High Prevalence of Enterobacterales in the Smear of Surface-Ripened Cheese with Contribution to Organoleptic Properties
Abstract
:1. Introduction
2. Materials and Methods
2.1. Cheese Samples
2.2. Sampling Procedure and Microbial Analysis by Culturing
2.3. Isolation of Gram-Negative Bacteria and Genotypic Identification by Partial 16S rRNA and gyrB Sequencing
2.4. Determination of Proteolytic and Lipolytic Activity of Gram-Negative Isolates
2.5. Determination of Extended Spectrum β–Lactamase (ESBL)-Mediated Resistance of Enterobacterales Isolated from the Cheese Surface Smear
2.6. Volatile Organic Flavor Compound Analysis of Cheese Surface Smear
3. Results
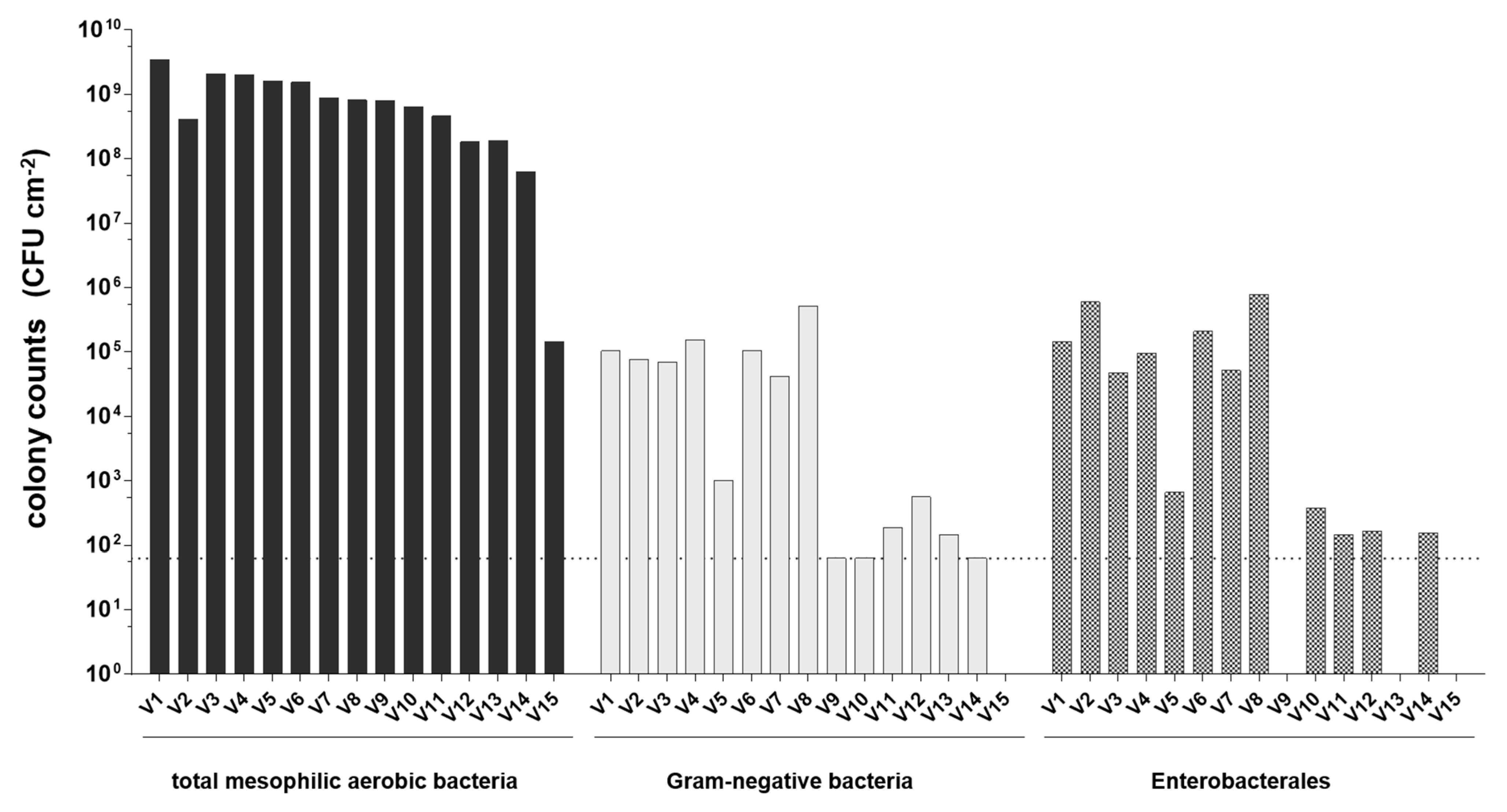
3.1. Abundance of Total Mesophilic Aerobic Bacteria, Gram-Negative Bacteria and Enterobacterales in Cheese Surface Smear
3.2. Genotypic Identification of Gram-Negative Isolates by Partial Sequencing of 16S rRNA and gyrB Genes
3.3. Proteolytic and Lipolytic Activities of Isolated Proteobacteria
3.4. Volatile Carbonic Flavor Compounds Produced by Gram-Negative Isolates of the Cheese Surface Smear
3.5. Extended Spectrum β–Lactamase (ESBL)-Mediated Resistances of Enterobacterales Isolated from the Cheese Surface Smear
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- McSweeney, P.L.H.; Ottogalli, G.; Fox, P.F. Diversity of cheese varieties: An overview. In Cheese: Chemistry, Physics and Microbiology; Fox, P.F., McSweeney, P.L.H., Cogan, T.M., Guinee, T.P., Eds.; Elsevier: Amsterdam, The Netherlands, 2004; Volume 2, pp. 1–23. [Google Scholar]
- Switzerlands Cheese Market in Figures 2020. Available online: https://www.cheesesfromswitzerland.com/fileadmin/content/SCM_Folder_2020_Taschenstatistik_300x234mm_E_RZ_lowres.pdf (accessed on 17 December 2021).
- Brennan, N.M.; Cogan, T.M.; Loessner, M.J.; Scherer, S. Bacterial surface-ripened cheeses. In Cheese: Chemistry, Physics and Microbiology; Fox, P.F., McSweeney, P.L.H., Cogan, T.M., Guinee, T.P., Eds.; Elsevier: Amsterdam, The Netherlands, 2004; Volume 2, pp. 199–225. [Google Scholar]
- Rademaker, J.; Peinhopf, M.; Rijnen, L.; Bockelmann, W.; Noordman, W. The surface microflora dynamics of bacterial smear-ripened Tilsit cheese determined by T-RFLP DNA population fingerprint analysis. Int. Dairy J. 2005, 15, 785–794. [Google Scholar] [CrossRef]
- Bockelmann, W.; Willems, K.; Neve, H.; Heller, K. Cultures for the ripening of smear cheeses. Int. Dairy J. 2005, 15, 719–732. [Google Scholar] [CrossRef]
