Optimizing Procedures for Antioxidant Phenolics Extraction from Skin and Kernel of Peanuts with Contrasting Levels of Drought Tolerance
Abstract
1. Introduction
2. Materials and Methods
2.1. Contrasting Germplasm of Arachis Hypogaea
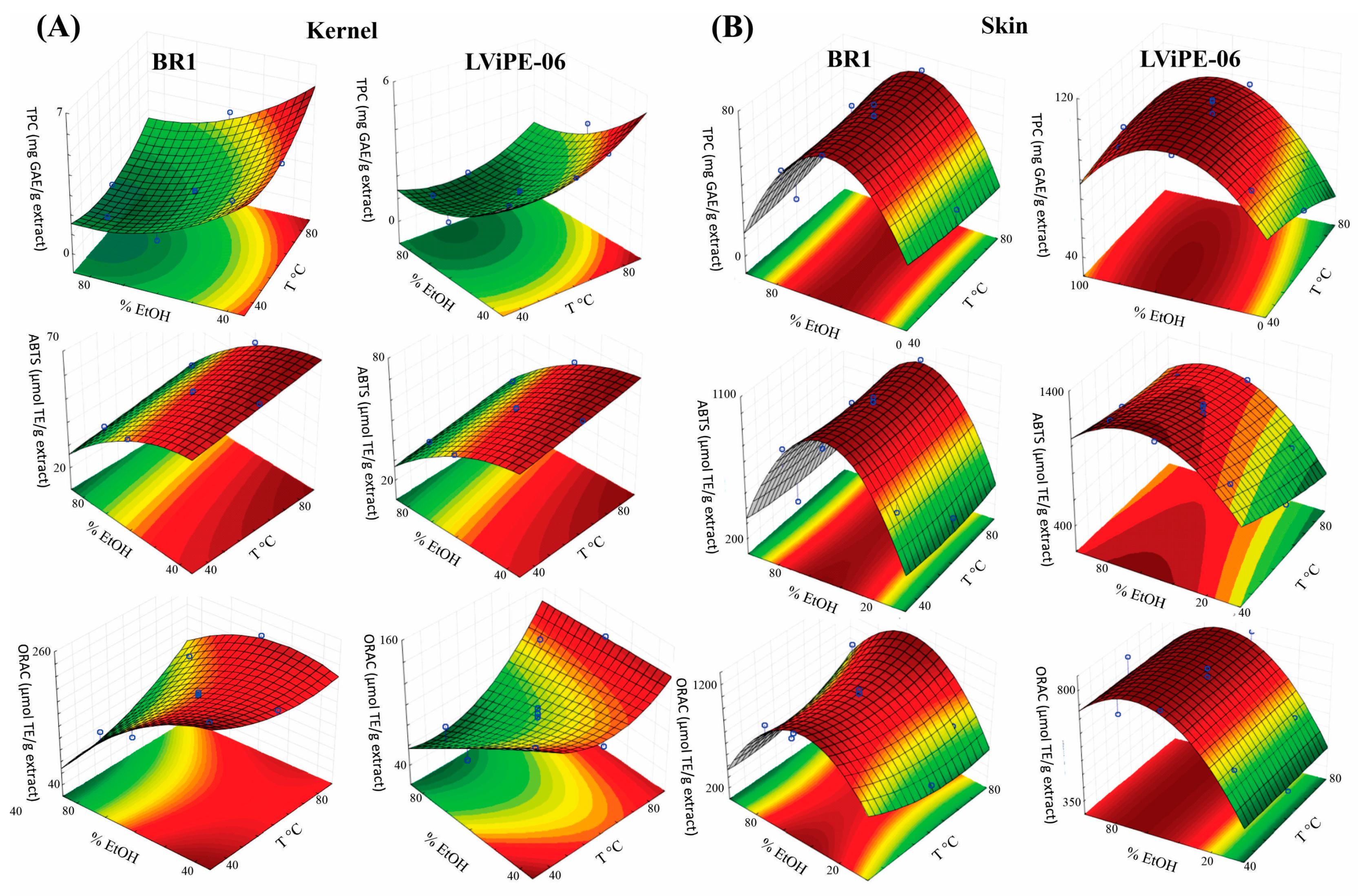
2.2. Experimental Design by Response Surface Methodology (RSM)
2.3. Total Phenolic Content (TPC)
2.4. Identification and Quantification of Phenolic Compounds Using High-Performance Liquid Chromatography Coupled with Electrospray Ioniza-Tion-Quadrupole-Time of Flight-Mass Spectrometry (HPLC-ESI-QTOF-MS) and Using High-Performance Liquid Chromatography with Photodiode Array Detector (HPLC-PDA)
2.5. Reactive Oxygen Species (ROS) Scavenging Activity
2.6. Statistical Analysis
3. Results
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
References
- Lozano, M.G.; de Oliveira Sartori, A.G.; Markowicz Bastos, D.H.; Bismara Regitano-d’Arce, M.A. Selected nutrients and antinutrients in peanut cultivars harvested in Brazil. J. Sci. Food Agric. 2019, 99, 5334–5340. [Google Scholar] [CrossRef] [PubMed]
- Juliano, F.F.; de Alvarenga, J.F.R.; Lamuela-Raventos, R.M.; Massarioli, A.P.; Lima, L.M.; Santos, R.C.; Alencar, S.M. Polyphenol analysis using high-resolution mass spectrometry allows differentiation of drought tolerant peanut genotypes. J. Sci. Food Agric. 2020, 100, 721–731. [Google Scholar] [CrossRef] [PubMed]
- Juliano, F.F.; Massarioli, A.P.; Lamuela-Raventos, R.M.; de Alvarenga, J.F.R.; de Lima, L.M.; dos Santos, R.C.; da Silva, C.F.; de Alencar, S.M. Do drought-adapted peanut genotypes have different bioactive compounds and ROS-scavenging activity? Eur. Food Res. Technol. 2021, 247, 1369–1378. [Google Scholar] [CrossRef]
- de Camargo, A.C.; Regitano-d’Arce, M.A.B.; Gallo, C.R.; Shahidi, F. Gamma-irradiation induced changes in microbiological status, phenolic profile and antioxidant activity of peanut skin. J. Funct. Foods 2015, 12, 129–143. [Google Scholar] [CrossRef]
- da Silva, R.A.; Ramos, J.P.C.; da Luz, L.N.; Cavalcanti, J.J.V.; de Lima, L.M.; Freire, R.M.M.; da Silva, C.R.C.; dos Santos, R.C. Assessment of genetic divergence in runner peanut genotypes grown in the Brazilian Northeast environments. African J. Agric. Res. 2016, 11, 1456–1462. [Google Scholar]
- Pereira, J.W.D.L.; Albuquerque, M.B.; Filho, P.A.M.; Nogueira, R.J.M.C.; de Lima, L.M.; Santos, R.C. Assessment of drought tolerance of peanut cultivars based on physiological and yield traits in a semiarid environment. Agric. Water Manag. 2016, 166, 70–76. [Google Scholar] [CrossRef]
