Microbial Ecology of Sheep Milk, Artisanal Feta, and Kefalograviera Cheeses. Part II: Technological, Safety, and Probiotic Attributes of Lactic Acid Bacteria Isolates
Abstract
:1. Introduction
2. Materials and Methods
2.1. Microbial Strains and Culture Conditions
2.2. Technological Properties
2.2.1. Milk Acidification and Coagulation
2.2.2. Caseinolytic and Lipolytic Capacity
2.2.3. Production of Exopolysaccharides (EPS)
2.3. Safety Properties
2.3.1. Production of Antimicrobial Compounds
2.3.2. Biogenic Amine Production
2.3.3. Hemolytic Activity
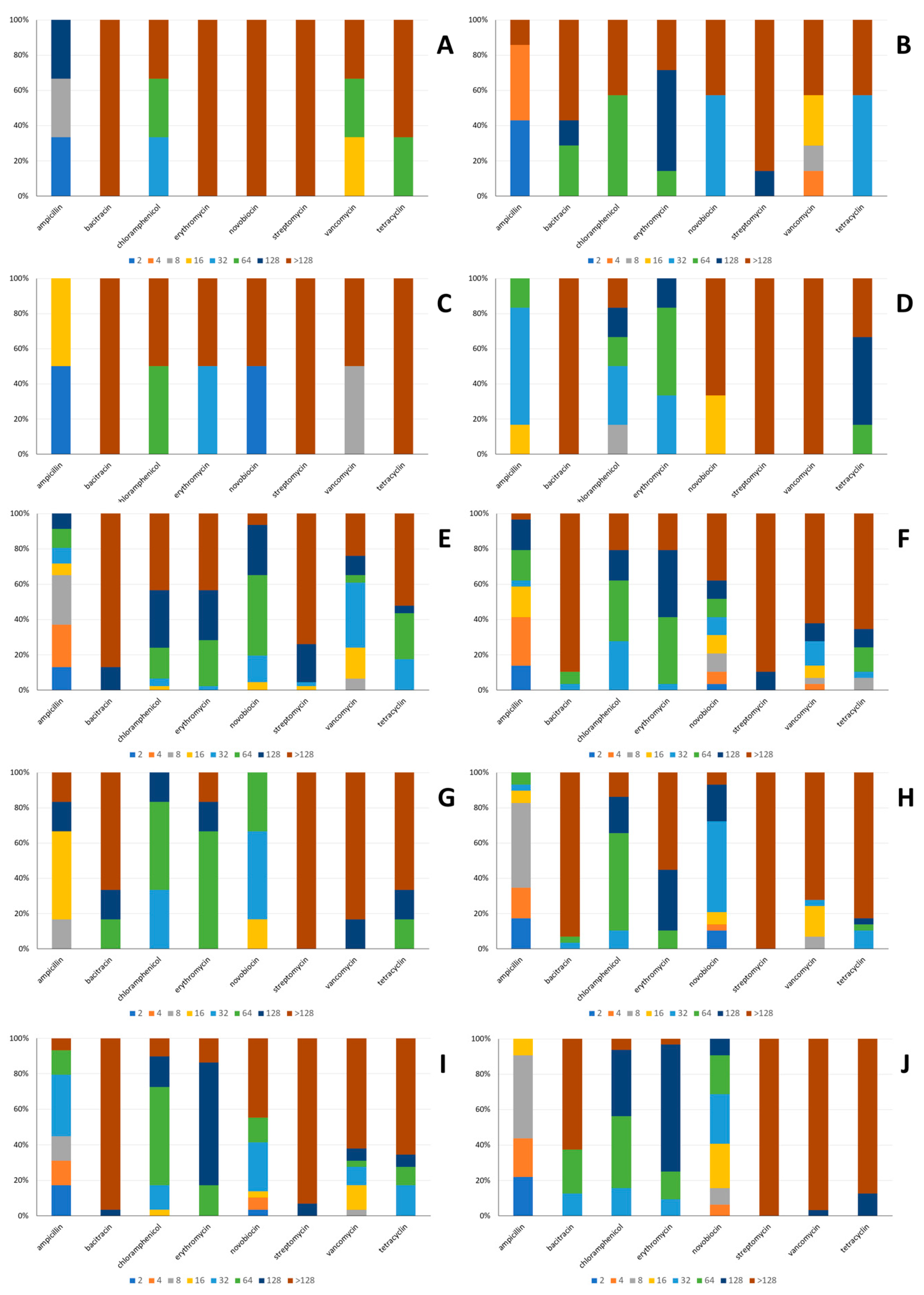
2.3.4. Antibiotic Susceptibility
2.3.5. Gelatinase Activity
2.4. Probiotic-Related Activities
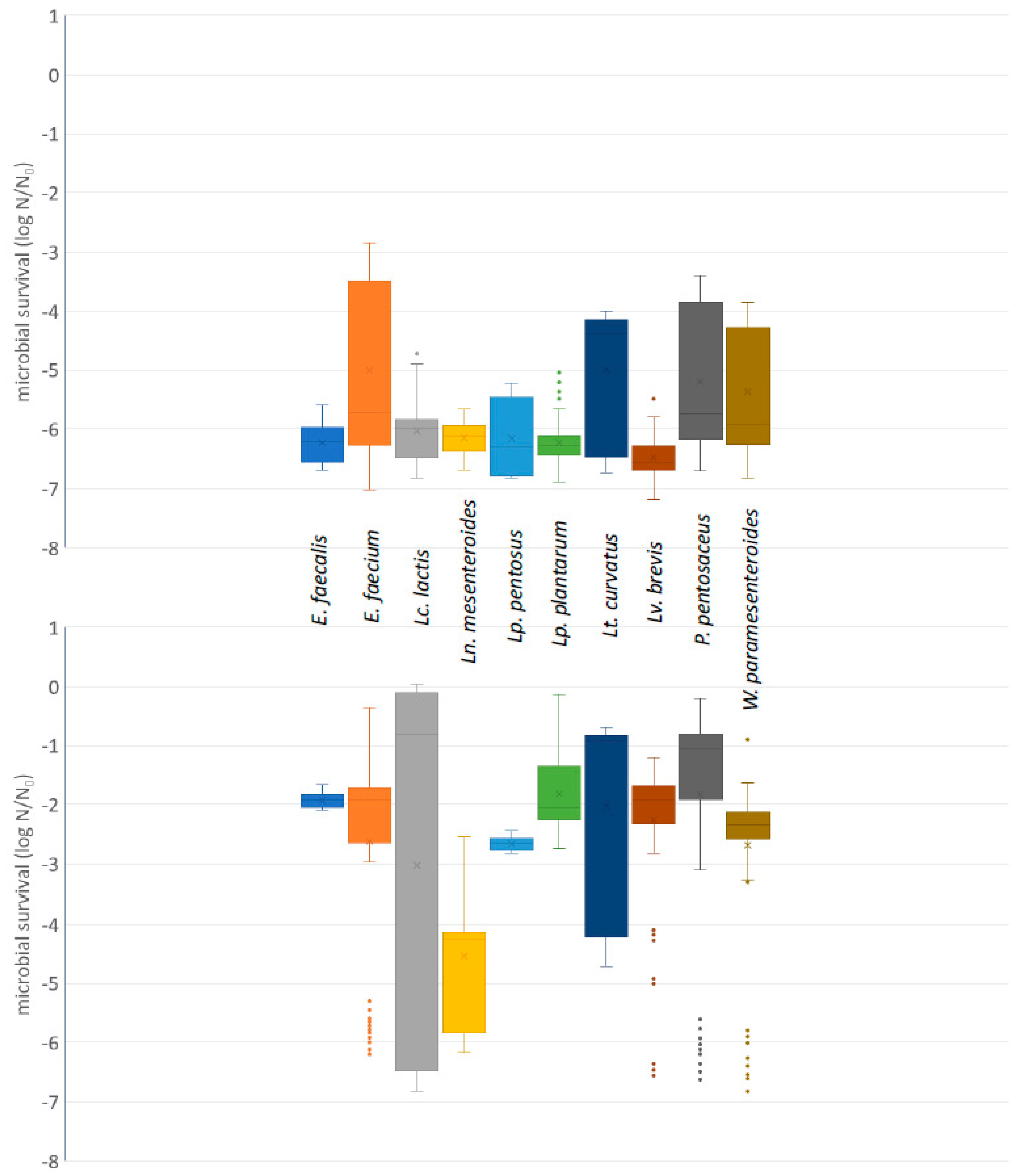
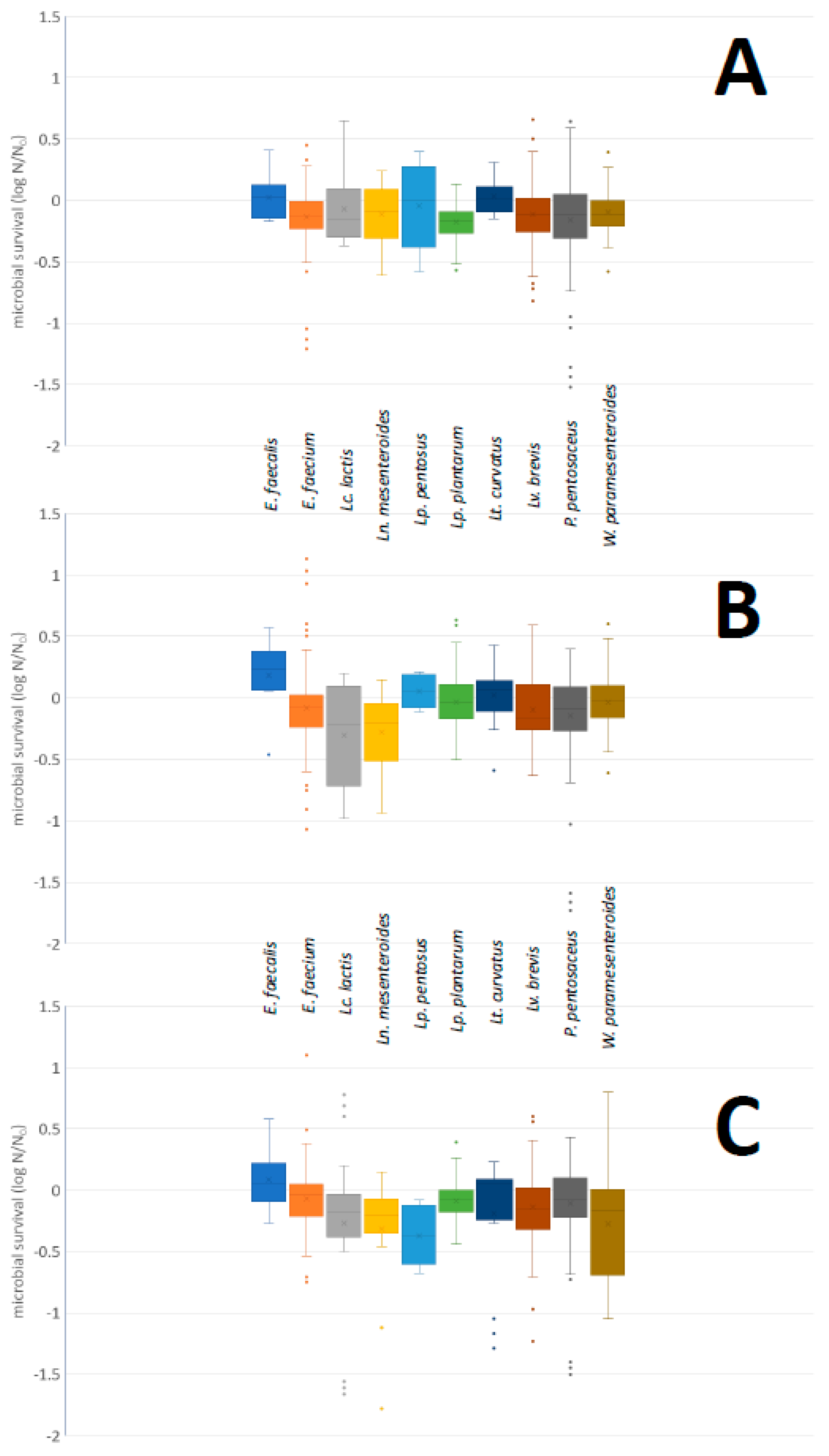
2.4.1. Acid and Bile Salt Tolerance
2.4.2. Bile Salt Hydrolase Activity
3. Results
3.1. Technological Properties
3.1.1. Milk Acidification and Coagulation
3.1.2. Caseinolytic and Lipolytic Capacity
3.1.3. Production of Exopolysaccharides (EPS)
3.2. Safety Properties
3.2.1. Production of Antimicrobial Compounds
3.2.2. Biogenic Amine Production
3.2.3. Hemolytic Activity
3.2.4. Antibiotic Susceptibility
3.2.5. Gelatinase Activity
3.3. Probiotic-Related Activities
3.3.1. Acid and Bile Salt Tolerance
3.3.2. Bile Salt Hydrolase Activity
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Fox, P.F.; Guinee, T.P.; Cogan, T.M.; McSweeney, P.L.H. Fundamentals of Cheese Science; Springer: New York, NY, USA, 2000; pp. 333–390. [Google Scholar]
- Kunji, E.R.S.; Mierau, I.; Hagting, A.; Poolman, B.; Konings, W.N. The proteolytic systems of lactic acid bacteria. Antonie Leeuwenhoek 1996, 70, 187–221. [Google Scholar] [CrossRef]
- Liu, M.; Bayjanov, J.R.; Renckens, B.; Nauta, A.; Siezen, R.J. The proteolytic system of lactic acid bacteria revisited: A genomic comparison. BMC Genom. 2010, 11, 36. [Google Scholar] [CrossRef] [Green Version]
- Liu, M.; Nauta, A.; Francke, C.; Siezen, R.J. Comparative genomics of enzymes in flavor-forming pathways from amino acids in lactic acid bacteria. Appl. Environ. Microbiol. 2008, 74, 4590–4600. [Google Scholar] [CrossRef] [Green Version]
