalle, C. Ceramides as mediators of oxidative stress and inflammation in cardiometabolic disease. Int. J. Mol. Sci. 2022, 23, 2719. [Google Scholar] [CrossRef] [PubMed]
| Item 1 | CPE Supplementation, g/d | SEM | p-Value 2 | |||||
|---|---|---|---|---|---|---|---|---|
| 0 | 50 | 100 | 150 | T | L | Q | ||
| IgA, mg/mL | 0.96 | 1.04 | 1.03 | 1.03 | 0.037 | 0.277 | 0.168 | 0.241 |
| IgG, mg/mL | 4.38 | 4.39 | 4.64 | 4.59 | 0.161 | 0.476 | 0.185 | 0.844 |
| TNF-α, pg/mL | 79.25 | 84.61 | 83.17 | 84.61 | 3.780 | 0.693 | 0.385 | 0.601 |
| IL-1β, pg/mL | 71.10 a | 65.87 b | 65.28 b | 67.20 b | 1.240 | 0.010 | 0.045 | 0.011 |
| IL-6, pg/mL | 165.32 a | 166.66 a | 153.22 b | 153.88 b | 3.241 | 0.008 | 0.006 | 0.925 |
| IL-8, pg/mL | 67.65 | 65.20 | 63.37 | 64.99 | 1.262 | 0.117 | 0.088 | 0.112 |
| IFN-γ, pg/mL | 47.33 | 44.31 | 46.22 | 45.00 | 1.050 | 0.178 | 0.314 | 0.422 |
| DPPH scavenging activity, % | 19.86 b | 20.27 b | 22.59 ab | 23.39 a | 0.954 | 0.030 | 0.006 | 0.841 |
| CAT, U/mL | 215.69 | 216.27 | 216.03 | 214.19 | 1.970 | 0.867 | 0.599 | 0.550 |
| MDA, ng/mL | 8.56 a | 8.23 ab | 8.02 b | 7.84 b | 0.164 | 0.031 | 0.017 | 0.714 |
| LZM, μg/mL | 10.87 b | 11.53 ab | 13.43 a | 13.54 a | 0.756 | 0.020 | 0.003 | 0.697 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhao, Y.; Yu, S.; Zhang, S.; Li, Y.; Tu, Y.; Liu, M.; Jiang, L. Microbiome-Metabolomics Insights into the Milk of Lactating Dairy Cows to Reveal the Health-Promoting Effects of Dietary Citrus Peel Extracts on the Mammary Metabolism. Foods 2022, 11, 4119. https://doi.org/10.3390/foods11244119
Zhao Y, Yu S, Zhang S, Li Y, Tu Y, Liu M, Jiang L. Microbiome-Metabolomics Insights into the Milk of Lactating Dairy Cows to Reveal the Health-Promoting Effects of Dietary Citrus Peel Extracts on the Mammary Metabolism. Foods. 2022; 11(24):4119. https://doi.org/10.3390/foods11244119
Chicago/Turabian StyleZhao, Yuchao, Shiqiang Yu, Shuyue Zhang, Yuqin Li, Yan Tu, Ming Liu, and Linshu Jiang. 2022. "Microbiome-Metabolomics Insights into the Milk of Lactating Dairy Cows to Reveal the Health-Promoting Effects of Dietary Citrus Peel Extracts on the Mammary Metabolism" Foods 11, no. 24: 4119. https://doi.org/10.3390/foods11244119