- Dutra, W.F.; Guerra, Y.L.; Ramos, J.P.C.; Fernandes, P.D.; Silva, C.R.C.; Bertioli, D.J.; Leal-Bertioli, S.C.M.; Santos, R.C. Introgression of wild alleles into the tetraploid peanut crop to improve water use efficiency, earliness and yield. PLoS ONE 2018, 13, e0198776. [Google Scholar] [CrossRef] [PubMed]
- Yadav, B.; Jogawat, A.; Rahman, M.S.; Narayan, O.P. Secondary metabolites in the drought stress tolerance of crop plants: A review. Gene Rep. 2021, 23, 101040. [Google Scholar] [CrossRef]
- Akcay, U.C.; Ercan, O.; Kavas, M.; Yildiz, L.; Yilmaz, C.; Oktem, H.A.; Yucel, M. Drought-induced oxidative damage and antioxidant responses in peanut (Arachis hypogaea L.) seedlings. Plant Growth Regul. 2010, 61, 21–28. [Google Scholar] [CrossRef]
- Neto, A.D.A.; Nogueira, R.J.M.C.; Filho, P.A.M.; Santos, R.C. Physiological and biochemical responses of peanut genotypes to water deficit. J. Plant Interact. 2010, 5, 1–10. [Google Scholar] [CrossRef]
- Aninbon, C.; Jogloy, S.; Vorasoot, N.; Patanothai, A.; Nuchadomrong, S.; Senawong, T. Effect of end of season water deficit on phenolic compounds in peanut genotypes with different levels of resistance to drought. Food Chem. 2016, 196, 123–129. [Google Scholar] [CrossRef] [PubMed]
- Chukwumah, Y.; Walker, L.; Vogler, B.; Verghese, M. Changes in the phytochemical composition and profile of raw, boiled, and roasted peanuts. J. Agric. Food Chem. 2007, 55, 9266–9273. [Google Scholar] [CrossRef] [PubMed]
- Mahatma, M.K.; Thawait, L.K.; Bishi, S.K.; Khatediya, N.; Rathnakumar, A.L.; Lalwani, H.B.; Misra, J.B. Nutritional composition and antioxidant activity of Spanish and Virginia groundnuts (Arachis hypogaea L.): A comparative study. J. Food Sci. Technol. 2016, 53, 2279–2286. [Google Scholar] [CrossRef] [PubMed]
- Ballard, T.S.; Mallikarjunan, P.; Zhou, K.; O’keefe, S.F. Optimizing the extraction of phenolic antioxidants from peanut skins using response surface methodology. J. Agric. Food Chem. 2009, 57, 3064–3072. [Google Scholar] [CrossRef] [PubMed]
- Attree, R.; Du, B.; Xu, B. Distribution of phenolic compounds in seed coat and cotyledon, and their contribution to antioxidant capacities of red and black seed coat peanuts (Arachis hypogaea L.). Ind. Crops Prod. 2015, 67, 448–456. [Google Scholar] [CrossRef]
- Food and Agriculture Organization of the United Nations. FAOSTAT. Available online: https://www.fao.org/faostat/en/#data/QCL (accessed on 13 January 2022).
- Chukwumah, Y.; Walker, L.T.; Verghese, M. Peanut skin color: A biomarker for total polyphenolic content and antioxidative capacities of peanut cultivars. Int. J. Mol. Sci. 2009, 10, 4941–4952. [Google Scholar] [CrossRef]