- Brennan, N.M.; Ward, A.C.; Beresford, T.P.; Fox, P.F.; Goodfellow, M.; Cogan, T.M. Biodiversity of the bacterial flora on the surface of a smear cheese. Appl. Environ. Microbiol. 2002, 68, 820–830. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mounier, J.; Goerges, S.; Gelsomino, R.; Vancanneyt, M.; Vandemeulebroecke, K.; Hoste, B.; Brennan, N.; Scherer, S.; Swings, J.; Fitzgerald, G. Sources of the adventitious microflora of a smear ripened cheese. J. Appl. Microbiol. 2006, 101, 668–681. [Google Scholar] [CrossRef]
- Mounier, J.; Gelsomino, R.; Goerges, S.; Vancanneyt, M.; Vandemeulebroecke, K.; Hoste, B.; Scherer, S.; Swings, J.; Fitzgerald, G.F.; Cogan, T.M. Surface microflora of four smear-ripened cheeses. Appl. Environ. Microbiol. 2005, 71, 6489–6500. [Google Scholar] [CrossRef] [Green Version]
- Roth, E.; Schwenninger, S.M.; Eugster-Meier, E.; Lacroix, C. Facultative anaerobic halophilic and alkaliphilic bacteria isolated from a natural smear ecosystem inhibit Listeria growth in early ripening stages. Int. J. Food Microbiol. 2011, 147, 26–32. [Google Scholar] [CrossRef]
- Delbès, C.; Ali-Mandjee, L.; Montel, M.C. Monitoring bacterial communities in raw milk and cheese by culture-dependent and-independent 16S rRNA gene-based analyses. Appl. Environ. Microbiol. 2007, 73, 1882–1891. [Google Scholar] [CrossRef] [Green Version]
- Ogier, J.-C.; Serror, P. Safety assessment of dairy microorganisms: The Enterococcus genus. Int. J. Food Microbiol. 2008, 126, 291–301. [Google Scholar] [CrossRef]
- Coton, M.; Delbès-Paus, C.; Irlinger, F.; Desmasures, N.; Le Fleche, A.; Stahl, V.; Montel, M.C.; Coton, E. Diversity and assessment of potential risk factors of Gram-negative isolates associated with French cheeses. Food Microbiol. 2011, 29, 88–98. [Google Scholar] [CrossRef]
- Amato, L.; Ritschard, J.S.; Kurtz, O.; Arias-Roth, E.; Lacroix, C.; Schuppler, M.; Meile, L. Microbial composition of defect smear—A problem evolving during foil-prepacked storage of red-smear cheeses. Int. Dairy J. 2012, 27, 77–85. [Google Scholar] [CrossRef]
- Deetae, P.; Mounier, J.; Bonnarme, P.; Spinnler, H.; Irlinger, F.; Helinck, S. Effects of Proteus vulgaris growth on the establishment of a cheese microbial community and on the production of volatile aroma compounds in a model cheese. J. Appl. Microbiol. 2009, 107, 1404–1413. [Google Scholar] [CrossRef] [PubMed]
- Larpin-Laborde, S.; Imran, M.; Bonaïti, C.; Bora, N.; Gelsomino, R.; Goerges, S.; Irlinger, F.; Goodfellow, M.; Ward, A.C.; Vancanneyt, M.; et al. Surface microbial consortia from Livarot, a French smear-ripened cheese. Can. J. Microbiol. 2011, 57, 651–660. [Google Scholar] [CrossRef] [PubMed]
- Chaves-López, C.; De Angelis, M.; Martuscelli, M.; Serio, A.; Paparella, A.; Suzzi, G. Characterization of the Enterobacteriaceae isolated from an artisanal Italian ewe’s cheese (Pecorino Abruzzese). J. Appl. Microbiol. 2006, 101, 353–360. [Google Scholar] [CrossRef] [PubMed]
