The Acrylamide Degradation by Probiotic Strain Lactobacillus acidophilus LA-5
Abstract
:1. Introduction
2. Materials and Methods
2.1. Chemicals
2.2. Bacteria Strain and Microbial Media
2.3. Bacteria Preparation
2.4. Measurement of Optical Density of Bacterial Suspension: Calibration
2.5. Preparation of Acrylamide “Stock” Solution
2.6. The Evaluation of Acrylamide Degradation by Lactobacillus acidophilus LA-5
2.6.1. The Degradation of Acrylamide in a Model Medium
2.6.2. The Impact of Basic Milk Components on the Degradation of Acrylamide
2.6.3. The Evaluation of Acrylamide Degradation by Lactobacillus acidophilus LA-5 in Real Solution
2.7. The Determination of Acrylamide Concentration Changes in Samples
2.7.1. The Preparation of Internal Standard (ITS) Solution
2.7.2. The Determination of Acrylamide Concentration by Gas Chromatography (GC)
2.8. The Impact of Acrylamide on Microbial Metabolism
2.8.1. Sample Preparation for HPLC Analyses
2.8.2. The Determination of Sugars Content by HPLC
2.8.3. The Determination of Lactic and Acetic Acids Content by HPLC
2.9. Statistical Analysis
3. Results and Discussion
3.1. Acrylamide Degradation in Carbon- and Nitrogen-Limited Conditions
3.2. The Impact of Milk Components on Acrylamide Degradation