- Sarnoski, P.J.; Johnson, J.V.; Reed, K.A.; Tanko, J.M.; O’Keefe, S.F. Separation and characterisation of proanthocyanidins in Virginia type peanut skins by LC-MSn. Food Chem. 2012, 131, 927–939. [Google Scholar] [CrossRef]
- Khuri, A.I.; Mukhopadhyay, S. Response surface methodology. Wiley Interdiscip. Rev. Comput. Stat. 2010, 2, 128–149. [Google Scholar] [CrossRef]
- de Vasconcelos, F.M.T.; de Vasconcelos, R.A.; da Luz, L.N.; Thiago Cabral, N.; de Oliveira Júnior, J.O.L.; Dias Santiago, A.; Sgrillo, E.; Correia Farias, F.J.; de Albuquerque Melo Filho, P.; dos Santos, R.C. Adaptabilidade e estabilidade de genótipos eretos de amendoim cultivados nas regiões Nordeste e Centro-Oeste. Cienc. Rural 2015, 45, 1375–1380. [Google Scholar] [CrossRef]
- Nogueira, R.J.M.C.; dos Santos, R.C. Alterações fisiológicas no amendoim submetido ao estresse hídrico. Rev. Bras. Eng. Agrícola e Ambient. 2000, 4, 41–45. [Google Scholar] [CrossRef]
- Gomes, L.d.R.; dos Santos, R.C.; da Anunciação Filho, C.J.; Melo Filho, P.d.A. Adaptabilidade e estabilidade fenotípica de genótipos de amendoim de porte ereto. Pesqui. Agropecuária Bras. 2007, 42, 985–989. [Google Scholar] [CrossRef]
- Hoffmann-ribani, R.; Rodriguez-amaya, D.B. Optimization of a method for determination of flavonols and flavones in fruits by HPLC using statistical design and response surface analysis. Química Nova 2008, 31, 1378–1384. [Google Scholar] [CrossRef]
- Melo, P.S.; Massarioli, A.; Denny, C.; dos Santos, L.F.; Franchin, M.; Pereira, G.E.; Vieira, T.M.F.D.S.; Rosalen, P.L.; de Alencar, S.M. Winery by-products: Extraction optimization, phenolic composition and cytotoxic evaluation to act as a new source of scavenging of reactive oxygen species. Food Chem. 2015, 181, 160–169. [Google Scholar] [CrossRef] [PubMed]
- Chisté, R.C.; Mercadante, A.Z.; Gomes, A.; Fernandes, E.; Lima, J.; Bragagnolo, N. In vitro scavenging capacity of annatto seed extracts against reactive oxygen and nitrogen species. Food Chem. 2011, 127, 419–426. [Google Scholar] [CrossRef] [PubMed]
- Mariutti, L.R.B.; Rodrigues, E.; Chisté, R.C.; Fernandes, E.; Mercadante, A.Z. The Amazonian fruit Byrsonima crassifolia effectively scavenges reactive oxygen and nitrogen species and protects human erythrocytes against oxidative damage. Food Res. Int. 2014, 64, 618–625. [Google Scholar] [CrossRef] [PubMed]
- Lilliefors, H.W. On the Kolmogorov-Smirnov Test for Normality with Mean and Variance Unknown. J. Am. Stat. Assoc. 1967, 62, 399–402. [Google Scholar] [CrossRef]