- Maifreni, M.; Frigo, F.; Bartolomeoli, I.; Innocente, N.; Biasutti, M.; Marino, M. Identification of the Enterobacteriaceae in Montasio cheese and assessment of their amino acid decarboxylase activity. J. Dairy Res. 2013, 80, 122–127. [Google Scholar] [CrossRef] [PubMed]
- Irlinger, F.; Layec, S.; Hélinck, S.; Dugat-Bony, E. Cheese rind microbial communities: Diversity, composition and origin. FEMS Microbiol. Let. 2014, 362, 1–11. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tornadijo, M.; García, M.; Fresno, J.; Carballo, J. Study of Enterobacteriaceae during the manufacture and ripening of San Simón cheese. Food Microbiol. 2001, 18, 499–509. [Google Scholar] [CrossRef]
- Quigley, L.; O’Sullivan, O.; Beresford, T.P.; Ross, R.P.; Fitzgerald, G.F.; Cotter, P.D. High-Throughput Sequencing for detection of subpopulations of bacteria not previously associated with artisanal cheeses. Appl. Environ. Microbiol. 2012, 78, 5717–5723. [Google Scholar] [CrossRef] [Green Version]
- Unno, R.; Suzuki, T.; Matsutani, M.; Ishikawa, M. Evaluation of the relationships between microbiota and metabolites in soft-type ripened cheese using an integrated omics approach. Front. Microbiol. 2021, 12, 681185. [Google Scholar] [CrossRef]
- Kamelamela, N.; Zalesne, M.; Morimoto, J.; Robbat, A.; Wolfe, B.E. Indigo- and indirubin-producing strains of Proteus and Psychrobacter are associated with purple rind defect in a surface-ripened cheese. Food Microbiol. 2018, 76, 543–552. [Google Scholar] [CrossRef]
- Peng, S.; Schafroth, K.; Jakob, E.; Stephan, R.; Hummerjohann, J. Behaviour of Escherichia coli strains during semi-hard and hard raw milk cheese production. Int. Dairy J. 2013, 31, 117–120. [Google Scholar] [CrossRef] [Green Version]
- Bokulich, N.A.; Mills, D.A. Facility-specific “house” microbiome drives microbial landscapes of artisan cheesemaking plants. Appl. Environ. Microbiol. 2013, 79, 5214–5223. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Didienne, R.; Defargues, C.; Callon, C.; Meylheuc, T.; Hulin, S.; Montel, M.-C. Characteristics of microbial biofilm on wooden vats (‘gerles’) in PDO Salers cheese. Int. J. Food Microbiol. 2012, 156, 91–101. [Google Scholar] [PubMed]
- Ksontini, H.; Kachouri, F.; Hamdi, M. Dairy biofilm: Impact of microbial community on raw milk quality. J. Food Qual. 2013, 36, 282–290. [Google Scholar] [CrossRef]
- Quigley, L.; O’Sullivan, O.; Stanton, C.; Beresford, T.P.; Ross, R.P.; Fitzgerald, G.F.; Cotter, P.D. The complex microbiota of raw milk. FEMS Microbiol. Rev. 2013, 37, 664–698. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Irlinger, F.; Monnet, C. Temporal differences in microbial composition of Époisses cheese rinds during ripening and storage. J. Dairy Sci. 2021, 104, 7500–7508. [Google Scholar] [CrossRef]
