A Review of Isothermal Amplification Methods and Food-Origin Inhibitors against Detecting Food-Borne Pathogens
Abstract
:1. Introduction
2. Isothermal Amplification Methods
3. Inhibitors That Originate from Food during the Isothermal Amplification Process
4. Inhibitors Originating from Isothermal Amplification Processes
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Osaili, T.M.; Hasan, F.; Dhanasekaran, D.K.; Obaid, R.S.; Al-Nabulsi, A.A.; Ayyash, M.; Karam, L.; Savvaidis, I.N.; Holley, R. Effect of active essential oils added to chicken tawook on the behaviour of Listeria monocytogenes, Salmonella spp. and Escherichia coli O157: H7 during storage. Int. J. Food Microbiol. 2021, 337, 108947. [Google Scholar] [CrossRef] [PubMed]
- Chai, C.; Oh, S.W. Electrochemical impedimetric biosensors for food safety. Food Sci. Biotechnol. 2020, 29, 879–887. [Google Scholar] [CrossRef] [PubMed]
- Kim, J.H.; Oh, S.W. Rapid and sensitive detection of E. coli O157: H7 and S. Typhimurium in iceberg lettuce and cabbage using filtration, DNA concentration, and qPCR without enrichment. Food Chem. 2020, 327, 127036. [Google Scholar] [CrossRef] [PubMed]
- Rajapaksha, P.; Elbourne, A.; Gangadoo, S.; Brown, R.; Cozzolino, D.; Chapman, J. A review of methods for the detection of pathogenic microorganisms. Analyst 2019, 144, 396–411. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Qi, W.; Wang, L.; Lin, J.; Liu, Y. Magnetic Bead Chain-Based Continuous-Flow DNA Extraction for Microfluidic PCR Detection of Salmonella. Micromachines 2021, 12, 384. [Google Scholar] [CrossRef]
- Tabatabaei, M.S.; Islam, R.; Ahmed, M. Applications of gold nanoparticles in ELISA, PCR, and immuno-PCR assays: A review. Anal. Chim. Acta 2021, 1143, 250–266. [Google Scholar] [CrossRef]
- Liu, Z.; Sun, J.; Zhao, G.; Xiong, S.; Ma, Y.; Zheng, M. Transient stem-loop structure of nucleic acid template may interfere with polymerase chain reaction through endonuclease activity of Taq DNA polymerase. Gene 2021, 764, 145095. [Google Scholar] [CrossRef]
- Zhang, H.; Li, H.; Zhu, H.; Pekárek, J.; Podešva, P.; Chang, H.; Neužil, P. Revealing the secrets of PCR. Sens. Actuators B 2019, 298, 126924. [Google Scholar] [CrossRef]
- Asadi, R.; Mollasalehi, H. The mechanism and improvements to the isothermal amplification of nucleic acids, at a glance. Anal. Biochem. 2021, 631, 114260. [Google Scholar] [CrossRef]
- Kumar, Y. Isothermal amplification-based methods for assessment of microbiological safety and authenticity of meat and meat products. Food Control 2021, 121, 107679. [Google Scholar] [CrossRef]
- Barreda-García, S.; Miranda-Castro, R.; de-Los-Santos-Álvarez, N.; Miranda-Ordieres, A.J.; Lobo-Castañón, M.J. Helicase-dependent isothermal amplification: A novel tool in the development of molecular-based analytical systems for rapid pathogen detection. Anal. Bioanal. Chem. 2018, 410, 679–693. [Google Scholar] [CrossRef] [PubMed]
- Hu, J.; Wang, Y.; Su, H.; Ding, H.; Sun, X.; Gao, H.; Geng, Y.; Wang, Z. Rapid analysis of Escherichia coli O157: H7 using isothermal recombinase polymerase amplification combined with triple-labeled nucleotide probes. Mol. Cell Probes 2020, 50, 101501. [Google Scholar] [CrossRef]
- Huang, M.; Yang, F.; Fu, J.; Xiao, P.; Tu, J.; Lu, Z. Reaction parameter comparison and optimization of multiple displacement amplification. Anal. Methods 2020, 12, 46–53. [Google Scholar] [CrossRef]
- Milton, A.A.P.; Momin, K.M.; Priya, G.B.; Ghatak, S.; Gandhale, P.N.; Angappan, M.; Das, S.; Sen, A. A novel in situ methodology for visual detection of Clostridium perfringens in pork harnessing saltatory rolling circle amplification. Anaerobe 2021, 69, 102324. [Google Scholar] [CrossRef] [PubMed]
- Vichaibun, V.; Kanchanaphum, P. Quantitative LAMP and PCR detection of Salmonella in chicken samples collected from local markets around Pathum Thani Province, Thailand. Int. J. Food Sci. 2020, 8833173. [Google Scholar] [CrossRef]
- Zhai, L.; Liu, H.; Chen, Q.; Lu, Z.; Zhang, C.; Lv, F.; Bie, X. Development of a real-time nucleic acid sequence–based amplification assay for the rapid detection of Salmonella spp. from food. Braz. J. Microbiol. 2019, 50, 255–261. [Google Scholar] [CrossRef]
- Srimongkol, G.; Ditmangklo, B.; Choopara, I.; Thaniyavarn, J.; Dean, D.; Kokpol, S.; Vilaivan, T.; Somboonna, N. Rapid colorimetric loop-mediated isothermal amplification for hypersensitive point-of-care Staphylococcus aureus enterotoxin A gene detection in milk and pork products. Sci. Rep. 2020, 10, 1–11. [Google Scholar]
- Chin, N.A.; Salihah, N.T.; Shivanand, P.; Ahmed, M.U. Recent trends and developments of PCR-based methods for the detection of food-borne Salmonella bacteria and Norovirus. J. Food Sci. Technol. 2021, 57, 1–13. [Google Scholar] [CrossRef]