- Bartlett, M.S. Properties of Sufficiency and Statistical Tests. Proc. R. Soc. Lond. Ser. A-Math. Phys. Sci. 1937, 160, 268–282. [Google Scholar] [CrossRef]
- Cruz, C.D. GENES—Software para análise de dados em estatística experimental e em genética quantitativa. Acta Sci.-Agron. 2013, 35, 271–276. [Google Scholar]
- Ma, Y.; Kosinska-Cagnazzo, A.; Kerr, W.L.; Amarowicz, R.; Swanson, R.B.; Pegg, R.B. Separation and characterization of phenolic compounds from dry-blanched peanut skins by liquid chromatography-electrospray ionization mass spectrometry. J. Chromatogr. A 2014, 1356, 64–81. [Google Scholar] [CrossRef] [PubMed]
- Cádiz-Gurrea, M.d.l.L.; Fernández-Arroyo, S.; Joven, J.; Segura-Carretero, A. Comprehensive characterization by UHPLC-ESI-Q-TOF-MS from an Eryngium bourgatii extract and their antioxidant and anti-inflammatory activities. Food Res. Int. 2013, 50, 197–204. [Google Scholar] [CrossRef]
- Pierini, G.D.; Maccio, S.A.; Robledo, S.N.; Ferrari, A.G.-M.; Banks, C.E.; Fernández, H.; Zon, M.A. Screen-printed electrochemical-based sensor for taxifolin determination in edible peanut oils. Microchem. J. 2020, 159, 105442. [Google Scholar] [CrossRef]
- Hassanein, A.; Ibrahim, E.; Ali, R.A.; Hashem, H. Differential metabolic responses associated with drought tolerance in egyptian rice. J. Appl. Biol. Biotechnol. 2021, 9, 37–46. [Google Scholar]
- Raval, S.S.; Mahatma, M.K.; Chakraborty, K.; Bishi, S.K.; Singh, A.L.; Rathod, K.J.; Jadav, J.K.; Sanghani, J.M.; Mandavia, M.K.; Gajera, H.; et al. Metabolomics of groundnut (Arachis hypogaea L.) genotypes under varying temperature regimes. Plant Growth Regul. 2018, 84, 493–505. [Google Scholar] [CrossRef]
- Rice-Evans, C.A.; Miller, N.J.; Paganga, G. Structure-antioxidant activity relationships of flavonoids and phenolic acids. Free Radic. Biol. Med. 1996, 20, 933–956. [Google Scholar] [CrossRef]
- Soares, J.C.; Rosalen, P.L.; Lazarini, J.G.; Massarioli, A.P.; da Silva, C.F.; Nani, B.D.; Franchin, M.; de Alencar, S.M. Comprehensive characterization of bioactive phenols from new Brazilian superfruits by LC-ESI-QTOF-MS, and their ROS and RNS scavenging effects and anti-inflammatory activity. Food Chem. 2019, 281, 178–188. [Google Scholar] [CrossRef]
- Vissotto, L.C.; Rodrigues, E.; Chisté, R.C.; Benassi, M.d.T.; Mercadante, A.Z. Correlation, by multivariate statistical analysis, between the scavenging capacity against reactive oxygen species and the bioactive compounds from frozen fruit pulps. Food Sci. Technol. 2013, 33, 57–65. [Google Scholar] [CrossRef]