- Sambrook, J.; Russell, D.W. Molecular Cloning; Cold Spring Harbor Laboratory Press: Woodbury, NY, USA, 2001; Volume 1–3. [Google Scholar]
- Brosius, J.; Palmer, M.L.; Kennedy, P.J.; Noller, H.F. Complete nucleotide sequence of a 16S ribosomal RNA gene from Escherichia coli. Proc. Natl. Acad. Sci. USA 1978, 75, 4801–4805. [Google Scholar] [CrossRef] [Green Version]
- Yin, H.; Cao, L.; Xie, M.; Chen, Q.; Qiu, G.; Zhou, J.; Wu, L.; Wang, D.; Liu, X. Bacterial diversity based on 16S rRNA and gyrB genes at Yinshan mine, China. Syst. Appl. Microbiol. 2008, 31, 302–311. [Google Scholar] [CrossRef]
- Altschul, S.F.; Madden, T.L.; Schäffer, A.A.; Zhang, J.; Zhang, Z.; Miller, W.; Lipman, D.J. Gapped BLAST and PSI-BLAST: A new generation of protein database search programs. Nucl. Acids Res. 1997, 25, 3389–3402. [Google Scholar] [CrossRef] [Green Version]
- European Committee on Antimicrobial Susceptibility Testing. EUCAST Guidelines for Detection of Resistance Mechanisms and Specific Resistances of Clinical and/or Epidemiological Importance; EUCAST: Basel, Switzerland, 2013. [Google Scholar]
- Morales, P.; Fernández-García, E.; Nuñez, M. Caseinolysis in cheese by Enterobacteriaceae strains of dairy origin. Lett. Appl. Microbiol. 2003, 37, 410–414. [Google Scholar] [CrossRef]
- Dahl, S.; Tavaria, F.K.; Xavier Malcata, F. Relationships between flavour and microbiological profiles in Serra da Estrela cheese throughout ripening. Int. Dairy J. 2000, 10, 255–262. [Google Scholar] [CrossRef]
- Deetae, P.; Bonnarme, P.; Spinnler, H.; Helinck, S. Production of volatile aroma compounds by bacterial strains isolated from different surface-ripened French cheeses. Appl. Microbiol. Biotechnol. 2007, 76, 1161–1171. [Google Scholar] [CrossRef] [PubMed]
- Irlinger, F.; In Yung, S.A.Y.; Sarthou, A.-S.; Delbès-Paus, C.; Montel, M.-C.; Coton, E.; Coton, M.; Helinck, S. Ecological and aromatic impact of two Gram-negative bacteria (Psychrobacter celer and Hafnia alvei) inoculated as part of the whole microbial community of an experimental smear soft cheese. Int. J. Food Microbiol. 2012, 153, 332–338. [Google Scholar] [CrossRef] [PubMed]
- Morales, P.; Feliu, I.; Fernández-García, E.; Nun, M. Volatile compounds produced in cheese by Enterobacteriaceae strains of dairy origin. J. Food Protect. 2004, 67, 567–573. [Google Scholar] [CrossRef]
- Tornadijo, E.; Fresno, J.M.; Carballo, J.; Martín-Sarmiento, R. Study of Enterobacteriaceae throughout the manufacturing and ripening of hard goats’ cheese. J. Appl. Bacteriol. 1993, 75, 240–246. [Google Scholar] [CrossRef] [PubMed]