- Schrader, C.; Schielke, A.; Ellerbroek, L.; Johne, R. PCR inhibitors–occurrence, properties and removal. J. Appl. Microbiol. 2012, 113, 1014–1026. [Google Scholar] [CrossRef]
- Kant, K.; Shahbazi, M.A.; Dave, V.P.; Ngo, T.A.; Chidambara, V.A.; Than, L.Q.; Bang, D.D.; Wolff, A. Microfluidic devices for sample preparation and rapid detection of foodborne pathogens. Biotechnol. Adv. 2018, 36, 1003–1024. [Google Scholar] [CrossRef] [Green Version]
- Sidstedt, M.; Rådström, P.; Hedman, J. PCR inhibition in qPCR, dPCR and MPS—mechanisms and solutions. Anal. Bioanal. Chem. 2020, 412, 2009–2023. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, X.; Lowe, S.B.; Gooding, J.J. Brief review of monitoring methods for loop-mediated isothermal amplification (LAMP). Biosens. Bioelectron. 2014, 61, 491–499. [Google Scholar] [CrossRef]
- Becherer, L.; Borst, N.; Bakheit, M.; Frischmann, S.; Zengerle, R.; von Stetten, F.V. Loop-mediated isothermal amplification (LAMP)–review and classification of methods for sequence-specific detection. Anal. Methods 2020, 12, 717–746. [Google Scholar] [CrossRef] [Green Version]
- Tao, Y.; Yun, J.; Wang, J.; Xu, P.; Li, C.; Liu, H.; Lan, Y.; Pan, J.; Du, W. High-performance detection of Mycobacterium bovis in milk using digital LAMP. Food Chem. 2020, 327, 126945. [Google Scholar] [CrossRef] [PubMed]
- Meagher, R.J.; Priye, A.; Light, Y.K.; Huang, C.; Wang, E. Impact of primer dimers and self-amplifying hairpins on reverse transcription loop-mediated isothermal amplification detection of viral RNA. Analyst 2018, 143, 1924–1933. [Google Scholar] [CrossRef] [PubMed]
- Fernández-Soto, P.; Mvoulouga, P.O.; Akue, J.P.; Abán, J.L.; Santiago, B.V.; Sánchez, M.C.; Muro, A. Development of a highly sensitive loop-mediated isothermal amplification (LAMP) method for the detection of Loa loa. PLoS ONE 2014, 9, e94664. [Google Scholar] [CrossRef]
- Shahbazi, E.; Mollasalehi, H.; Minai-Tehrani, D. Development and evaluation of an improved quantitative loop-mediated isothermal amplification method for rapid detection of Morganella morganii. Talanta 2019, 191, 54–58. [Google Scholar] [CrossRef]
- Chen, Y.; Li, H.; Yang, L.; Wang, L.; Sun, R.; Shearer, J.E.; Sun, F. Rapid Detection of Clostridium botulinum in Food Using Loop-Mediated Isothermal Amplification (LAMP). Int. J. Environ. Res. Public Health 2021, 18, 4401. [Google Scholar] [CrossRef]
- Ahuja, A.; Somvanshi, V.S. Diagnosis of plant-parasitic nematodes using loop-mediated isothermal amplification (LAMP): A review. J. Crop. Prot. 2021, 147, 105459. [Google Scholar] [CrossRef]
- Leonardo, S.; Toldrà, A.; Campàs, M. Biosensors based on isothermal DNA amplification for bacterial detection in food safety and environmental monitoring. Sensors 2021, 21, 602. [Google Scholar] [CrossRef]
- Lobato, I.M.; O’Sullivan, C.K. Recombinase polymerase amplification: Basics, applications and recent advances. Trends Anal. Chem. 2018, 98, 19–35. [Google Scholar] [CrossRef] [PubMed]
- Lillis, L.; Siverson, J.; Lee, A.; Cantera, J.; Parker, M.; Piepenburg, O.; Lehman, D.A.; Boyle, D.S. Factors influencing recombinase polymerase amplification (RPA) assay outcomes at point of care. Mol. Cell Probes 2016, 30, 74–78. [Google Scholar] [CrossRef] [Green Version]
- Zhai, J.; Wang, L.; Qiao, X.; Zhao, J.; Wang, X.; He, X. Detection of Neisseria gonorrhoeae and Chlamydia trachomatis infections in pregnant women by multiplex recombinase polymerase amplification. PLoS ONE 2021, 16, e0251119. [Google Scholar] [CrossRef] [PubMed]
- Mayboroda, O.; Gonzalez Benito, A.; Sabaté del Rio, J.; Svobodova, M.; Julich, S.; Tomaso, H.; O’Sullivan, C.K.; Katakis, I. Isothermal solid-phase amplification system for detection of Yersinia pestis. Anal. Bioanal. Chem. 2016, 408, 671–676. [Google Scholar] [CrossRef]
- Li, J.; Macdonald, J.; von Stetten, F. Review: A comprehensive summary of a decade development of the recombinase polymerase amplification. Analyst 2018, 144, 31–67. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Du, X.J.; Zhou, T.J.; Li, P.; Wang, S. A rapid Salmonella detection method involving thermophilic helicase dependent amplification and a lateral flow assay. Mol. Cell Probes 2017, 34, 37–44. [Google Scholar] [CrossRef] [PubMed]
- Glökler, J.; Lim, T.S.; Ida, J.; Frohme, M. Isothermal amplifications–a comprehensive review on current methods. Crit Rev. Biochem. Mol. Biol. 2021, 56, 543–586. [Google Scholar] [CrossRef]
- Kumar, A.A.; Hennek, J.W.; Smith, B.S.; Kumar, S.; Beattie, P.; Jain, S.; Rolland, J.P.; Stossel, T.P.; Chunda-Liyoka, C.; Whitesides, G.M. From the bench to the field in low-cost diagnostics: Two case studies. Angew. Chem. Int. Ed. Engl. 2015, 54, 5836–5853. [Google Scholar] [CrossRef]