- Samak, G.; Shenoy, R.P.; Manjunatha, S.M.; Vinayak, K.S. Superoxide and hydroxyl radical scavenging actions of botanical extracts of Wagatea spicata. Food Chem. 2009, 115, 631–634. [Google Scholar] [CrossRef]
- Santos, R.C. Novas Cultivares Brs 151 L-7: Nova Cultivar De Amendoim. Pesq. Agropec. Bras. 2000, 35, 665–670. [Google Scholar] [CrossRef]
- Santos, R.C.; Rêgo, G.M.; da Silva, A.P.; Vasconcelos, J.O.; Coutinho, J.L.; Melo Filho, P.D.A. Produtividade de linhagens avançadas de amendoim em condições de sequeiro no Nordeste brasileiro. Rev. Bras. Eng. Agrícola Ambient. 2010, 14, 589–593. [Google Scholar] [CrossRef][Green Version]
- dos Santos, R.C.; Silva, A.F.; Gondim, T.M.S.; de Oliveira Junior, J.O.L.; de Araujo Neto, R.B.; Sagrilo, E.; de Vasconcelos, R.A.; Melo Filho, P.d.A.; da Silva Filho, J.L. Stability and adaptability of runner peanut genotypes based on nonlinear regression and AMMI analysis. Pesqui. Agropecu. Bras. 2012, 47, 1118–1124. [Google Scholar] [CrossRef]

| Botanic Type | Genotype | Origin/Genetic Base | Skin Color | Seed Size | Cycle (Days) | Drought Tolerance |
|---|---|---|---|---|---|---|
| Spanish | Senegal 55437 | Africa/Cultivar | Tan | Small | 75–80 | Tolerant |
| L7 Bege | Brazil/Top line | Tan | Large | 85–90 | Tolerant | |
| Senegal 57422 | Africa/Cultivar | Tan | Average | 85–90 | Tolerant | |
| L50 | Africa/Top line | Tan | Average | 85–90 | Tolerant | |
| Virginia | LViPE-06 | Brazil/Top line | Tan | Extra large | 125–130 | Sensitive |
| LGoPE-06 | Brazil/Top line | Tan | Extra large | 125–130 | Sensitive | |
| F.M407B | Brazil/Top line | Tan | Large | 110–115 | Sensitive | |
| M407.424B | Brazil/Top line | Red | Large | 110–115 | Sensitive | |
| F.M424B | Brazil/Top line | Tan | Large | 115–125 | Mid tolerant | |
| Florunner | USA/Cultivar | Tan | Large | 120–125 | Sensitive | |
| Valencia | BR1 | Brazil/Cultivar | Red | Average | 85–89 | Tolerant |
| Tatu | Argentina/Cultivar | Red | Average | 95–100 | Sensitive | |
| Porto Alegre | Brazil/Accession | Tan | Average | 118–120 | Sensitive | |
| BRS151 L7 | Brazil/Cultivar | Red | Large | 85–89 | Tolerant |
| Genotype | TPC (mg GAE/g Extract) * | Phenolic Profile (mg/g Extract) | ||
|---|---|---|---|---|
| Caffeic Acid | p-Coumaric Acid | Rutin | ||
| Senegal 55437 (DT) | 21.03 ± 1.23 b | 0.329 ± 0.009 | 0.192 ± 0.001 | n.d. |
| L7 Bege (DT) | 18.82 ± 0.30 d | 0.203 ± 0.028 | 0.220 ± 0.018 | n.d. |
| Senegal 57422 (DT) | 22.29 ± 0.62 b | 0.215 ± 0.008 | 0.101 ± 0.004 | n.d. |
| L50 (DT) | 19.87 ± 0.69 c | 0.170 ± 0.016 | 0.180 ± 0.016 | n.d. |