- Deetae, P.; Spinnler, H.-E.; Bonnarme, P.; Helinck, S. Growth and aroma contribution of Microbacterium foliorum, Proteus vulgaris and Psychrobacter sp. during ripening in a cheese model medium. Appl. Microbiol. Biotechnol. 2009, 82, 169–177. [Google Scholar] [CrossRef] [PubMed]
- Montel, M.-C.; Buchin, S.; Mallet, A.; Delbès–Paus, C.; Vuitton, D.A.; Desmasures, N.; Berthier, F. Traditional cheeses: Rich and diverse microbiota with associated benefits. Int. J. Food Microbiol. 2014, 177, 136–154. [Google Scholar] [CrossRef]
- Mounier, J.; Monnet, C.; Jacques, N.; Antoinette, A.; Irlinger, F. Assessment of the microbial diversity at the surface of Livarot cheese using culture-dependent and independent approaches. Int. J. Food Microbiol. 2009, 133, 31–37. [Google Scholar] [CrossRef] [Green Version]
- Ogier, J.-C.; Lafarge, V.; Girard, V.; Rault, A.; Maladen, V.; Gruss, A.; Leveau, J.-Y.; Delacroix-Buchet, A. Molecular fingerprinting of dairy microbial ecosystems by use of Temporal Temperature and Denaturing Gradient Gel Electrophoresis. Appl. Env. Microbiol. 2004, 70, 5628–5643. [Google Scholar] [CrossRef] [Green Version]
- Leriche, F.; Bordessoules, A.; Fayolle, K.; Karoui, R.; Laval, K.; Leblanc, L.; Dufour, E. Alteration of raw-milk cheese by Pseudomonas spp.: Monitoring the sources of contamination using fluorescence spectroscopy and metabolic profiling. J. Microbiol. Meth. 2004, 59, 33–41. [Google Scholar] [CrossRef]
- Schornsteiner, E.; Mann, E.; Bereuter, O.; Wagner, M.; Schmitz-Esser, S. Cultivation-independent analysis of microbial communities on Austrian raw milk hard cheese rinds. Int. J. Food Microbiol. 2014, 180, 88–97. [Google Scholar] [CrossRef]
- Fritsche, K.; Auling, G.; Andreesen, J.R.; Lechner, U. Defluvibacter lusatiae gen. nov., sp. nov., a new chlorophenol-degrading member of the α-2 subgroup of Proteobacteria. Syst. Appl. Microbiol. 1999, 22, 197–204. [Google Scholar] [CrossRef]
- Pence, M.A.; Sharon, J.; McElvania Tekippe, E.; Pakalniskis, B.L.; Ford, B.A.; Burnham, C.-A.D. Two cases of Kerstersia gyiorum isolated from sites of chronic infection. J. Clin. Microbiol. 2013, 51, 2001–2004. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Silipo, A.; Sturiale, L.; De Castro, C.; Lanzetta, R.; Parrilli, M.; Garozzo, D.; Molinaro, A. Structure of the lipopolysaccharide isolated from the novel species Uruburuella suis. Carbohydr. Res. 2012, 357, 75–82. [Google Scholar] [CrossRef]
- Wessels, D.; Jooste, P.J.; Mostert, J.F. Psychrotrophic, proteolytic and lipolytic properties of Enterobacteriaceae isolated from milk and dairy products. Int. J. Food Microbiol. 1989, 9, 79–83. [Google Scholar] [CrossRef]