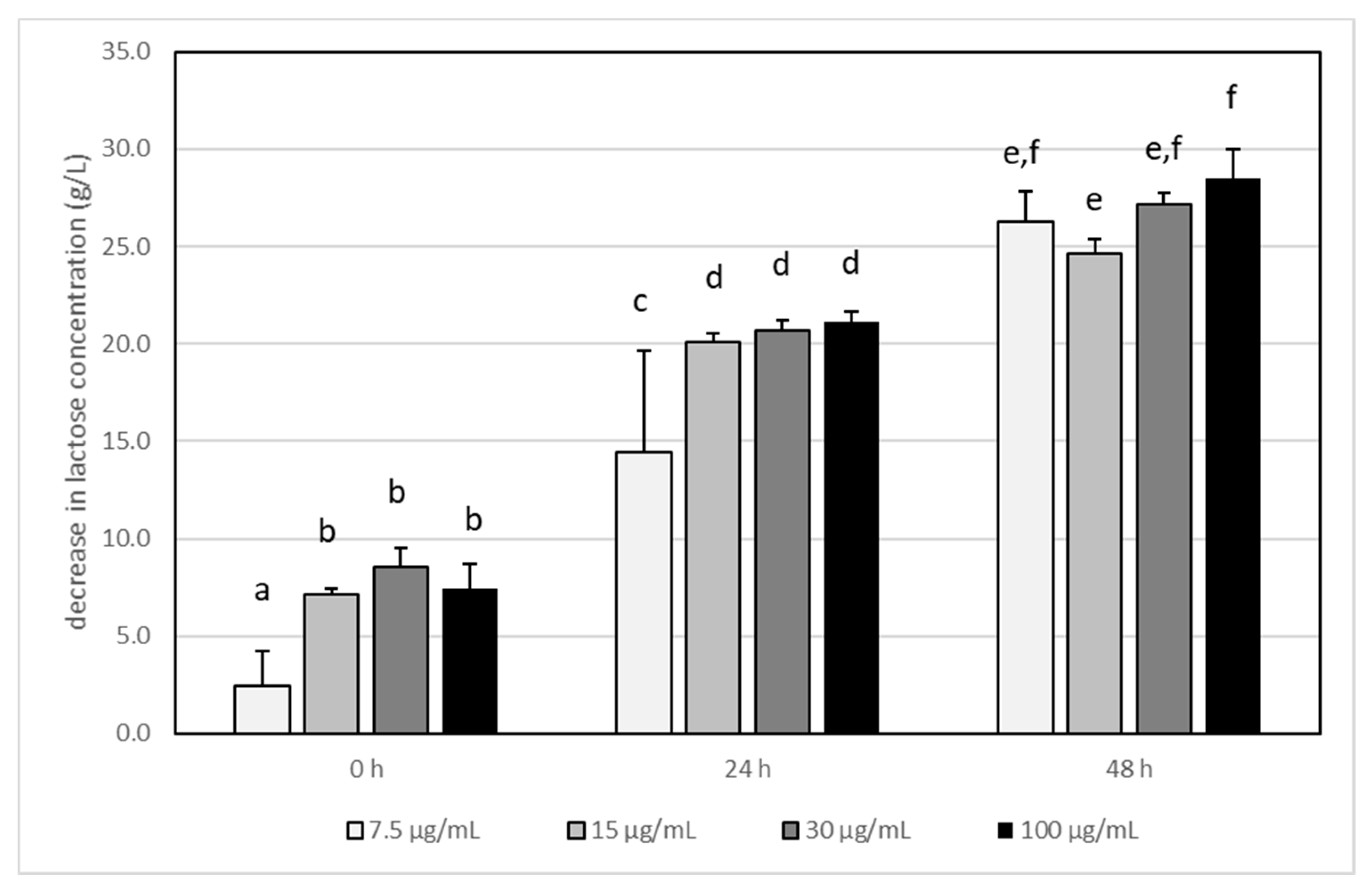
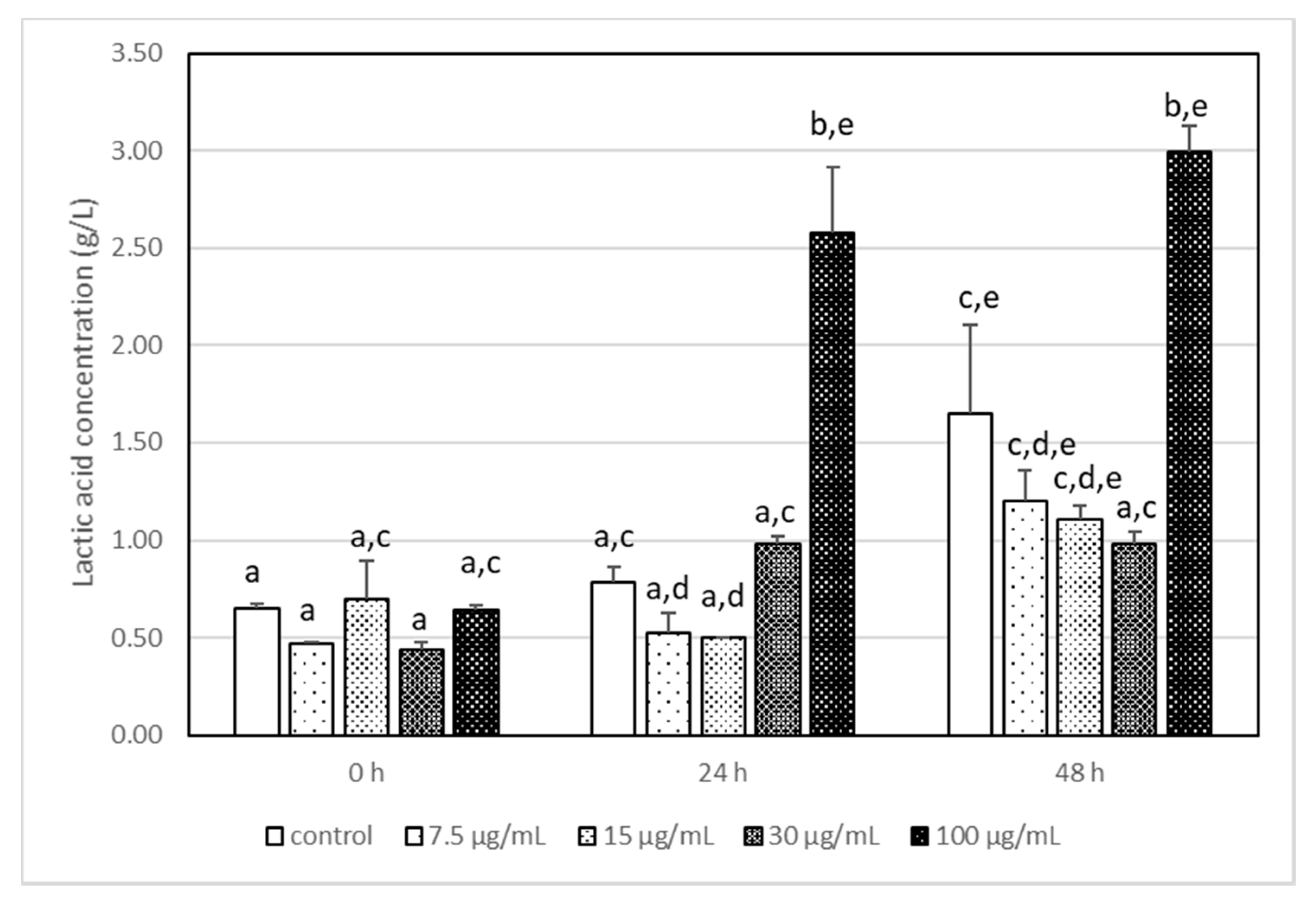
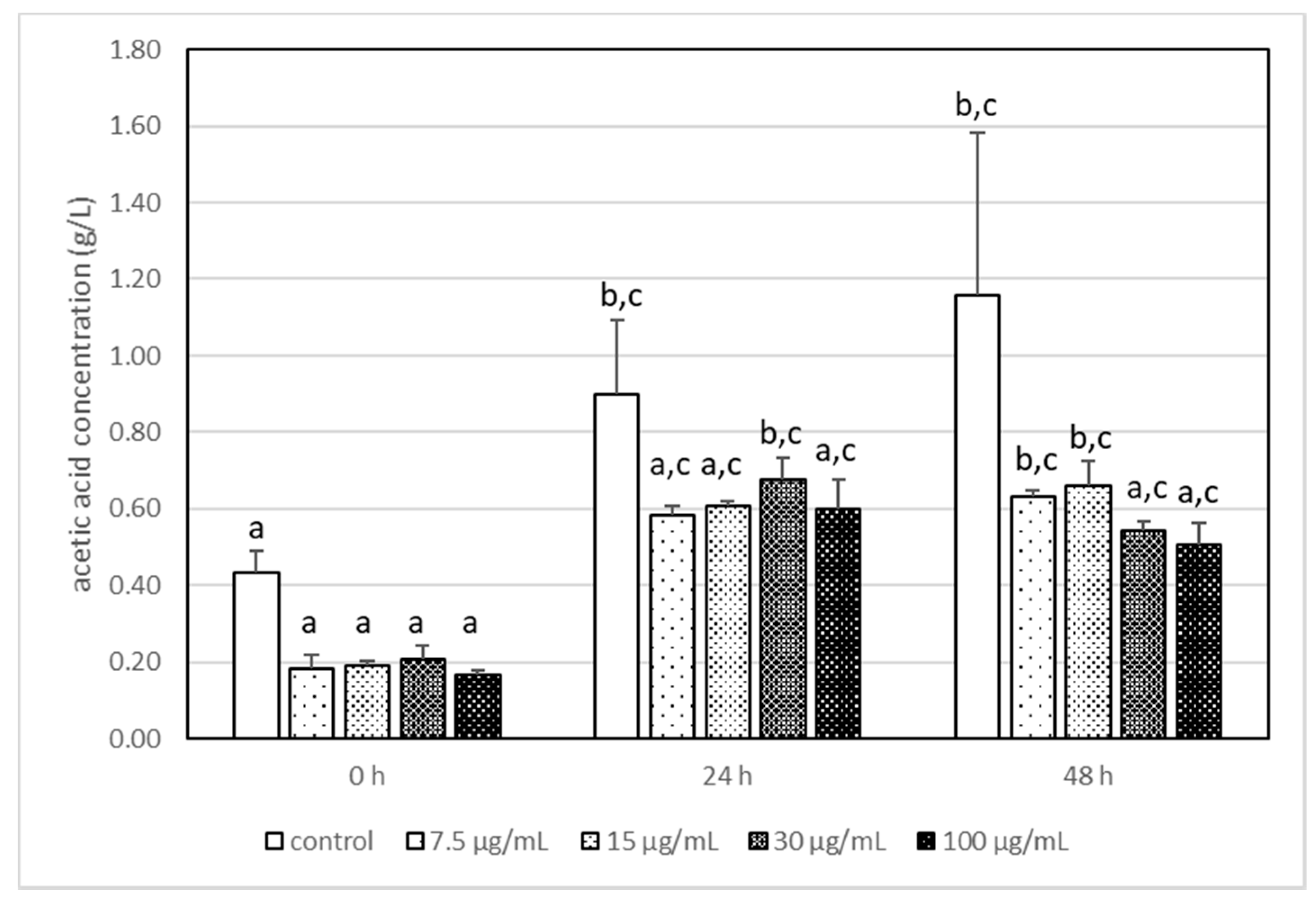
3.3. The Impact of Acrylamide on Microbial Metabolism–Analysis of Produced Metabolites
4. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- International Agency on Research on Cancer. IARC Monographs on the Evaluation of Carcinogenic Risks to Humans. 1994, Volume 60: Some Industrial Chemicals. Available online: https://publications.iarc.fr/Book-And-Report-Series/Iarc-Monographs-On-The-Identification-Of-Carcinogenic-Hazards-To-Humans/Some-Industrial-Chemicals-1994 (accessed on 23 October 2021).