- Yang, Z.; McLendon, C.; Hutter, D.; Bradley, K.M.; Hoshika, S.; Frye, C.; Benner, S.A. Helicase dependent isothermal amplification of DNA and RNA using self-avoiding molecular recognition systems. ChemBioChem 2015, 16, 1365–1370. [Google Scholar] [CrossRef] [PubMed]
- Thekisoe, O.M.; Bazie, R.S.; Coronel-Servian, A.M.; Sugimoto, C.; Kawazu, S.I.; Inoue, N. Stability of loop-mediated isothermal amplification (LAMP) reagents and its amplification efficiency on crude trypanosome DNA templates. J. Vet. Med. Sci. 2009, 71, 471–475. [Google Scholar] [CrossRef] [Green Version]
- Xu, L.; Duan, J.; Chen, J.; Ding, S.; Cheng, W. Recent advances in rolling circle amplification-based biosensing strategies—A review. Anal. Chim. Acta 2021, 1148, 238187. [Google Scholar] [CrossRef]
- Hønsvall, B.K.; Robertson, L.J. From research lab to standard environmental analysis tool: Will NASBA make the leap? Water Res. 2017, 109, 389–397. [Google Scholar] [CrossRef] [PubMed]
- Brennan, D.; Coughlan, H.; Clancy, E.; Dimov, N.; Barry, T.; Kinahan, D.; Galvin, P. Development of an on-disc isothermal in vitro amplification and detection of bacterial RNA. Sens. Actuators B 2017, 239, 235–242. [Google Scholar] [CrossRef] [Green Version]
- Takahashi, M.K.; Tan, X.; Dy, A.J.; Braff, D.; Akana, R.T.; Furuta, Y.; Collins, J.J. A low-cost paper-based synthetic biology platform for analyzing gut microbiota and host biomarkers. Nat. Commun. 2018, 9, 1–12. [Google Scholar] [CrossRef] [PubMed]
- Li, J.; Macdonald, J. Advances in isothermal amplification: Novel strategies inspired by biological processes. Biosens. Bioelectron. 2015, 64, 196–211. [Google Scholar] [CrossRef] [PubMed]
- Kellner, M.J.; Koob, J.G.; Gootenberg, J.S.; Abudayyeh, O.O.; Zhang, F. SHERLOCK: Nucleic acid detection with CRISPR nucleases. Nat. Protoc. 2019, 14, 2986–3012. [Google Scholar] [CrossRef] [PubMed]
- Pardee, K.; Green, A.A.; Takahashi, M.K.; Braff, D.; Lambert, G.; Lee, J.W.; Ferrante, T.; Ma, D.; Donghia, D.; Fan, M.; et al. Rapid, low-cost detection of Zika virus using programmable biomolecular components. Cell 2016, 165, 1255–1266. [Google Scholar] [CrossRef] [Green Version]
- van Emmerik, C.L.; Gachulincova, I.; Lobbia, V.R.; Daniëls, M.A.; Heus, H.A.; Soufi, A.; Nelissen, F.H.T.; van Ingen, H. Ramified Rolling Circle Amplification for efficient and flexible synthesis of nucleosomal DNA sequences. bioRxiv 2019, 676528. [Google Scholar] [CrossRef] [Green Version]
- Wang, Z.; Yang, Q.; Zhang, Y.; Meng, Z.; Ma, X.; Zhang, W. Saltatory rolling circle amplification (SRCA): A novel nucleic acid isothermal amplification technique applied for rapid detection of Shigella spp. in vegetable salad. Food Anal. Methods 2018, 11, 504–513. [Google Scholar] [CrossRef]
- Spits, C.; Le Caignec, C.; De Rycke, M.; Van Haute, L.; Van Steirteghem, A.; Liebaers, I.; Sermon, K. Optimization and evaluation of single-cell whole-genome multiple displacement amplification. Hum. Mutat. 2006, 27, 496–503. [Google Scholar] [CrossRef]
- Tian, H.C.; Benitez, J.J.; Craighead, H.G. Single cell on-chip whole genome amplification via micropillar arrays for reduced amplification bias. PLoS ONE 2018, 13, e0191520. [Google Scholar] [CrossRef] [Green Version]
- Forghani, F.; Li, S.; Zhang, S.; Mann, D.A.; Deng, X.; den Bakker, H.C.; Diez-Gonzalez, F. Detection and Serotyping of Salmonella and Escherichia coli in Wheat Flour by a Quasimetagenomic Approach Assisted by Magnetic Capture, Multiple Displacement Amplification and Real-Time Sequencing. Appl. Environ. Microbiol. 2020, 86, e00097-20. [Google Scholar] [CrossRef] [PubMed]
- Wang, W.; Ren, Y.; Lu, Y.; Xu, Y.; Crosby, S.D.; Di Bisceglie, A.M.; Fan, X. Template-dependent multiple displacement amplification for profiling human circulating RNA. Biotechniques 2017, 63, 21–27. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jamal, R.; Li, X.; Weidhaas, J. Template length, concentration and guanidine and cytosine content influence on multiple displacement amplification efficiency. J. Microbiol. Methods 2021, 181, 106146. [Google Scholar] [CrossRef] [PubMed]
- Long, N.; Qiao, Y.; Xu, Z.; Tu, J.; Lu, Z. Recent advances and application in whole-genome multiple displacement amplification. Quant. Biol. 2020, 8, 1–16. [Google Scholar] [CrossRef]
- Smith, T.J.; O’Connor, L.; Glennon, M.; Maher, M. Molecular diagnostics in food safety: Rapid detection of food-borne pathogens. Ir. J. Agric. Food Res. 2000, 39, 309–319. [Google Scholar]
- Santiago-Felipe, S.; Tortajada-Genaro, L.A.; Morais, S.; Puchades, R.; Maquieira, Á. Isothermal DNA amplification strategies for duplex microorganism detection. Food Chem. 2015, 174, 509–515. [Google Scholar] [CrossRef]
- Kong, M.; Chen, X.G.; Xing, K.; Park, H.J. Antimicrobial properties of chitosan and mode of action: A state of the art review. Int. J. Food Microbiol. 2010, 144, 51–63. [Google Scholar] [CrossRef]