- Park, Y.W.; Nam, M.S. Bioactive peptides in milk and dairy products: A review. Korean J. Food Sci. Anim. Resour. 2015, 35, 831–840. [Google Scholar] [CrossRef] [Green Version]
- McSweeney, P.; Sousa, M. Biochemical pathways for the production of flavour compounds in cheeses during ripening: A review. Lait 2000, 80, 293–324. [Google Scholar] [CrossRef]
- Ruas-Madiedo, P.; Reyes-Gavilan, C.G. Invited review: Methods for the screening, isolation, and characterization of exopolysaccharides produced by lactic acid bacteria. J. Dairy Sci. 2005, 88, 843–856. [Google Scholar] [CrossRef] [Green Version]
- Chabot, S.; Yu, H.L.; De Leseleuc, L.; Cloutier, D.; van Calsteren, M.R.; Lessard, M.; Roy, D.; Lacroix, M.; Oth, D. Exopolysaccharide from Lactobacillus rhamnosus RW-9595M stimulate TNF, IL6 and IL-12 in human and mouse cultured immunocompetent cells, and IFN-g in mouse splenocytes. Lait 2001, 81, 683–697. [Google Scholar] [CrossRef] [Green Version]
- Hu, S.-M.; Zhou, J.-M.; Zhou, Q.-Q.; Li, P.; Xie, Y.-Y.; Zhou, T.; Gu, Q. Purification, characterization and biological activities of exopolysaccharides from Lactobacillus rhamnosus ZFM231 isolated from milk. LWT—Food Sci. Technol. 2021, 147, 111561. [Google Scholar] [CrossRef]
- Li, J.; Li, Q.; Gao, N.; Wang, Z.; Li, F.; Lia, J.; Shan, A. Exopolysaccharides produced by Lactobacillus rhamnosus GG alleviate hydrogen peroxide-induced intestinal oxidative damage and apoptosis through the Keap1/Nrf2 and Bax/Bcl-2 pathways in vitro. Food Funct. 2021, 12, 9632–9641. [Google Scholar] [CrossRef]
- von Wintersdorff, C.J.H.; Penders, J.; van Niekerk, J.M.; Mills, N.D.; Majumder, S.; van Alphen, L.B.; Savelkoul, P.H.M.; Wolffs, P.F.G. Dissemination of antimicrobial resistance in microbial ecosystems through horizontal gene transfer. Front. Microbiol. 2016, 7, 173. [Google Scholar] [CrossRef] [Green Version]
- Stefanska, I.; Kwiecien, E.; Józwiak-Piasecka, K.; Garbowska, M.; Binek, M.; Rzewuska, M. Antimicrobial susceptibility of lactic acid bacteria strains of potential use as feed additives—The basic safety and usefulness criterion. Front. Vet. Sci. 2021, 8, 687071. [Google Scholar] [CrossRef]