| LViPE-06 (DS) | 21.77 ± 0.80 b | 0.134 ± 0.004 | 0.190 ± 0.005 | n.d. |
| LGoPE-06 (DS) | 20.33 ± 0.40 c | 0.163 ± 0.002 | 0.163 ± 0.004 | n.d. |
| FM407B (DS) | 20.00 ± 0.14 c | 0.187 ± 0.013 | 0.240 ± 0.009 | n.d. |
| M.407.424B (DS) | 21.43 ± 0.58 b | 0.136 ± 0.006 | 0.268 ± 0.017 | n.d. |
| FM.424B (MDT) | 21.55 ± 0.77 b | 0.176 ± 0.015 | 0.125 ± 0.001 | n.d. |
| Florunner (DS) | 21.42 ± 1.41 b | 0.208 ± 0.013 | 0.157 ± 0.005 | n.d. |
| BR1 (DT) | 28.72 ± 0.90 a | 0.239 ± 0.003 | 0.555 ± 0.018 | 0.061 ± 0.002 |
| Tatu (DS) | 20.80 ± 0.88 b | 0.143 ± 0.037 | 0.176 ± 0.027 | 0.054 ± 0.004 |
| Porto Alegre (DS) | 20.38 ± 0.48 c | 0.261 ± 0.028 | 0.168 ± 0.011 | n.d. |
| BRS151 L7 (DT) | 18.56 ± 0.72 d | 0.143 ± 0.005 | 0.187 ± 0.013 | n.d. |
| LOD (µg/mL) | 0.089 | 0.040 | 0.014 | |
| LOQ (µg/mL) | 0.274 | 0.123 | 0.019 | |
| Linearity (R2) | 0.9998 | 0.9999 | 0.9998 | |
| Genotype | TPC (mg GAE/g Extract) * | Phenolic Profile (mg/g Extract) | ||||||
|---|---|---|---|---|---|---|---|---|
| Protocatechuic Acid | (+)-Catechin | Procyanidin A2 | Quercetin | Rutin | Quercetin-3-β-Glucoside | Kaempferol-3-Glucoside | ||
| Senegal 55437 (DT) | 612.87 ± 5.91 b | 0.212 ± 0.007 | 3.33 ± 0.33 | 6.15 ± 0.84 | 0.076 ± 0.006 | 0.102 ± 0.004 | n.d. | n.d. |
| L7 Bege (DT) | 555.84 ± 18.17 c | 0.176 ± 0.009 | 2.41 ± 0.19 | 7.93 ± 0.31 | 0.089 ± 0.006 | 0.055 ± 0.002 | n.d. | n.d. |
| Senegal 57422 (DT) | 608.21 ± 55.39 b | 0.235 ± 0.011 | 0.59 ± 0.08 | 2.42 ± 0.76 | 0.024 ± 0.006 | 0.076 ± 0.004 | n.d. | 0.041 ± 0.001 |
| L50 (DT) | 555.89 ± 26.02 c | 0.218 ± 0.026 | 0.79 ± 0.05 | 2.62 ± 0.13 | 0.040 ± 0.004 | 0.046 ± 0.002 | n.d. | n.d. |
| LViPE-06 (DS) | 467.57 ± 47.67 d | 0.092 ± 0.012 | n.d. | n.d. | 0.036 ± 0.001 | 0.043 ± 0.011 | n.d. | 0.077 ± 0.007 |
| LGoPE-06 (DS) | 506.89 ± 15.41 d | 0.059 ± 0.006 | n.d. | n.d. | 0.036 ± 0.001 | 0.042 ± 0.013 | n.d. | n.d. |
| FM407B (DS) | 673.67 ± 52.64 a | 0.206 ± 0.005 | 1.44 ± 0.04 | 5.37 ± 0.70 | 0.103 ± 0.003 | 0.060 ± 0.007 | n.d. | n.d. |
| M.407.424B (DS) | 552.83 ± 24.99 c | 0.675 ± 0.027 | 0.96 ± 0.03 | 3.89 ± 0.24 | 0.761 ± 0.032 | 0.379 ± 0.033 | 1.02 ± 0.05 | 0.099 ± 0.002 |
| FM.424B (MDT) | 538.14 ± 12.72 c | 0.210 ± 0.004 | n.d. | 1.26 ± 0.48 | 0.055 ± 0.001 | 0.178 ± 0.006 | n.d. | n.d. |
| Florunner (DS) | 512.30 ± 3.87 d | 0.252 ± 0.033 | n.d. | 1.30 ± 0.27 | 0.019 ± 0.001 | 0.171 ± 0.006 | n.d. | n.d. |
| BR1 (DT) | 545.62 ± 6.84 c | 0.687 ± 0.078 | 1.61 ± 0.20 | 3.70 ± 0.72 | 0.580 ± 0.063 | 0.439 ± 0.025 | 0.82 ± 0.05 | 0.061 ± 0.007 |