- Narushima, J.; Kimata, S.; Soga, K.; Sugano, Y.; Kishine, M.; Takabatake, R.; Mano, J.; Kitta, K.; Kanamaru, S.; Shirakawa, N.; et al. Rapid DNA template preparation directly from a rice sample without purification for loop-mediated isothermal amplification (LAMP) of rice genes. Biosci. Biotechnol. Biochem. 2020, 84, 670–677. [Google Scholar] [CrossRef]
- Kalendar, R.; Boronnikova, S.; Seppänen, M. Isolation and purification of DNA from complicated biological samples. Methods Mol. Biol. 2021, 2222, 57–67. [Google Scholar]
- Ramírez, M.H.; Salazar Duque, H.J.; Urrea Trujillo, A.I. Quality of cocoa (Theobroma cacao L.) DNA from foliar tissue at different stages of development. Acta Agron. 2018, 67, 311–318. [Google Scholar] [CrossRef]
- Demeke, T.; Ratnayaka, I.; Phan, A. Effects of DNA extraction and purification methods on real-time quantitative PCR analysis of Roundup Ready soybean. J. AOAC Int. 2009, 92, 1136–1144. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Demeke, T.; Malabanan, J.; Holigroski, M.; Eng, M. Effect of source of DNA on the quantitative analysis of genetically engineered traits using digital PCR and real-time PCR. J. AOAC Int. 2017, 100, 492–498. [Google Scholar] [CrossRef] [PubMed]
- Xu, W. Adaptable methods to extract nucleic acid targets and evaluate quality. In Functional Nucleic Acids Detection in Food Safety; Springer: Singapore, 2016; pp. 17–36. [Google Scholar]
- Suther, C.; Moore, M.D. Quantification and discovery of PCR inhibitors found in food matrices commonly associated with foodborne viruses. Food Sci. Hum. Wellness 2019, 8, 351–355. [Google Scholar] [CrossRef]
- Pang, B.; Yao, S.; Xu, K.; Wang, J.; Song, X.; Mu, Y.; Zhao, C.; Li, J. A novel visual-mixed-dye for LAMP and its application in the detection of foodborne pathogens. Anal. Biochem. 2019, 574, 1–6. [Google Scholar] [CrossRef]
- Jin, Z.; Ding, G.; Li, G.; Yang, G.; Han, Y.; Hao, N.; Deng, J.; Zhang, Y.; Zhang, W.; Li, W. Rapid detection of foodborne bacterial pathogens using visual high-throughput microfluidic chip. J. Chem. Technol. Biotechnol. 2020, 95, 1460–1466. [Google Scholar] [CrossRef]
- Arunrut, N.; Kiatpathomchai, W.; Ananchaipattana, C. Development and evaluation of real-time loop mediated isothermal amplification assay for rapid and sensitive detection of Salmonella spp. in chicken meat products. J. Food Saf. 2018, 38, e12564. [Google Scholar] [CrossRef]
- Fang, J.; Wu, Y.; Qu, D.; Ma, B.; Yu, X.; Zhang, M.; Han, J. Propidium monoazide real-time loop-mediated isothermal amplification for specific visualization of viable Salmonella in food. Lett. Appl. Microbiol. 2018, 67, 79–88. [Google Scholar] [CrossRef]
- Mei, X.; Zhai, X.; Lei, C.; Ye, X.; Kang, Z.; Wu, X.; Xiang, R.; Wang, Y.; Wang, H. Development and application of a visual loop-mediated isothermal amplification combined with lateral flow dipstick (LAMP-LFD) method for rapid detection of Salmonella strains in food samples. Food Control 2019, 104, 9–19. [Google Scholar] [CrossRef]
- Kim, J.Y.; Lee, J.L. Rapid detection of Salmonella enterica serovar enteritidis from eggs and chicken meat by real-time recombinase polymerase amplification in comparison with the two-step real-time PCR. J. Food Saf. 2016, 36, 402–411. [Google Scholar] [CrossRef]
- Chen, J.; Liu, X.; Chen, J.; Guo, Z.; Wang, Y.; Chen, G.; Chen, X.; Yan, Q.; Yang, P.; Li, R.; et al. Development of a rapid test method for Salmonella enterica detection-based on fluorescence probe-based recombinase polymerase amplification. Food Anal. Methods 2019, 12, 1791–1798. [Google Scholar] [CrossRef]
- Zhao, L.; Wang, J.; Sun, X.X.; Wang, J.; Chen, Z.; Xu, X.; Dong, M.; Guo, Y.N.; Wang, Y.; Chen, P.; et al. Development and evaluation of the rapid and sensitive RPA assays for specific detection of Salmonella spp. in food samples. Front. Cell Infect. Microbiol. 2021, 11, 631921. [Google Scholar] [CrossRef]
- D’souza, D.H.; Jaykus, L.A. Nucleic acid sequence-based amplification for the rapid and sensitive detection of Salmonella enterica from foods. J. Appl. Microbiol. 2003, 95, 1343–1350. [Google Scholar] [CrossRef]
- Lv, X.; Wang, L.; Zhang, J.; Zeng, H.; Chen, X.; Shi, L.; Cui, H.; He, X.; Zhao, L. Rapid and sensitive detection of VBNC Escherichia coli O157: H7 in beef by PMAxx and real-time LAMP. Food Control 2020, 115, 107292. [Google Scholar] [CrossRef]
- Kim, J.H.; Oh, S.W. Development of a filtration-based LAMP–LFA method as sensitive and rapid detection of E. coli O157: H7. J. Food Sci. Technol. 2019, 56, 2576–2583. [Google Scholar] [CrossRef] [PubMed]
- Azinheiro, S.; Carvalho, J.; Prado, M.; Garrido-Maestu, A. Correction: Azinheiro et al. Multiplex Detection of Salmonella spp., E. coli O157 and L. monocytogenes by qPCR Melt Curve Analysis in Spiked Infant Formula. Microorganisms 2020, 8, 1359. [Google Scholar] [CrossRef] [PubMed]