- Perez-Ramos, A.; Madi-Moussa, D.; Coucheney, F.; Drider, D. Current knowledge of the mode of action and immunity mechanisms of LAB-bacteriocins. Microorganisms 2021, 9, 2107. [Google Scholar] [CrossRef]
- Soltani, S.; Hammami, R.; Cotter, P.D.; Rebuffat, S.; Said, L.B.; Gaudreau, H.; Bedard, F.; Biron, E.; Drider, D.; Fliss, I. Bacteriocins as a new generation of antimicrobials: Toxicity aspects and regulations. FEMS Microbiol. Rev. 2021, 45, fuaa039. [Google Scholar] [CrossRef]
- Lopes, M.F.S.; Simões, A.P.; Tenreiro, R.; Marques, J.J.F.; Crespo, M.T.B. Activity and expression of a virulence factor, gelatinase, in dairy enterococci. Int. J. Food Microbiol. 2006, 112, 208–214. [Google Scholar] [CrossRef]
- Semedo-Lemsaddek, T.; Cota, J.B.; Ribeiro, T.; Pimentel, A.; Tavares, L.; Bernando, F.; Oliveira, M. Resistance and virulence distribution in enterococci isolated from broilers reared in two farming systems. Ir. Vet. J. 2021, 74, 22. [Google Scholar] [CrossRef]
- Molenaar, D.; Bosscher, J.S.; Ten Brink, B.; Driessen, A.J.M.; Konings, W.N. Generation of a proton motive force by histidine decarboxylation and electrogenic histidine/histamine antiport in Lactobacillus buchneri. J. Bacteriol. 1993, 175, 2864–2870. [Google Scholar] [CrossRef] [Green Version]
- Ladero, V.; Calles-Enriquez, M.; Fernandez, M.; Alvarez, M. Toxicological effects of dietary biogenic amines. Curr. Res. Nutr. Food Sci. 2010, 6, 145–156. [Google Scholar] [CrossRef]
- Reale, A.; Di Renzo, T.; Rossi, F.; Zotta, T.; Iacumin, L.; Preziuso, M.; Parente, E.; Sorrentino, E.; Coppola, R. Tolerance of Lactobacillus casei, Lactobacillus paracasei and Lactobacillus rhamnosus strains to stress factors encountered in food processing and in the gastro-intestinal tract. LWT—Food Sci. Technol. 2015, 60, 721–728. [Google Scholar] [CrossRef]