- Duda-Chodak, A.; Wajda, Ł.; Tarko, T.; Sroka, P.; Satora, P. A review of the interactions between acrylamide, microorganisms and food components. Food Funct. 2016, 7, 1282–1295. [Google Scholar] [CrossRef]
- Maan, A.A.; Anjum, M.A.; Khan, M.K.I.; Nazir, A.; Saeed, F.; Afzaal, M.; Aadil, R.M. Acrylamide formation and different mitigation strategies during food processing–A review. Food Rev. Int. 2020. [Google Scholar] [CrossRef]
- Buranasilp, K.; Charoenpanich, J. Biodegradation of acrylamide by Enterobacter aerogenes isolated from wastewater in Thailand. J. Environ. Sci. 2011, 23, 396–403. [Google Scholar] [CrossRef]
- Petka, K.; Tarko, T.; Duda-Chodak, A. Is acrylamide as harmful as we think? A new look at the impact of acrylamide on the viability of beneficial intestinal bacteria of the genus Lactobacillus. Nutrients 2020, 12, 1157. [Google Scholar] [CrossRef] [Green Version]
- Serrano-Niño, J.C.; Cavazos-Garduño, A.; Cantú-Cornelio, F.; González-Córdova, A.F.; Vallejo-Córdoba, B.; Hernández-Mendoza, A.; García, H.S. In vitro reduced availability of aflatoxin B1 and acrylamide by bonding interactions with teichoic acids from Lactobacillus strains. LWT-Food Sci. Technol. 2015, 64, 1334–1341. [Google Scholar] [CrossRef]
- Koszucka, A.; Nowak, A.; Nowak, I.; Motyl, I. Acrylamide in human diet, its metabolism, toxicity, inactivation and the associated European Union legal regulations in food industry. Crit. Rev. Food Sci. Nutr. 2020, 60, 1677–1692. [Google Scholar] [CrossRef]
- Hill, C.; Guarner, F.; Reid, G.; Gibson, G.R.; Merenstein, D.J.; Pot, B.; Morelli, L.; Canani, R.B.; Flint, H.J.; Salminen, S.; et al. The international scientific association for probiotics and prebiotics consensus statement on the scope and appropriate use of the term probiotic. Nat. Rev. Gastroenterol. Hepatol. 2014, 11, 506–514. [Google Scholar] [CrossRef] [Green Version]
- Ayivi, R.D.; Gyawali, R.; Krastanov, A.; Aljaloud, S.O.; Worku, M.; Tahergorabi, R.; da Silva, R.C.; Ibrahim, S.A. Lactic acid bacteria: Food safety and human health applications. Dairy 2020, 1, 202–232. [Google Scholar] [CrossRef]
- Quinto, E.J.; Jiménez, P.; Caro, I.; Tejero, J.; Mateo, J.; Girbés, T. Probiotic lactic acid bacteria: A review. Food Nutr. Sci. 2014, 5, 1765–1775. [Google Scholar] [CrossRef] [Green Version]
- Cizeikiene, D.; Gaide, I.; Basinskiene, L. Effect of Lactic Acid Fermentation on Quinoa Characteristics and Quality of Quinoa-Wheat Composite Bread. Foods 2021, 10, 171. [Google Scholar] [CrossRef]
- Hadidi, M.; Majidiyan, N.; Jelyani, A.Z.; Moreno, A.; Hadian, Z.; Mousavi Khanegah, A. Alginate/Fish Gelatin-Encapsulated Lactobacillus acidophilus: A Study on Viability and Technological Quality of Bread during Baking and Storage. Foods 2021, 10, 2215. [Google Scholar] [CrossRef] [PubMed]
- Petka, K.; Wajda, Ł.; Duda-Chodak, A. The utilisation of acrylamide by selected microorganisms used for fermentation of food. Toxics 2021, 9, 295. [Google Scholar] [CrossRef] [PubMed]