- Wachiralurpan, S.; Sriyapai, T.; Areekit, S.; Sriyapai, P.; Augkarawaritsawong, S.; Santiwatanakul, S.; Chansiri, K. Rapid colorimetric assay for detection of Listeria monocytogenes in food samples using LAMP formation of DNA concatemers and gold nanoparticle-DNA probe complex. Front. Chem. 2018, 6, 90. [Google Scholar] [CrossRef] [Green Version]
- Gao, W.; Huang, H.; Zhang, Y.; Zhu, P.; Yan, X.; Fan, J.; Chen, X. Recombinase polymerase amplification-based assay for rapid detection of Listeria monocytogenes in food samples. Food Anal. Methods 2017, 10, 1972–1981. [Google Scholar] [CrossRef]
- Du, X.J.; Zang, Y.X.; Liu, H.B.; Li, P.; Wang, S. Recombinase polymerase amplification combined with lateral flow strip for Listeria monocytogenes detection in food. J. Food Sci. 2018, 83, 1041–1047. [Google Scholar] [CrossRef] [PubMed]
- Wang, L.; Zhao, P.; Si, X.; Li, J.; Dai, X.; Zhang, K.; Gao, S.; Dong, J. Rapid and specific detection of Listeria monocytogenes with an isothermal amplification and lateral flow strip combined method that eliminates false-positive signals from primer-dimers. Front. Microbiol. 2020, 10, 2959. [Google Scholar] [CrossRef] [PubMed]
- Uyttendaele, M.; Schukkink, R.; Van Gemen, B.; Debevere, J. Development of NASBA®, a nucleic acid amplification system, for identification of Listeria monocytogenes and comparison to ELISA and a modified FDA method. Int. J. Food Microbiol. 1995, 27, 77–89. [Google Scholar] [CrossRef]
- Prompamorn, P.; Sithigorngul, P.; Rukpratanporn, S.; Longyant, S.; Sridulyakul, P.; Chaivisuthangkura, P. The development of loop-mediated isothermal amplification combined with lateral flow dipstick for detection of Vibrio parahaemolyticus. Appl. Microbiol. 2011, 52, 344–351. [Google Scholar] [CrossRef] [PubMed]
- Simpkins, S.A.; Chan, A.B.; Hays, J.; Pöpping, B.; Cook, N. An RNA transcription-based amplification technique (NASBA) for the detection of viable Salmonella enterica. Lett. Appl. Microbiol. 2000, 30, 75–79. [Google Scholar] [CrossRef] [PubMed]
- Chen, X.; Wu, X.; Gan, M.; Xu, F.; He, L.; Yang, D.; Xu, H.; Shah, N.P.; Wei, H. Rapid detection of Staphylococcus aureus in dairy and meat foods by combination of capture with silica-coated magnetic nanoparticles and thermophilic helicase-dependent isothermal amplification. J. Dairy Sci. 2015, 98, 1563–1570. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gao, W.; Huang, H.; Zhu, P.; Yan, X.; Fan, J.; Jiang, J.; Xu, J. Recombinase polymerase amplification combined with lateral flow dipstick for equipment-free detection of Salmonella in shellfish. Bioprocess Biosyst. Eng. 2018, 41, 603–611. [Google Scholar] [CrossRef]
- Ma, B.; Li, J.; Chen, K.; Yu, X.; Sun, C.; Zhang, M. Multiplex recombinase polymerase amplification assay for the simultaneous detection of three foodborne pathogens in seafood. Foods 2020, 9, 278. [Google Scholar] [CrossRef] [Green Version]
- Garrido-Maestu, A.; Azinheiro, S.; Carvalho, J.; Prado, M. Combination of immunomagnetic separation and real-time recombinase polymerase amplification (IMS-qRPA) for specific detection of Listeria monocytogenes in smoked salmon samples. J. Food Sci. 2019, 84, 1881–1887. [Google Scholar] [CrossRef]
- Jiang, W.; Ren, Y.; Han, X.; Xue, J.; Shan, T.; Chen, Z.; Liu, Y.; Wang, Q. Recombinase polymerase amplification-lateral flow (RPA-LF) assay combined with immunomagnetic separation for rapid visual detection of Vibrio parahaemolyticus in raw oysters. Anal. Bioanal. Chem. 2020, 412, 2903–2914. [Google Scholar] [CrossRef]
- Cao, X.; Zhao, L.; Zhang, J.; Chen, X.; Shi, L.; Fang, X.; Xie, H.; Chang, Y.; Wang, L. Detection of viable but nonculturable Vibrio parahaemolyticus in shrimp samples using improved real-time PCR and real-time LAMP methods. Food Control 2019, 103, 145–152. [Google Scholar] [CrossRef]
- Zhu, P.; Gao, W.; Huang, H.; Jiang, J.; Chen, X.; Fan, J.; Yan, X. Rapid detection of Vibrio parahaemolyticus in shellfish by real-time recombinase polymerase amplification. Food Anal. Methods 2018, 11, 2076–2084. [Google Scholar] [CrossRef]
- Zhang, M.; Liu, J.; Shen, Z.; Liu, Y.; Song, Y.; Liang, Y.; Li, Z.; Nie, L.; Fang, Y.; Zhao, Y. A newly developed paper embedded microchip based on LAMP for rapid multiple detections of foodborne pathogens. BMC Microbiol. 2021, 21, 197. [Google Scholar] [CrossRef]
- Wang, Y.; Li, H.; Li, D.; Li, K.; Wang, Y.; Xu, J.; Ye, C. Multiple cross displacement amplification combined with gold nanoparticle-based lateral flow biosensor for detection of Vibrio parahaemolyticus. Front. Microbiol. 2016, 7, 2047. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Anupama, K.P.; Nayak, A.; Karunasagar, I.; Maiti, B. Rapid visual detection of Vibrio parahaemolyticus in seafood samples by loop-mediated isothermal amplification with hydroxynaphthol blue dye. World J. Microbiol. Biotechnol. 2020, 36, 76. [Google Scholar] [CrossRef] [PubMed]