- Tsigkrimani, M.; Bakogianni, M.; Paramithiotis, S.; Bosnea, L.; Pappa, E.; Kondyli, E.; Drosinos, E.H.; Skandamis, P.Ν.; Mataragas, M. Microbial ecology of artisanal Feta and Kefalograviera cheeses. Part I: Bacterial community and its functional characteristics with focus on lactic acid bacteria as determined by culture-dependent methods and phenotype microarrays. Microorganisms 2022, 10, 161. [Google Scholar] [CrossRef]
- Bancalari, E.; Sardaro, M.L.S.; Levante, A.; Marseglia, A.; Caligiani, A.; Lazzi, C.; Neviani, E.; Gatti, M. An integrated strategy to discover Lactobacillus casei group strains for their potential use as aromatic starters. Food Res. Int. 2017, 100, 682–690. [Google Scholar] [CrossRef]
- Carrazco-Palafox, J.; Rivera-Chavira, B.E.; Ramírez-Baca, N.; Manzanares-Papayanopoulos, L.I.; Nevárez-Moorillón, G.V. Improved method for qualitative screening of lipolytic bacterial strains. MethodsX 2018, 5, 68–74. [Google Scholar] [CrossRef]
- Smitinont, T.; Tansakul, C.; Tanasupawat, S.; Keeratipibul, S.; Navarini, L.; Bosco, M.; Cescutti, P. Exopolysaccharide-producing lactic acid bacteria strains from traditional thai fermented foods: Isolation, identification and exopolysaccharide characterization. Int. J. Food Microbiol. 1991, 51, 105–111. [Google Scholar] [CrossRef]
- Syrokou, M.K.; Tziompra, S.; Psychogiou, E.-E.; Mpisti, S.-D.; Paramithiotis, S.; Bosnea, L.; Mataragas, M.; Skandamis, P.N.; Drosinos, E.H. Technological and safety attributes of lactic acid bacteria and yeasts isolated from spontaneously fermented Greek wheat sourdoughs. Microorganisms 2021, 9, 671. [Google Scholar] [CrossRef]
- Bover-Cid, S.; Holzapfel, W.H. Improved screening procedure for biogenic amine production by lactic acid bacteria. Int. J. Food Microbiol. 1999, 53, 33–41. [Google Scholar] [CrossRef]
- Hudzicki, J. Kirby-Bauer Disk Diffusion Susceptibility Test Protocol. Available online: https://asm.org/Protocols/Kirby-Bauer-Disk-Diffusion-Susceptibility-Test-Pro (accessed on 13 September 2020).
- Cruz, T.E.E.; Torres, J.M.O. Gelatin Hydrolysis Test Protocol. 2012. Available online: https://asm.org/Protocols/Gelatin-Hydrolysis-Test-Protocol (accessed on 13 September 2020).