- Deetae, P.; Saint-Eve, A.; Spinnler, H.E.; Helinck, S. Critical effect of oxygen on aroma compound production by Proteus vulgaris. Food Chem. 2011, 126, 134–139. [Google Scholar] [CrossRef]
- Duthoit, F.; Callon, C.; Tessier, L.; Montel, M.-C. Relationships between sensorial characteristics and microbial dynamics in “Registered Designation of Origin” Salers cheese. Int. J. Food Microbiol. 2005, 103, 259–270. [Google Scholar] [CrossRef] [PubMed]
- Marilley, L.; Casey, M. Flavours of cheese products: Metabolic pathways, analytical tools and identification of producing strains. Int. J. Food Microbiol. 2004, 90, 139–159. [Google Scholar] [CrossRef]
- Westlinga, M.; Danielsson-Thama, M.-L.; Jassb, J.; Nilsena, A.; Öströma, A.; Thama, W. Contribution of Enterobacteriaceae to sensory characteristics in soft cheeses made from raw milk. Proc. Food Sci. 2016, 7, 17–20. [Google Scholar] [CrossRef] [Green Version]
- Bourdichon, F.; Casaregola, S.; Farrokh, C.; Frisvad, J.C.; Gerds, M.L.; Hammes, W.P.; Harnett, J.; Huys, G.; Laulund, S.; Ouwehand, A.; et al. Food fermentations: Microorganisms with technological beneficial use. Int. J. Food Microbiol. 2012, 154, 87–97. [Google Scholar] [CrossRef]
- Ritschard, J.S.; Amato, L.; Kumar, Y.; Müller, B.; Meile, L.; Schuppler, M. The role of the surface smear microbiome in the development of defective smear on surface ripened red smear cheese. AIMS Microbiol. 2018, 4, 622–641. [Google Scholar] [CrossRef]
- Martín, M.C.; Martínez, N.; del Rio, B.; Ladero, V.; Fernández, M.; Alvarez, M.A. A novel real-time polymerase chain reaction-based method for the detection and quantification of lactose-fermenting Enterobacteriaceae in the dairy and other food industries. J. Dairy Sci. 2010, 93, 860–867. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- EFSA Panel on Biological Hazards (BIOHAZ). Scientific Opinion on the update of the list of QPS-recommended biological agents intentionally added to food and feed as notified to EFSA (2017–2019). EFSA J. 2020, 18, 5966. [Google Scholar]
- De Buyser, M.-L.; Dufour, B.; Maire, M.; Lafarge, V. Implication of milk and milk products in food-borne diseases in France and in different industrialised countries. Int. J. Food Microbiol. 2001, 67, 1–17. [Google Scholar] [CrossRef]
- Imran, M.; Bré, J.-M.; Guéguen, M.; Vernoux, J.-P.; Desmasures, N. Reduced growth of Listeria monocytogenes in two model cheese microcosms is not associated with individual microbial strains. Food Microbiol. 2013, 33, 30–39. [Google Scholar] [CrossRef]