- Choi, G.; Jung, J.H.; Park, B.H.; Oh, S.J.; Seo, J.H.; Choi, J.S.; Seo, T.S. A centrifugal direct recombinase polymerase amplification (direct-RPA) microdevice for multiplex and real-time identification of food poisoning bacteria. Lab Chip 2016, 16, 2309–2316. [Google Scholar] [CrossRef] [PubMed]
- Cai, Q.; Wang, R.; Qiao, Z.; Yang, W. Single-digit Salmonella detection with the naked eye using bio-barcode immunoassay coupled with recombinase polymerase amplification and a CRISPR-Cas12a system. Analyst 2021, 146, 5271–5279. [Google Scholar] [CrossRef]
- Rani, A.; Ravindran, V.B.; Surapaneni, A.; Shahsavari, E.; Haleyur, N.; Mantri, N.; Ball, A.S. Evaluation and comparison of recombinase polymerase amplification coupled with lateral-flow bioassay for Escherichia coli O157: H7 detection using different genes. Sci. Rep. 2021, 11, 1881. [Google Scholar] [CrossRef] [PubMed]
- Garrido-Maestu, A.; Azinheiro, S.; Fuciños, P.; Carvalho, J.; Prado, M. Comparative study of multiplex real-time recombinase polymerase amplification and ISO 11290-1 methods for the detection of Listeria monocytogenes in dairy products. Food Microbiol. 2020, 92, 103570. [Google Scholar] [CrossRef]
- O’Grady, J.; Lacey, K.; Glynn, B.; Smith, T.J.; Barry, T.; Maher, M. tmRNA—A novel high-copy-number RNA diagnostic target–its application for Staphylococcus aureus detection using real-time NASBA. FEMS Microbiol. Lett. 2009, 301, 218–223. [Google Scholar] [CrossRef] [Green Version]
- Clancy, E.; Higgins, O.; Forrest, M.S.; Boo, T.W.; Cormican, M.; Barry, T.; Piepenburg, O.; Smith, T.J. Development of a rapid recombinase polymerase amplification assay for the detection of Streptococcus pneumoniae in whole blood. BMC Infect. Dis. 2015, 15, 481. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Raghavendra, M.P.; Kumar, P.R.; Prakash, V. Mechanism of inhibition of rice bran lipase by polyphenols: A case study with chlorogenic acid and caffeic acid. J. Food Sci. 2007, 72, E412–E419. [Google Scholar] [CrossRef]
- Japelaghi, R.H.; Haddad, R.; Garoosi, G.A. Rapid and efficient isolation of high-quality nucleic acids from plant tissues rich in polyphenols and polysaccharides. Mol. Biotechnol. 2011, 49, 129–137. [Google Scholar] [CrossRef] [PubMed]
- Opel, K.L.; Chung, D.; McCord, B.R. A study of PCR inhibition mechanisms using real time PCR. J. Forensic. Sci. 2010, 55, 25–33. [Google Scholar] [CrossRef] [PubMed]
- Krause, D.O.; Smith, W.J.; Mcsweeney, C.S. Extraction of microbial DNA from rumen contents containing plant tannins. Biotechniques 2001, 31, 294–298. [Google Scholar] [PubMed] [Green Version]
- Sharma, P.; Purohit, S.D. An improved method of DNA isolation from polysaccharide rich leaves of Boswellia serrata Roxb. Indian J. Biotechnol. 2012, 11, 67–71. [Google Scholar]
- Moser, C.; Gatto, P.; Moser, M.; Pindo, M.; Velasco, R. Isolation of functional RNA from small amounts of different grape and apple tissues. Mol. Biotechnol. 2004, 26, 95–100. [Google Scholar] [CrossRef]
- Barreda-García, S.; Miranda-Castro, R.; de-Los-Santos-Álvarez, N.; Miranda-Ordieres, A.J.; Lobo-Castañón, M.J. Comparison of isothermal helicase-dependent amplification and PCR for the detection of Mycobacterium tuberculosis by an electrochemical genomagnetic assay. Anal. Bioanal. Chem. 2016, 408, 8603–8610. [Google Scholar] [CrossRef]
- Munawar, M.A.; Martin, F.; Toljamo, A.; Kokko, H.; Oksanen, E. RPA-PCR couple: An approach to expedite plant diagnostics and overcome PCR inhibitors. BioTechniques 2020, 69, 270–280. [Google Scholar] [CrossRef]
- Doseeva, V.; Forbes, T.; Wolff, J.; Khripin, Y.; O’Neil, D.; Rothmann, T.; Nazarenko, I. Multiplex isothermal helicase-dependent amplification assay for detection of Chlamydia trachomatis and Neisseria gonorrhoeae. Diagn. Microbiol. Infect. Dis 2011, 71, 354–365. [Google Scholar] [CrossRef]
- Murakami, T.; Sumaoka, J.; Komiyama, M. Sensitive isothermal detection of nucleic-acid sequence by primer generation-rolling circle amplification. Nucleic Acids Res. 2009, 37, e19. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Paez, J.G.; Lin, M.; Beroukhim, R.; Lee, J.C.; Zhao, X.; Richter, D.J.; Gabriel, S.; Herman, P.; Sasaki, H.; Altshuler, D.; et al. Genome coverage and sequence fidelity of ϕ29 polymerase-based multiple strand displacement whole genome amplification. Nucleic Acids Res. 2004, 32, e71. [Google Scholar] [CrossRef]
- Sidoti, F.; Bergallo, M.; Terlizzi, M.E.; Piasentin Alessio, E.P.; Astegiano, S.; Gasparini, G.; Cavallo, R. Development of a quantitative real-time nucleic acid sequence-based amplification assay with an internal control using molecular beacon probes for selective and sensitive detection of human rhinovirus serotypes. Mol. Biotechnol. 2012, 50, 221–228. [Google Scholar] [CrossRef] [PubMed]