- Paramithiotis, S.; Melissari, Ι.; Drosinos, Ε.H. In vitro assessment of properties associated with the survival through the gastrointestinal tract of staphylococci isolated from traditional sausage fermentation. Food Microbiol. 2006, 23, 663–671. [Google Scholar] [CrossRef]
- Roberfroid, M.B. What is beneficial for health? The concept of functional food. Food Chem. Toxicol. 1999, 37, 1039–1041. [Google Scholar] [CrossRef] [Green Version]
- Araújo-Rodrigues, H.; dos Santos, M.T.P.G.; Ruiz-Moyano, S.; Tavaria, F.K.; Martins, A.P.L.; Alvarenga, N.; Pintado, M.E. Technological and protective performance of LAB isolated from Serpa PDO cheese: Towards selection and development of an autochthonous starter culture. LWT—Food Sci. Technol. 2021, 150, 112079. [Google Scholar] [CrossRef]
- Londoño-Zapata, A.F.; Durango-Zuleta, M.M.; Sepúlveda-Valencia, J.U.; Moreno Herrera, C.X. Characterization of lactic acid bacterial communities associated with a traditional Colombian cheese: Double cream cheese. LWT—Food Sci. Technol. 2017, 82, 39–48. [Google Scholar] [CrossRef]
- Albayrak, Ç.B.; Duran, M. Isolation and characterization of aroma producing lactic acid bacteria from artisanal white cheese for multifunctional properties. LWT—Food Sci. Technol. 2021, 150, 112053. [Google Scholar] [CrossRef]
- Ribeiro, S.C. Technological properties of bacteriocin-producing lactic acid bacteria isolated from Pico cheese an artisanal cow’s milk cheese. J. Appl. Microbiol. 2014, 116, 573–585. [Google Scholar] [CrossRef]
- Kask, S.; Adamberg, K.; Orłowski, A.; Vogensen, F.K.; Møller, P.L.; Ardö, Y.; Paalme, T. Physiological properties of Lactobacillus paracasei, L. danicus and L. curvatus strains isolated from Estonian semi-hard cheese. Food Res. Int. 2003, 36, 1037–1046. [Google Scholar] [CrossRef]
- Ayad, E.H.E.; Nashat, S.; El-Sadek, N.; Metwaly, H.; El-Soda, M. Selection of wild lactic acid bacteria isolated from traditional Egyptian dairy products according to production and technological criteria. Food Microbiol. 2004, 21, 715–725. [Google Scholar] [CrossRef]
- Beresford, T.P.; Fitzsimons, N.A.; Brennan, N.L.; Cogan, T.M. Recent advances in cheese microbiology. Int. Dairy J. 2001, 11, 259–274. [Google Scholar] [CrossRef]
- Fusieger, A.; Martins, M.C.F.; Freitas, R.; Nero, L.A.; Carvalho, A.F. Technological properties of Lactococcus lactis subsp. lactis bv. diacetylactis obtained from dairy and non-dairy niches. Braz. J. Microbiol. 2020, 51, 313–321. [Google Scholar] [PubMed]
- Carafa, I.; Nardin, T.; Larcher, R.; Viola, R.; Tuohy, K.; Franciosi, E. Identification and characterization of wild lactobacilli and pediococci from spontaneously fermented Mountain Cheese. Food Microbiol. 2015, 48, 123–132. [Google Scholar] [CrossRef]
- Teixeira, C.G.; Fusieger, A.; Martins, E.; Freitas, R.; Vakarelova, M.; Nero, L.A.; Carvalho, A.F. Biodiversity and technological features of Weissella isolates obtained from Brazilian artisanal cheese-producing regions. LWT—Food Sci. Technol. 2021, 147, 111474. [Google Scholar] [CrossRef]
- Picon, A.; Garde, S.; Ávila, M.; Nuñez, M. Microbiota dynamics and lactic acid bacteria biodiversity in raw goat milk cheeses. Int. Dairy J. 2016, 58, 14–22. [Google Scholar] [CrossRef]
- Ma, C.L.; Zhang, L.W.; Yi, H.X.; Du, M.; Han, X.; Zhang, L.L.; Feng, Z.; Zhang, Y.C.; Li, Q. Technological characterization of lactococci isolated from traditional Chinese fermented milks. J. Dairy Sci. 2011, 94, 1691–1696. [Google Scholar] [CrossRef] [Green Version]
- Sarantinopoulos, P.; Andrighetto, C.; Georgalaki, M.D.; Rea, M.C.; Lombardi, A.; Cogan, T.M.; Kalantzopoulos, G.; Tsakalidou, E. Biochemical properties of enterococci relevant to their technological performance. Int. Dairy J. 2001, 11, 621–647. [Google Scholar] [CrossRef]
- Nieto-Arribas, P.; Poveda, J.M.; Seseña, S.; Palop, L.; Cabezas, L. Technological characterization of Lactobacillus isolates from traditional Manchego cheese for potential use as adjunct starter cultures. Food Control 2009, 20, 1092–1098. [Google Scholar] [CrossRef]
- González, L.; Sacristán, N.; Arenas, R.; Fresno, J.M.; Tornadijo, M.E. Enzymatic activity of lactic acid bacteria (with antimicrobial properties) isolated from a traditional Spanish cheese. Food Microbiol. 2010, 27, 592–597. [Google Scholar] [CrossRef] [PubMed]