- Amador, P.; Fernandes, R.; Prudêncio, C.; Brito, L. Resistance to β-lactams in bacteria isolated from different types of Portuguese cheese. Int. J. Mol. Sci. 2009, 10, 1538–1551. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hanson, N.D. AmpC β-lactamases: What do we need to know for the future? J. Antimicrob. Chemother. 2003, 52, 2–4. [Google Scholar] [CrossRef] [Green Version]
- Cantón, R.; Novais, A.; Valverde, A.; Machado, E.; Peixe, L.; Baquero, F.; Coque, T.M. Prevalence and spread of extended-spectrum β-lactamase-producing Enterobacteriaceae in Europe. Clin. Microbiol. Infect. 2008, 14, 144–153. [Google Scholar] [CrossRef] [Green Version]
| Sample | Cheese Type | Weight a | Milk Type b | Fat Content c | Production Type |
|---|---|---|---|---|---|
| V1 | Trappist-like | 0.4 kg | pasteurized | 45% | artisanal |
| V2 | Tilsit-like | 4 kg | thermized | 45% | traditional |
| V3 | Tilsit-like | 4 kg | pasteurized | 45% | traditional |
| V4 | Tilsit-like (mild/creamy) | 4 kg | pasteurized | 45% | traditional |
| V5 | Trappist-like | 0.8 kg | thermized | 51% | traditional |
| V6 | Raclette-like | 4 kg | thermized | 25% | artisanal |
| V7 | Raclette-like | 5 kg | pasteurized | 30% | artisanal |
| V8 | Tilsit-like (mild/creamy) | 3 kg | thermized | 53% | artisanal |
| V9 | Mountain cheese-like | 5 kg | thermized | 45% | artisanal |
| V10 | Tilsit-like (mild/creamy) | 6 kg | pasteurized | 31% | traditional |
| V11 | Tilsit-like | 7 kg | raw milk | 48% | traditional |
| V12 | Tilsit-like (mild/creamy) | 4 kg | pasteurized | 55% | industrial |
| V13 | Tilsit-like | 7 kg | raw milk | 45% | traditional |
| V14 | Raclette-like | 8 kg | raw milk | 51–54% | traditional |
| V15 | Tilsit-like (mild/creamy) | 2 kg | pasteurized | 45% | industrial |
| Occurrence in Surface Smear of Cheese Sample | ||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| V1 | V2 | V3 | V4 | V5 | V6 | V7 | V8 | V9 | V10 | V11 | V12 | V13 | V14 | V15 | ||
| Bacterial Species | Isolation Media | |||||||||||||||
| Advenella sp. | − | + | − | − | − | − | − | − | − | − | − | − | + | − | − | VRBG/PCAI |
| Advenella kashmirensis | − | − | − | − | − | − | − | − | − | − | − | − | + | − | − | Endo |
| Brevundimonas diminuta | − | − | − | − | − | − | − | − | − | − | − | − | + | − | − | PCAI |
| Citrobacter sp. | + | − | − | + | − | − | + | − | − | − | − | − | − | − | − | VRBG/PCAI/PCAIan/Endo |
| Defluvibacter sp. | − | − | − | − | − | − | − | − | − | − | − | − | + | − | − | VRBG |
| Enterobacter spp. | + | − | − | + | − | − | + | − | − | − | − | − | − | − | − | VRBG/PCAI/PCAIan/Endo |
| Hafnia alvei | − | − | − | − | − | + | − | − | − | − | − | − | − | − | − | PCAIan |
| Kerstersia gyiorum | − | − | − | − | − | − | − | − | − | − | + | − | − | − | − | Endo |
| Morganella spp. | + | − | + | − | + | − | − | − | − | − | − | − | − | − | − | VRBG/PCAI/Endo |