- Feng, J.; Dai, Z.; Tian, X.; Jiang, X. Detection of Listeria monocytogenes based on combined aptamers magnetic capture and loop-mediated isothermal amplification. Food Control 2018, 85, 443–452. [Google Scholar] [CrossRef]
- Zhang, X.; Qu, K.; Li, Q.; Cui, Z.; Zhao, J.; Sun, X. Recording the reaction process of loop-mediated isothermal amplification (LAMP) by monitoring the voltammetric response of 2′-deoxyguanosine 5′-triphosphate. Electroanalysis 2011, 23, 2438–2445. [Google Scholar] [CrossRef]
- Wastling, S.L.; Picozzi, K.; Kakembo, A.S.; Welburn, S.C. LAMP for human African trypanosomiasis: A comparative study of detection formats. PLoS Negl. Trop. Dis. 2010, 4, e865. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Deféver, T.; Druet, M.; Evrard, D.; Marchal, D.; Limoges, B. Real-time electrochemical PCR with a DNA intercalating redox probe. Anal. Chem. 2011, 83, 1815–1821. [Google Scholar] [CrossRef] [PubMed]
- Sun, Y.; Quyen, T.L.; Hung, T.Q.; Chin, W.H.; Wolff, A.; Bang, D.D. A lab-on-a-chip system with integrated sample preparation and loop-mediated isothermal amplification for rapid and quantitative detection of Salmonella spp. in food samples. Lab Chip 2015, 15, 1898–1904. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nadal, A.; Coll, A.; Cook, N.; Pla, M. A molecular beacon-based real time NASBA assay for detection of Listeria monocytogenes in food products: Role of target mRNA secondary structure on NASBA design. J. Microbiol. Methods 2007, 68, 623–632. [Google Scholar] [CrossRef]
- Tao, Z.Y.; Zhou, H.Y.; Xia, H.; Xu, S.; Zhu, H.W.; Culleton, R.L.; Han, E.T.; Lu, F.; Fang, Q.; Gu, Y.P.; et al. Adaptation of a visualized loop-mediated isothermal amplification technique for field detection of Plasmodium vivax infection. Parasit Vectors 2011, 4, 115. [Google Scholar] [CrossRef] [Green Version]
- Quyen, T.L.; Ngo, T.A.; Bang, D.D.; Madsen, M.; Wolff, A. Classification of multiple DNA dyes based on inhibition effects on real-time loop-mediated isothermal amplification (LAMP): Prospect for point of care setting. Front. Microbiol. 2019, 10, 2234. [Google Scholar] [CrossRef]
- Dixit, K.K.; Verma, S.; Singh, O.P.; Singh, D.; Singh, A.P.; Gupta, R.; Negi, N.S.; Das, P.; Sundar, S.; Singh, R.; et al. Validation of SYBR Green I-based closed tube loop mediated isothermal amplification (LAMP) assay and simplified direct-blood-lysis (DBL)-LAMP assay for diagnosis of visceral leishmaniasis (VL). PLoS Negl. Trop. Dis. 2018, 12, e0006922. [Google Scholar] [CrossRef] [Green Version]
- Ramalingam, N.; Liu, H.B.; Dai, C.C.; Jiang, Y.; Wang, H.; Wang, Q.M.; Hui, K.M.; Gong, H.Q. Real-time PCR array chip with capillary-driven sample loading and reactor sealing for point-of-care applications. Biomed. Microdevices 2009, 11, 1007–1020. [Google Scholar] [CrossRef] [PubMed]
- Hongwarittorrn, I.; Chaichanawongsaroj, N.; Laiwattanapaisal, W. Semi-quantitative visual detection of loop mediated isothermal amplification (LAMP)-generated DNA by distance-based measurement on a paper device. Talanta 2017, 175, 135–142. [Google Scholar] [CrossRef] [PubMed]
- Mori, Y.; Hirano, T.; Notomi, T. Sequence specific visual detection of LAMP reactions by addition of cationic polymers. BMC Biotechnol. 2006, 6, 3. [Google Scholar] [CrossRef] [Green Version]
- Steel, A.B.; Herne, T.M.; Tarlov, M.J. Electrostatic interactions of redox cations with surface-immobilized and solution DNA. Bioconjug. Chem. 1999, 10, 419–423. [Google Scholar] [CrossRef]
- Vidic, J.; Vizzini, P.; Manzano, M.; Kavanaugh, D.; Ramarao, N.; Zivkovic, M.; Radonic, V.; Knezevic, N.; Giouroudi, I.; Gadjanski, I. Point-of-need DNA testing for detection of foodborne pathogenic bacteria. Sensors 2019, 19, 1100. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ahmed, M.U.; Nahar, S.; Safavieh, M.; Zourob, M. Real-time electrochemical detection of pathogen DNA using electrostatic interaction of a redox probe. Analyst 2013, 138, 907–915. [Google Scholar] [CrossRef] [PubMed]
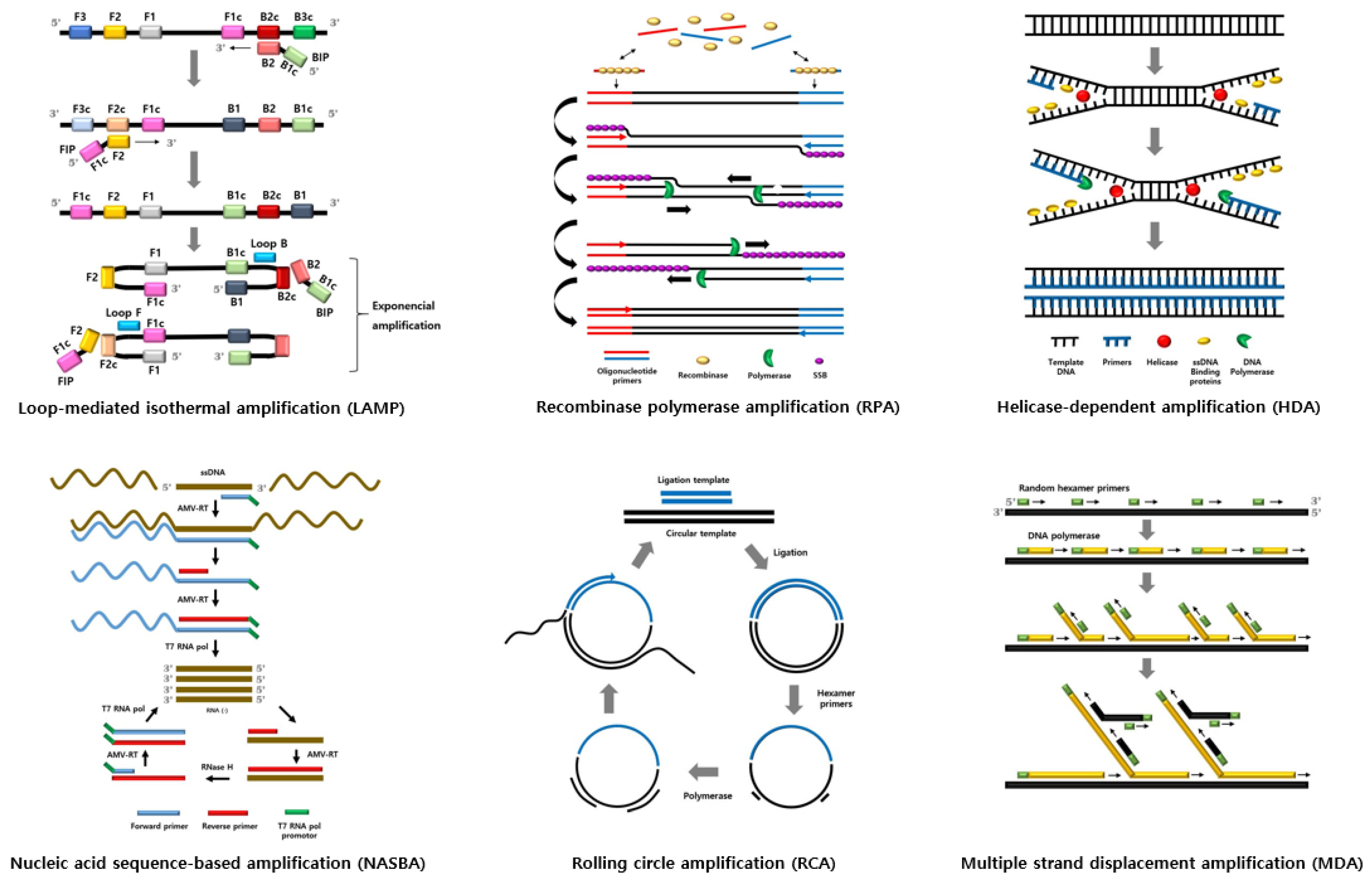
| Isothermal Amplification Methods | Number of Primers | Number of Enzymes | Pre-Heating | Working Temperature (°C) | Reaction Time (min) | Target Template | Amplicon | Resistance to Inhibitor | Reference |
|---|---|---|---|---|---|---|---|---|---|
| LAMP | 4–6 | 1 | No | 60–65 | 40–60 | DNA | DNA | High | [29] |
| RPA | 2 | 2 | No | 37–42 | 20–40 | DNA | DNA | Low | [12] |
| HDA | 2 | 1 (mHDA), 3 (tHDA) | No | 37 (mHDA), 60–65 (tHDA) | 100–120 | DNA | DNA | High | [11] |
| NASBA | 2 | 2–3 | Yes | 41 | 90–120 | RNA | RNA, DNA | Low | [43] |
| RCA | 1 | 1 | Yes | 30–65 | 60–90 | Circular DNA | DNA | Low | [42] |