- Domingos-Lopes, M.F.P.; Stanton, C.; Ross, P.R.; Dapkevicius, M.L.E.; Silva, C.C.G. Genetic diversity, safety and technological characterization of lactic acid bacteria isolated from artisanal Pico cheese. Food Microbiol. 2017, 63, 178–190. [Google Scholar] [CrossRef] [PubMed]
- Camara, S.P.; Dapkevicius, A.; Riquelme, C.; Elias, R.B.; Silva, C.C.G.; Malcata, F.X.; Dapkevicius, M.L.N.E. Potential of lactic acid bacteria from Pico cheese for starter culture development. Food Sci. Technol. Int. 2019, 25, 303–317. [Google Scholar] [CrossRef] [PubMed]
- Mohammed, S.; Çon, A.H. Isolation and characterization of potential probiotic lactic acid bacteria from traditional cheese. LWT—Food Sci. Technol. 2021, 152, 112319. [Google Scholar] [CrossRef]
- Islam, M.Z.; Uddin, M.E.; Rahman, M.T.; Islam, M.A.; Harun-ur-Rashid, M. Isolation and characterization of dominant lactic acid bacteria from raw goat milk: Assessment of probiotic potential and technological properties. Small Rumin. Res. 2021, 205, 106532. [Google Scholar] [CrossRef]
- Yerlikaya, O. Probiotic potential and biochemical and technological properties of Lactococcus lactis ssp. lactis strains isolated from raw milk and kefir grains. J. Dairy Sci. 2019, 102, 124–134. [Google Scholar]
- Nezhad, S.J.E.; Dovom, M.R.E.; Najafi, M.B.H.; Yavarmanesh, M.; Mayo, B. Technological characteristics of Lactobacillus spp. isolated from Iranian raw milk Motal cheese. LWT—Food Sci. Technol. 2020, 133, 110070. [Google Scholar] [CrossRef]
- Asteri, I.A.; Robertson, N.; Kagkli, D.M.; Andrewes, P.; Nychas, G.; Coolbear, T.; Holland, R.; Crow, V.; Tsakalidou, E. Technological and flavour potential of cultures isolated from traditional Greek cheeses—A pool of novel species and starters. Int. Dairy J. 2009, 19, 595–604. [Google Scholar] [CrossRef]
- Margalho, L.P.; Feliciano, M.E.; Silva, C.E.; Abreu, J.S.; Piran, M.V.F.; Sant’Ana, A.S. Brazilian artisanal cheeses are rich and diverse sources of nonstarter lactic acid bacteria regarding technological, biopreservative, and safety properties—Insights through multivariate analysis. J. Dairy Sci. 2020, 103, 7908–7926. [Google Scholar] [CrossRef]
- Franciosi, E.; Settanni, L.; Cavazza, A.; Poznanski, E. Biodiversity and technological potential of wild lactic acid bacteria from raw cows’ milk. Int. Dairy J. 2009, 19, 3–11. [Google Scholar] [CrossRef]
- Chajęcka-Wierzchowska, W.; Zadernowska, A.; Łaniewska-Trokenheim, Ł. Virulence factors of Enterococcus spp. presented in food. LWT—Food Sci. Technol. 2017, 75, 670–676. [Google Scholar] [CrossRef]
- Landeta, G.; Curiel, J.A.; Carrascosa, A.V.; Muñoz, R.; De las Rivas, B. Technological and safety properties of lactic acid bacteria isolated from Spanish dry-cured sausages. Meat Sci. 2013, 95, 272–280. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zoumpopoulou, G.; Papadimitriou, K.; Alexandraki, V.; Mavrogonatou, E.; Alexopoulou, K.; Anastasiou, R.; Georgalaki, M.; Kletsas, D.; Tsakalidou, E.; Giaouris, E. The microbiota of Kalathaki and Melichloro Greek artisanal cheeses comprises functional lactic acid bacteria. LWT—Food Sci. Technol. 2020, 130, 109570. [Google Scholar] [CrossRef]
- Popovic, N.; Dinic, M.; Tolinacki, M.; Mihajlovic, S.; Terzic-Vidojevic, A.; Bojic, S.; Djokic, J.; Golic, N.; Veljovic, K. New insight into biofilm formation ability, the presence of virulence genes and probiotic potential of Enterococcus sp. dairy isolates. Front. Microbiol. 2018, 9, 1–10. [Google Scholar] [CrossRef] [Green Version]
- EFSA Panel on Additives and Products or Substances used in Animal Feed (FEEDAP). Guidance on the assessment of bacterial susceptibility to antimicrobials of human and veterinary importance. EFSA J. 2012, 10, 2740. [Google Scholar]
- Clinical and Laboratory Standards Institute (CLSI). M45 Methods for Antimicrobial Dilution and Disk Susceptibility Testing of Infrequently Isolated or Fastidious Bacteria, 3rd ed.; CLSI Document M45-A; Clinical and Laboratory Standards Institute: Wayne, PA, USA, 2006; ISBN 1-56238-607-7. [Google Scholar]