- Barłowska, J.; Szwajkowska, M.; Litwińczuk, Z.; Król, J. Nutritional value and technological suitability of milk from various animal species used for dairy production. Compr. Rev. Food Sci. Food Saf. 2011, 10, 291–302. [Google Scholar] [CrossRef]
- Tarko, T.; Januszek, M.; Duda-Chodak, A.; Sroka, P. How keeving determines oenological parameters and concentration of volatile compounds in ciders? J. Food Anal. 2021, 100, 103897. [Google Scholar] [CrossRef]
- Wampler, D.A.; Ensign, S.A. Photoheterotrophic metabolism of acrylamide by a newly isolated strain of Rhodopseudomonas palustris. Appl. Environ. Microbiol. 2005, 71, 5850–5857. [Google Scholar] [CrossRef] [Green Version]
- Thanyacharoen, U.; Tani, A.; Charoenpanich, J. Isolation and characterization of Kluyvera georgiana strain with the potential for acrylamide biodegradation. J. Environ. Sci. Health Part A 2012, 47, 1491–1499. [Google Scholar] [CrossRef]
- Liu, S.-Q.; Holland, R.; McJarrow, P.; Crow, V.L. Serine metabolism in Lactobacillus plantarum. Int. J. Food Microbiol. 2003, 89, 265–273. [Google Scholar] [CrossRef]
- Hamzalioğlu, A.; Gökmen, V. Investigation of the reactions of acrylamide during in vitro multistep enzymatic digestion of thermally processed foods. Food Funct. 2015, 6, 108–113. [Google Scholar] [CrossRef]
- Jia, R.; Wan, X.; Geng, X.; Xue, D.; Xie, Z.; Chen, C. Microbial L-asparaginase for application in acrylamide mitigation from food: Current research status and future perspectives. Microorganisms 2021, 9, 1659. [Google Scholar] [CrossRef]
- Sun, Z.; Qin, R.; Li, D.; Ji, K.; Wang, T.; Cui, Z.; Huang, Y. A novel bacterial type II L-asparaginase and evaluation of its enzymatic acrylamide reduction in French fries. Int. J. Biol. Macromol. 2016, 92, 232–239. [Google Scholar] [CrossRef]
- Chi, H.; Chen, M.; Jiao, L.; Lu, Z.; Bie, X.; Zhao, H.; Lu, F. Characterization of a Novel L-Asparaginase from Mycobacterium gordonae with Acrylamide Mitigation Potential. Foods 2021, 10, 2819. [Google Scholar] [CrossRef] [PubMed]
- Rivas-Jimenez, L.; Ramírez-Ortiz, K.; González-Córdova, A.F.; Vallejo-Córdoba, B.; García, H.S.; Hernández-Mendoza, A. Evaluation of acrylamide-removing properties of two Lactobacillus strains under simulated gastrointestinal conditions using a dynamic system. Microbiol. Res. 2016, 190, 19–26. [Google Scholar] [CrossRef] [PubMed]
- Serrano-Niño, J.C.; Cavazos-Garduño, A.; González-Córdova, A.F.; Vallejo-Cordoba, B.; Hernández-Mendoza, A.; García, H.S. In vitro study of the potential protective role of Lactobacillus strains by acrylamide binding. J. Food Saf. 2014, 34, 62–68. [Google Scholar] [CrossRef]
- Bintsis, T. Lactic acid bacteria as starter cultures: An update in their metabolism and genetics. AIMS Microbiol. 2018, 4, 665–684. [Google Scholar] [CrossRef]
- Zalán, Z.; Hudáček, J.; Štětina, J.; Chumchalová, J.; Halász, A. Production of organic acids by Lactobacillus strains in three different media. Eur. Food Res. Technol. 2010, 230, 395–404. [Google Scholar] [CrossRef]