| Tatu (DS) | 568.70 ± 3.84 c | 0.826 ± 0.051 | 1.63 ± 0.12 | 4.66 ± 0.38 | 0.776 ± 0.048 | 0.440 ± 0.063 | 1.33 ± 0.19 | 0.104 ± 0.007 |
| Porto Alegre (DS) | 558.54 ± 26.72 c | 0.229 ± 0.010 | n.d. | n.d. | 0.038 ± 0.001 | 0.074 ± 0.010 | n.d. | n.d. |
| BRS151 L7 (DT) | 538.33 ± 5.95 c | 0.220 ± 0.003 | 3.13 ± 0.17 | 4.43 ± 0.40 | 0.088 ± 0.003 | 0.051 ± 0.005 | n.d. | n.d. |
| LOD (µg/mL) | 0.009 | 0.03 | 0.23 | 0.014 | 0.008 | 0.04 | 0.010 | |
| LOQ (µg/mL) | 0.045 | 0.09 | 0.71 | 0.041 | 0.024 | 0.11 | 0.039 | |
| Linearity (R2) | 0.9991 | 0.9999 | 0.9999 | 0.9999 | 0.9999 | 0.9999 | 0.9998 | |
| Seed Part/Genotype | O2•− | H2O2 | HOCl | •OH | ROO• |
|---|---|---|---|---|---|
| % Inhibition for Kernels or IC50 (μg/mL) for Skins | IC25 (μg/mL) for Kernels or IC50 (μg/mL) for Skins | IC50 (μg/mL) | IC50 (μg/mL) | (μmol TE/g) | |
| Kernel | |||||
| Senegal 55437 (DT) | 4.36 ± 0.79 h | 443.23 ± 5.57 f | 19.11 ± 0.57 c | 2.76 ± 0.14 d | 242.77 ± 12.87 e |
| L7 Bege (DT) | 18.47 ± 0.25 d | 505.32 ± 4.23 e | 18.71 ± 0.10 c | 5.02 ± 0.17 c | 296.09 ± 1.80 d |
| Senegal 57422 (DT) | 24.49 ± 1.91 b | 397.13 ± 15.09 f | 15.38 ± 0.19 d | 5.79 ± 0.43 b | 246.30 ± 28.41 e |
| L50 (DT) | 5.96 ± 0.82 h | 777.65 ± 32.98 c | 24.24 ± 4.96 b | 5.45 ± 0.44 b | 163.97 ± 3.22 g |
| LViPE-06 (DS) | 19.98 ± 1.33 c | 228.32 ± 15.08 h | 17.30 ± 0.61 c | 4.82 ± 0.17 c | 354.60 ± 32.17 c |
| LGoPE-06 (DS) | 8.46 ± 1.15 g | 656.18 ± 19.98 d | 13.90 ± 1.24 d | 5.03 ± 0.29 c | 325.44 ± 0.52 d |
| FM407B (DS) | 15.39 ± 0.95 e | 870.23 ± 23.34 a | 11.09 ± 0.73 e | 4.36 ± 0.32 c | 160.37 ± 26.54 g |
| M.407.424B (DS) | 13.66 ± 0.80 f | 760.11 ± 12.53 c | 16.03 ± 0.65 d | 3.93 ± 0.40 c | 279.57 ± 21.62 e |
| FM.424B (MDT) | 21.43 ± 0.21 c | 631.92 ± 46.76 d | 15.59 ± 0.76 d | 4.58 ± 0.46 c | 241.88 ± 25.16 e |
| Florunner (DS) | 17.90 ± 0.83 d | 802.06 ± 42.28 c | 16.50 ± 0.48 d | 6.86 ± 0.77 a | 249.97 ± 9.29 e |
| BR1 (DT) | 28.65 ± 0.18 a | 304.61 ± 28.50 g | 18.44 ± 0.46 c | 4.69 ± 0.15 c | 738.97 ± 6.89 a |
| Tatu (DS) | 21.04 ± 3.08 c | 430.76 ± 36.16 f | 33.63 ± 2.10 a | 4.42 ± 0.06 c | 401.49 ± 22.24 b |
| Porto Alegre (DS) | 7.57 ± 0.14 g | 820.01 ± 16.95 b | 20.16 ± 1.57 c | 6.70 ± 0.69 a | 205.20 ± 28.74 f |
| BRS151 L7 (DT) | 12.68 ± 0.83 f | 771.94 ± 13.44 c | 21.22 ± 0.01 c | 4.15 ± 0.67 c | 234.08 ± 20.92 e |
| Skin | |||||
| Senegal 55437 (DT) | 12.23 ± 0.49 h | 38.96 ± 1.93 b | 1.68 ± 0.16 c | 0.050 ± 0.011 e | 5151.17 ± 3.76 a |
| L7 Bege (DT) | 17.95 ± 1.52 e | 38.66 ± 1.34 b | 1.67 ± 0.09 c | 0.060 ± 0.001 e | 4542.50 ± 313.92 c |
| Senegal 57422 (DT) | 19.48 ± 0.94 d | 31.79 ± 2.15 c | 2.12 ± 0.19 b | 0.067 ± 0.003 d | 4072.05 ± 44.26 c |