| MDA | Random hexamer primers | 1 | No | 35 | 270 | Circular or linear DNAs | Ramified double–stranded DNAs | High | [57] |
| Type of Food | Target Bacterial | Isothermal Amplification Method | Reference | |
|---|---|---|---|---|
| Nucleic Acid Amplification | Detection Method | |||
| Meat | Salmonella spp. | LAMP | Intercalating dye | [5,66,67,68] |
| Real-time | [69,70] | |||
| LFA | [24,71] | |||
| RPA | Real-time | [72,73,74] | ||
| HDA | LFA | [37] | ||
| NASBA | ECL | [75] | ||
| Real-time | [16] | |||
| Escherichia coli O157:H7 | LAMP | Real-time | [76] | |
| LFA | [77] | |||
| RPA | DNA-binding dye | [78] | ||
| Listeria monocytogenes | LAMP | Intercalating dye | [79] | |
| RPA | LFA | [29,80,81,82] | ||
| NASBA | ELISA | [83] | ||
| Real-time | [84] | |||
| MDA | LFA | [54] | ||
| Vibrio parahaemolyticus | LAMP | LFA | [85] | |
| Staphylococcus aureus | LAMP | Intercalating dye | [67] | |
| HDA | Fluorescence | [86] | ||
| Seafood | Salmonella spp. | RPA | LFA | [87,88] |
| NASBA | ECL | [75] | ||
| Escherichia coli O157:H7 | RPA | LFA | [29] | |
| Listeria monocytogenes | RPA | Real-time | [89] | |
| LFA | [29,80,82] | |||
| NASBA | ELISA | [75] | ||
| Real-time | [84] | |||
| Vibrio parahaemolyticus | LAMP | Intercalating dye | [67,90] | |
| Real-time | [91] | |||
| RPA | Real-time | [92] | ||
| LFA | [29,88,93] | |||
| MDA | LFA | [94] | ||
| Staphylococcus aureus | RPA | LFA | [88] | |
| Vegetable | Salmonella spp. | LAMP | Real-time | [70] |
| RPA | Real-time | [74] | ||
| Dairy produce | Salmonella spp. | LAMP | Intercalating dye | [68,95] |
| LFA | [71] | |||
| RPA | Real-time | [96] | ||
| LFA | [29] | |||
| CRISPR/Cas12a | [97] | |||
| HDA | LFA | [37] | ||
| NASBA | ECL | [75] | ||
| Real-time | [16] | |||
| Escherichia coli O157:H7 | LAMP | Intercalating dye | [95] | |
| RPA | Real-time | [96] | ||
| LFA | [29,98] | |||
| Listeria monocytogenes | RPA | Real-time | [99] | |
| LFA | [29,80,81,82] | |||
| NASBA | ELISA | [83] | ||
| Vibrio parahaemolyticus | LAMP | Intercalating dye | [95] | |
| RPA | Real-time | [96] | ||
| LFA | [29] | |||
| Staphylococcus aureus | LAMP | Intercalating dye | [95] | |
| HDA | Fluorescence | [86] | ||
| NASBA | Real-time | [100] | ||
| Reaction Process | Inhibitors | Alleviation Strategies for Inhibition | Amplification Methods | Reference | |
|---|---|---|---|---|---|
| Sample preparation and DNA extraction | Residual food metrix | Use the nucleic acid sample after dilution | NASBA | [75] | |
| CTAB used as extraction buffer | Use direct PCR buffers | RPA | [96] | ||
| Nucleic acid amplification | Concentration | Magnesium ions | Increase the concentration of magnesium ions | tHDA | [108,109] |
| Add betaine, DMSO, and sorbitol to the reaction mixture | [37] | ||||
| Use 4–6 mM MgSO4, which is the optimal concentration for magnesium ions | RCA | [110] | |||
| Primer | Optimize concentration of primer | RPA | [32] | ||
| Multi-RPA | [111] | ||||
| Template or background DNA | Treat RNase A with pasteurization and 15 min incubation process before nucleic acid extraction | Real-time NASBA | [112] | ||
| Add the primer stability enhancer to the primer and beacon mixture | NASBA | [113] | |||
| Temperature | Temperature fluctuations | Optimize reaction temperature | MDA | [107] | |
| Heat denaturation | Substitute alkaline denaturation | [114] | |||
| Detection method | Colorimetric detection | SYBR Green I | Add fluorescent dyes after amplification | LAMP | [97] |
| Use wax capsules containing the dye, which react after amplification | [114] | ||||
| PEI | Add PEI after amplification | LAMP | [115] | ||
| Calcein | - | - | |||
| Electrochemical detection | Redox active compounds (e.g., MB and Hoechst 33258) | Use other redox molecules (e.g., osmium redox and RuHex) | LAMP | [116] | |
| Use voltammeric mode | [97] | ||||
| Use polydopamine-doped paper disks | [117] | ||||
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Moon, Y.-J.; Lee, S.-Y.; Oh, S.-W. A Review of Isothermal Amplification Methods and Food-Origin Inhibitors against Detecting Food-Borne Pathogens. Foods 2022, 11, 322. https://doi.org/10.3390/foods11030322
Moon Y-J, Lee S-Y, Oh S-W. A Review of Isothermal Amplification Methods and Food-Origin Inhibitors against Detecting Food-Borne Pathogens. Foods. 2022; 11(3):322. https://doi.org/10.3390/foods11030322
Chicago/Turabian StyleMoon, Ye-Ji, So-Young Lee, and Se-Wook Oh. 2022. "A Review of Isothermal Amplification Methods and Food-Origin Inhibitors against Detecting Food-Borne Pathogens" Foods 11, no. 3: 322. https://doi.org/10.3390/foods11030322
APA StyleMoon, Y.-J., Lee, S.-Y., & Oh, S.-W. (2022). A Review of Isothermal Amplification Methods and Food-Origin Inhibitors against Detecting Food-Borne Pathogens. Foods, 11(3), 322. https://doi.org/10.3390/foods11030322