| Morganella morganii | + | + | + | + | − | − | + | + | − | + | + | − | − | − | − | VRBG/PCAI/PCAIan/Endo |
| Proteus spp. | − | + | − | + | + | + | + | + | − | + | + | + | − | + | − | VRBG/PCAI/Endo |
| Proteus hauseri | − | + | + | − | − | − | − | − | − | − | − | − | − | − | − | VRBG/PCAI |
| Proteus vulgaris | − | + | + | − | − | + | − | − | − | − | − | − | − | − | − | VRBG/PCAI |
| Providencia sp. | − | + | − | − | − | − | − | − | − | − | − | − | − | − | − | VRBG |
| Providencia heimbachae | − | − | − | − | + | − | − | − | − | − | − | − | − | − | − | VRBG |
| Providencia rettgeri | − | + | − | − | − | − | − | − | − | − | − | − | − | − | − | VRBG |
| Pusillimonas spp. | − | + | − | − | − | − | − | − | − | − | − | − | + | + | − | VRBG/Endo |
| Serratia spp. | − | − | − | + | − | − | − | + | − | − | − | − | − | − | − | PCAI/PCAIan |
| Serratia proteamaculans | − | − | − | + | − | − | − | − | − | − | − | − | − | − | − | PCAIan |
| Stenotrophomonas sp. | − | − | − | − | − | − | − | − | − | − | − | − | + | − | − | PCAI |
| S. maltophilia | − | − | − | − | − | − | − | − | + | − | − | − | − | − | − | PCAI |
| Uruburuella suis | − | − | − | − | − | − | − | + | − | − | − | − | − | − | − | PCAI |
| Proteolysis Zone (mm) a | |||||||
|---|---|---|---|---|---|---|---|
| Aerobic Conditions | |||||||
| RT | 30 °C | ||||||
| Isolate No. | Species | 2 d | 5 d | 12 d | 2 d | 5 d | 12 d |
| V2.4 | Proteus sp. | 29 ± 2 | >40 | >40 | 27 ± 5 | >40 | >40 |
| V2.5 | Proteus sp. | 4 ± 2 | 3 ± 1 | >40 | 7 ± 1 | 2 | >40 |
| V2.6 | Proteus sp. | - | 5 ± 1 | 12 ± 4 | - | 2 ± 1 | 11 ±6 |
| V2.7 | Proteus sp. | 2 ± 1 | 19 ± 2 | >40 | 3 ± 1 | >40 | >40 |
| V2.8 | Proteus sp. | 38 ± 4 | >40 | >40 | - | >40 | >40 |
| V2.9 | Proteus sp. | - | 6 ± 2 | 20 ± 5 | 2 ± 1 | 7 ± 2 | 25 ± 2 |
| V5.3 | Proteus sp. | >40 | >40 | >40 | >40 | >40 | >40 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ritschard, J.S.; Van Loon, H.; Amato, L.; Meile, L.; Schuppler, M. High Prevalence of Enterobacterales in the Smear of Surface-Ripened Cheese with Contribution to Organoleptic Properties. Foods 2022, 11, 361. https://doi.org/10.3390/foods11030361
Ritschard JS, Van Loon H, Amato L, Meile L, Schuppler M. High Prevalence of Enterobacterales in the Smear of Surface-Ripened Cheese with Contribution to Organoleptic Properties. Foods. 2022; 11(3):361. https://doi.org/10.3390/foods11030361
Chicago/Turabian StyleRitschard, Jasmine S., Hanne Van Loon, Lea Amato, Leo Meile, and Markus Schuppler. 2022. "High Prevalence of Enterobacterales in the Smear of Surface-Ripened Cheese with Contribution to Organoleptic Properties" Foods 11, no. 3: 361. https://doi.org/10.3390/foods11030361