| L50 (DT) | 16.31 ± 0.43 f | 37.76 ± 2.91 b | 1.88 ± 0.02 c | 0.077 ± 0.005 c | 4371.66 ± 11.63 b |
| LViPE-06 (DS) | 32.89 ± 2.24 a | 40.14 ± 1.92 a | 2.45 ± 0.19 a | 0.104 ± 0.018 a | 3108.93 ± 237.57 f |
| LGoPE-06 (DS) | 27.99 ± 0.87 b | 42.84 ± 2.08 a | 2.00 ± 0.08 b | 0.103 ± 0.024 a | 3166.66 ± 118.59 f |
| FM407B (DS) | 14.40 ± 0.16 g | 38.23 ± 2.14 b | 1.81 ± 0.11 c | 0.064 ± 0.004 d | 4906.55 ± 94.01 a |
| M.407.424B (DS) | 16.26 ± 1.40 f | 29.07 ± 0.93 c | 2.17 ± 0.08 b | 0.056 ± 0.002 e | 5093.12 ± 171.13 a |
| FM.424B (MDT) | 16.52 ± 0.86 f | 36.46 ± 2.19 b | 2.01 ± 0.19 b | 0.071 ± 0.007 c | 4465.02 ± 219.20 b |
| Florunner (DS) | 21.10 ± 0.36 d | 42.18 ± 0.17 a | 1.73 ± 0.09 c | 0.095 ± 0.002 b | 3754.35 ± 64.26 d |
| BR1 (DT) | 17.83 ± 1.13 e | 25.89 ± 2.75 d | 2.08 ± 0.04 b | 0.077 ± 0.002 c | 4666.06 ± 125.27 b |
| Tatu (DS) | 14.41 ± 0.25 g | 24.63 ± 1.90 d | 1.70 ± 0.01 c | 0.073 ± 0.001 c | 5230.78 ± 65.56 a |
| Porto Alegre (DS) | 23.64 ± 1.08 c | 38.58 ± 0.11 b | 2.27 ± 0.27 a | 0.089 ± 0.005 b | 3444.22 ± 35.54 e |
| BRS151 L7 (DT) | 15.39 ± 0.84 f | 37.44 ± 0.97 b | 1.74 ± 0.04 c | 0.065 ± 0.001 d | 4604.19 ± 225.37 b |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Massarioli, A.P.; Sartori, A.G.d.O.; Juliano, F.F.; Santos, R.C.d.; Ramos, J.P.C.; Lima, L.M.d.; Alencar, S.M.d. Optimizing Procedures for Antioxidant Phenolics Extraction from Skin and Kernel of Peanuts with Contrasting Levels of Drought Tolerance. Foods 2022, 11, 449. https://doi.org/10.3390/foods11030449
Massarioli AP, Sartori AGdO, Juliano FF, Santos RCd, Ramos JPC, Lima LMd, Alencar SMd. Optimizing Procedures for Antioxidant Phenolics Extraction from Skin and Kernel of Peanuts with Contrasting Levels of Drought Tolerance. Foods. 2022; 11(3):449. https://doi.org/10.3390/foods11030449
Chicago/Turabian StyleMassarioli, Adna P., Alan G. de O. Sartori, Fernanda F. Juliano, Roseane C. dos Santos, Jean Pierre C. Ramos, Liziane Maria de Lima, and Severino Matias de Alencar. 2022. "Optimizing Procedures for Antioxidant Phenolics Extraction from Skin and Kernel of Peanuts with Contrasting Levels of Drought Tolerance" Foods 11, no. 3: 449. https://doi.org/10.3390/foods11030449
APA StyleMassarioli, A. P., Sartori, A. G. d. O., Juliano, F. F., Santos, R. C. d., Ramos, J. P. C., Lima, L. M. d., & Alencar, S. M. d. (2022). Optimizing Procedures for Antioxidant Phenolics Extraction from Skin and Kernel of Peanuts with Contrasting Levels of Drought Tolerance. Foods, 11(3), 449. https://doi.org/10.3390/foods11030449