- Clinical and Laboratory Standards Institute (CLSI). Performance Standards for Antimicrobial Susceptibility Testing, 27th ed.; CLSI Supplement M100; Clinical and Laboratory Standards Institute: Wayne, PA, USA, 2017. [Google Scholar]
- Sohanang, F.S.N.; Coton, M.; Debaets, S.; Coton, E.; Tatsadjieu, L.N.; Mohammadou, B.A. Bacterial diversity of traditional fermented milks from Cameroon and safety and antifungal activity assessment for selected lactic acid bacteria. LWT—Food Sci. Technol. 2021, 138, 110635. [Google Scholar] [CrossRef]
- Caro, I.; Quinto, E.J.; Fuentes, L.; Alessandria, V.; Cocolin, L.S.; Redondo-del-Río, M.P.; Mayo, B.; Flórez, A.B.; Mateo, J. Characterization of Lactococcus strains isolated from artisanal Oaxaca cheese. LWT—Food Sci. Technol. 2020, 122, 109041. [Google Scholar] [CrossRef]
- Ruiz-Moyano, S.; Gonçalves dos Santos, M.T.P.; Galván, A.I.; Merchán, A.V.; González, E.; Córdoba, M.G.; Benito, M.J. Screening of autochthonous lactic acid bacteria strains from artisanal soft cheese: Probiotic characteristics and prebiotic metabolism. LWT—Food Sci. Technol. 2019, 114, 108388. [Google Scholar] [CrossRef]
- Mokoena, M.P. Lactic acid bacteria and their bacteriocins: Classification, biosynthesis and applications against uropathogens: A mini-review. Molecules 2017, 22, 1255. [Google Scholar] [CrossRef]
- İspirli, H.; Demirbaş, F.; Dertli, E. Characterization of functional properties of Enterococcus spp. isolated from Turkish white cheese. LWT—Food Sci. Technol. 2017, 75, 358–365. [Google Scholar] [CrossRef]
- Zoumpopoulou, G.; Tzouvanou, A.; Mavrogonatou, E.; Alexandraki, V.; Georgalaki, M.; Anastasiou, R.; Papadelli, M.; Manolopoulou, E.; Kazou, M.; Kletsas, D.; et al. Probiotic features of lactic acid bacteria isolated from a diverse pool of traditional Greek dairy products regarding specific strain–host interactions. Probiotics Antimicrob. Proteins 2018, 10, 313–322. [Google Scholar] [CrossRef] [PubMed]
| Strain (DRD) | Incubation Time (h) | Coagulation 2 | |||||||
|---|---|---|---|---|---|---|---|---|---|
| 3 | 6 | 9 | 12 | 15 | 18 | 21 | 24 | ||
| E. faecalis | |||||||||
| 2602 | 0.08 (0.017) | 0.21 (0.011) | 0.38 (0.020) | 0.50 (0.010) | 0.57 (0.036) | 0.66 (0.034) | 0.74 (0.066) | 0.76 (0.047) | 24 h |
| 2651 | 0.06 (0.005) | 0.27 (0.026) | 0.38 (0.025) | 0.54 (0.061) | 0.99 (0.065) | 1.10 (0.050) | 0.94 (0.615) | 1.37 (0.028) | 24 h |
| 2722 | 0.08 (0.043) | 0.31 (0.057) | 0.50 (0.029) | 0.64 (0.015) | 0.80 (0.025) | 0.88 (0.032) | 0.95 (0.053) | 0.98 (0.040) | 24 h |
| Lc. lactis | |||||||||
| 2649 | 0.05 (0.076) | 0.28 (0.096) | 0.20 (0.049) | 1.74 (0.021) | 1.92 (0.032) | 2.17 (0.006) | 2.16 (0.036) | 2.13 (0.025) | 12 h |
| 2658 | 0.07 (0.070) | 0.61 (0.095) | 1.81 (0.025) | 2.05 (0.030) | 2.22 (0.015) | 2.23 (0.006) | 2.16 (0.140) | 2.27 (0.035) | 6 h |
| 2692 | 0.08 (0.031) | 0.47 (0.146) | 1.35 (0.311) | 1.91 (0.066) | 2.11 (0.036) | 2.13 (0.043) | 2.17 (0.026) | 2.18 (0.030) | 6 h |
| 2699 | 0.08 (0.040) | 0.12 (0.050) | 0.16 (0.032) | 0.17 (0.032) | 0.11 (0.049) | 0.19 (0.01) | 0.19 (0.038) | 0.21 (0.041) | NC |
| Lp. pentosus | |||||||||
| 2662 | 0.10 (0.043) | 0.53 (0.217) | 1.38 (0.361) | 1.94 (0.085) | 2.16 (0.057) | 2.16 (0.063) | 2.20 (0.028) | 2.18 (0.066) | 6 h |
| 2693 | 0.09 (0.034) | 0.10 (0.045) | 0.14 (0.025) | 0.16 (0.030) | 0.15 (0.066) | 0.19 (0.02) | 0.19 (0.023) | 0.18 (0.020) | NC |
| Lt. curvatus | |||||||||
| 2708 | 0.03 (0.005) | 0.10 (0.015) | 0.24 (0.040) | 0.31 (0.020) | 0.45 (0.010) | 0.50 (0.011) | 0.54 (0.010) | 0.59 (0.023) | 24 h |
| 2709 | 0.05 (0.020) | 0.19 (0.026) | 0.33 (0.051) | 0.48 (0.034) | 0.73 (0.055) | 0.78 (0.049) | 0.85 (0.041) | 0.89 (0.050) | 3 d |
| 2716 | 0.03 (0.015) | 0.13 (0.020) | 0.25 (0.020) | 0.34 (0.035) | 0.64 (0.058) | 0.74 (0.035) | 0.89 (0.015) | 0.96 (0.030) | 2 d |
| 2728 | 0.04 (0.025) | 0.10 (0.025) | 0.16 (0.035) | 0.20 (0.041) | 0.24 (0.017) | 0.23 (0.020) | 0.24 (0.015) | 0.23 (0.047) | NC |
| E. faecium | |||||||||
| 2579 | 0.08 (0.02) | 0.24 (0.03) | 0.36 (0.037) | 0.40 (0.045) | 0.50 (0.065) | 0.52 (0.070) | 0.55 (0.075) | 0.58 (0.089) | 4 d |
| 2581 | 0.07 (0.020) | 0.19 (0.030) | 0.29 (0.036) | 0.32 (0.035) | 0.39 (0.047) | 0.40 (0.020) | 0.40 (0.015) | 0.43 (0.015) | NC |