- Kim, W.S.; Perl, L.; Park, J.H.; Tandianus, J.E.; Dunn, N.W. Assessment of stress response of the probiotic Lactobacillus acidophilus. Curr. Microbiol. 2001, 43, 346–350. [Google Scholar] [CrossRef]
- Papadimitriou, K.; Alegría, Á.; Bron, P.A.; de Angelis, M.; Gobbetti, M.; Kleerebezem, M.; Lemos, J.A.; Linares, D.M.; Ross, P.; Stanton, C.; et al. Stress physiology of lactic acid bacteria. Microbiol. Mol. Biol. Rev. 2016, 80, 837–890. [Google Scholar] [CrossRef] [Green Version]
- Van de Guchte, M.; Serror, P.; Chervaux, C.; Smokvina, T.; Ehrlich, S.D.; Maguin, E. Stress responses in lactic acid bacteria. Antonie Leeuwenhoek 2002, 82, 187–216. [Google Scholar] [CrossRef]
- Hickey, M.W.; Hillier, A.J.; Jago, G.R. Metabolism of pyruvate and citrate in lactobacilli. Aust. J. Biol. Sci. 1983, 36, 487–796. [Google Scholar] [CrossRef] [Green Version]
- Meremäe, K.; Roasto, M.; Tamme, T.; Ivanova, M.; Hänninen, M.L.; Elias, P. In vitro study of the antimicrobial effect of selected probiotics combined with prebiotics on Campylobacter jejuni. Arch. Lebensm. 2010, 61, 132–138. [Google Scholar] [CrossRef]
- Zhai, Z.; Douillard, F.P.; An, H.; Wang, G.; Guo, X.; Luo, Y.; Hao, Y. Proteomic characterization of the acid tolerance response in Lactobacillus delbrueckii subsp. bulgaricus CAUH1 and functional identification of a novel acid stress-related transcriptional regulator Ldb0677. Environ. Microbiol. 2014, 16, 1524–1537. [Google Scholar] [CrossRef] [PubMed]
- Li, J.; Ma, M.; Sarker, M.R.; McClane, B.A. CodY is a global regulator of virulence-associated properties for Clostridium perfringens type D strain CN3718. mBio 2013, 4, e00770-13. [Google Scholar] [CrossRef] [PubMed] [Green Version]
| Time (h) | Initial Acrylamide Concentration (µg/mL) | Changes in Acrylamide Concentration (Positive Control vs. Test Sample) |
|---|---|---|
| 0 | 7.5 | 0.1 ± 0.28 a |
| 24 | 0.2 ± 0.32 a | |
| 48 | 0.4 ± 0.16 a | |
| 0 | 15.0 | 4.4 ± 0.40 a |
| 24 | 5.1 ± 0.22 b | |
| 48 | 5.7 ± 0.22 c | |
| 0 | 30.0 | 0.2 ± 0,59 a |
| 24 | 0.3 ± 0,18 a | |
| 48 | 0.7 ± 0.92 a | |
| 0 | 100.0 | 0.6 ± 0.39 a |
| 24 | 4.6 ± 0.89 b | |
| 48 | 7.0 ± 2.25 c |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Petka, K.; Sroka, P.; Tarko, T.; Duda-Chodak, A. The Acrylamide Degradation by Probiotic Strain Lactobacillus acidophilus LA-5. Foods 2022, 11, 365. https://doi.org/10.3390/foods11030365
Petka K, Sroka P, Tarko T, Duda-Chodak A. The Acrylamide Degradation by Probiotic Strain Lactobacillus acidophilus LA-5. Foods. 2022; 11(3):365. https://doi.org/10.3390/foods11030365
Chicago/Turabian StylePetka, Katarzyna, Paweł Sroka, Tomasz Tarko, and Aleksandra Duda-Chodak. 2022. "The Acrylamide Degradation by Probiotic Strain Lactobacillus acidophilus LA-5" Foods 11, no. 3: 365. https://doi.org/10.3390/foods11030365
APA StylePetka, K., Sroka, P., Tarko, T., & Duda-Chodak, A. (2022). The Acrylamide Degradation by Probiotic Strain Lactobacillus acidophilus LA-5. Foods, 11(3), 365. https://doi.org/10.3390/foods11030365