| 2585 | 0.06 (0.023) | 0.17 (0.028) | 0.29 (0.030) | 0.33 (0.045) | 0.35 (0.015) | 0.47 (0.015) | 0.46 (0.020) | 0.43 (0.055) | NC |
| 2588 | 0.07 (0.028) | 0.15 (0.030) | 0.30 (0.026) | 0.34 (0.035) | 0.39 (0.052) | 0.51 (0.017) | 0.48 (0.055) | 0.46 (0.061) | 4 d |
| 2667 | 0.05 (0.045) | 0.22 (0.025) | 0.58 (0.085) | 0.91 (0.005) | 1.21 (0.023) | 1.37 (0.011) | 1.46 (0.01) | 1.52 (0.026) | 12 h |
| 2670 | 0.07 (0.036) | 0.22 (0.036) | 0.60 (0.020) | 0.85 (0.045) | 1.10 (0.025) | 1.29 (0.023) | 1.37 (0.02) | 1.39 (0.026) | 24 h |
| 2587 | 0.05 (0.026) | 0.07 (0.037) | 0.07 (0.011) | 0.07 (0.005) | 0.08 (0.030) | 0.13 (0.02) | 0.12 (0.02) | 0.11 (0.075) | NC |
| 2600 | 0.08 (0.017) | 0.24 (0.037) | 0.46 (0.025) | 0.59 (0.020) | 0.64 (0.015) | 0.70 (0.015) | 0.72 (0.037) | 0.72 (0.015) | 3 d |
| 2695 | 0.04 (0.083) | 0.08 (0.115) | 0.12 (0.110) | 0.21 (0.173) | 0.21 (0.023) | 0.26 (0.025) | 0.42 (0.191) | 0.42 (0.026) | 3 d |
| 2711 | 0.04 (0.032) | 0.12 (0.045) | 0.25 (0.055) | 0.33 (0.100) | 0.53 (0.040) | 0.72 (0.03) | 0.81 (0.030) | 0.83 (0.126) | 3 d |
| 2720 | 0.07 (0.030) | 0.19 (0.025) | 0.36 (0.026) | 0.38 (0.030) | 0.49 (0.015) | 0.56 (0.017) | 0.56 (0.030) | 0.55 (0.126) | 5 d |
| 2723 | 0.05 (0.005) | 0.21 (0.025) | 0.30 (0.026) | 0.32 (0.030) | 0.44 (0.015) | 0.43 (0.017) | 0.45 (0.030) | 0.51 (0.126) | NC |
| 2691 | 0.07 (0.030) | 0.16 (0.076) | 0.49 (0.080) | 0.75 (0.076) | 1.00 (0.017) | 1.23 (0.030) | 1.35 (0.005) | 1.39 (0.02) | 24 h |
| 2686 | 0.05 (0.001) | 0.19 (0.003) | 0.27 (0.020) | 0.31 (0.015) | 0.39 (0.030) | 0.36 (0.030) | 0.40 (0.030) | 0.41 (0.052) | NC |
| 2724 | 0.07 (0.036) | 0.19 (0.010) | 0.35 (0.020) | 0.44 (0.040) | 0.54 (0.037) | 0.63 (0.005) | 0.68 (0.035) | 0.78 (0.181) | 6 d |
| P. pentosaceus | |||||||||
| 2616 | 0.1 (0.032) | 0.15 (0.005) | 0.23 (0.020) | 0.29 (0.026) | 0.30 (0.02) | 0.32 (0.051) | 0.38 (0.040) | 0.41 (0.040) | NC |
| 2635 | 0.11 (0.025) | 0.20 (0.040) | 0.32 (0.026) | 0.39 (0.005) | 0.48 (0.041) | 0.52 (0.023) | 0.53 (0.025) | 0.54 (0.021) | NC |
| 2681 | 0.07 (0.066) | 0.11 (0.055) | 0.21 (0.010) | 0.42 (0.051) | 0.50 (0.032) | 0.77 (0.121) | 1.00 (0.115) | 1.13 (0.085) | 3 d |
| 2689 | 0.06 (0.02) | 0.14 (0.049) | 0.32 (0.047) | 0.71 (0.060) | 1.08 (0.041) | 1.30 (0.005) | 1.44 (0.041) | 1.47 (0.026) | 24 h |
| 2694 | 0.01 (0.017) | 0.03 (0.036) | 0.06 (0.07) | 0.13 (0.090) | 0.73 (0.936) | 0.83 (0.990) | 0.87 (1.013) | 0.88 (0.962) | 3 d |
| 2753 | 0.09 (0.062) | 0.16 (0.130) | 0.21 (0.225) | 0.23 (0.239) | 0.15 (0.015) | 0.18 (0.023) | 0.24 (0.032) | 0.31 (0.106) | NC |
| 2634 | 0.08 (0.034) | 0.12 (0.045) | 0.16 (0.025) | 0.17 (0.011) | 0.15 (0.068) | 0.22 (0.02) | 0.22 (0.025) | 0.23 (0.001) | NC |
| 2652 | 0.10 (0.040) | 0.30 (0.045) | 0.50 (0.055) | 0.73 (0.051) | 1.10 (0.015) | 1.30 (0.001) | 1.44 (0.011) | 1.49 (0.023) | 12 h |
| 2655 | 0.11 (0.025) | 0.58 (0.050) | 1.12 (0.015) | 1.53 (0.025) | 1.78 (0.050) | 1.86 (0.025) | 1.92 (0.005) | 1.96 (0.069) | 6 h |
| 2657 | 0.11 (0.026) | 0.66 (0.070) | 1.85 (0.020) | 2.00 (0.037) | 2.21 (0.011) | 2.22 (0.020) | 2.23 (0.015) | 2.22 (0.064) | 6 h |
| 2761 | 0.08 (0.011) | 0.18 (0.036) | 0.26 (0.043) | 0.35 (0.030) | 0.42 (0.005) | 0.48 (0.001) | 0.58 (0.020) | 0.65 (0.005) | 2 d |
| 2679 | 0.01 (0.023) | 0.06 (0.032) | 0.33 (0.020) | 0.44 (0.023) | 0.01 (0.034) | 0.01 (0.017) | 0.13 (0.199) | 0.18 (0.238) | NC |
| 2676 | 0.07 (0.061) | 0.12 (0.086) | 0.22 (0.097) | 0.33 (0.070) | 0.45 (0.011) | 0.63 (0.075) | 0.66 (0.126) | 0.65 (0.188) | 4 d |
| 2748 | 0.10 (0.015) | 0.24 (0.028) | 0.34 (0.036) | 0.42 (0.020) | 0.54 (0.020) | 0.60 (0.028) | 0.67 (0.02) | 0.72 (0.043) | 24 h |
| 2749 | 0.08 (0.005) | 0.21 (0.02) | 0.29 (0.032) | 0.4 (0.043) | 0.52 (0.028) | 0.58 (0.036) | 0.72 (0.005) | 0.89 (0.026) | 2 d |
| 2752 | 0.09 (0.011) | 0.18 (0.020) | 0.25 (0.030) | 0.30 (0.010) | 0.31 (0.011) | 0.31 (0.015) | 0.36 (0.026) | 0.42 (0.030) | 6 d |
| Ln. mesenteroides | |||||||||
| 2576 | 0.09 (0.020) | 0.22 (0.011) | 0.32 (0.015) | 0.36 (0.020) | 0.40 (0.011) | 0.42 (0.005) | 0.39 (0.030) | 0.38 (0.01) | NC |
| 2604 | 0.12 (0.02) | 0.43 (0.026) | 0.93 (0.026) | 1.29 (0.055) | 1.50 (0.075) | 1.65 (0.060) | 1.72 (0.034) | 1.74 (0.060) | 12 h |
| 2703 | 0.06 (0.080) | 0.18 (0.189) | 0.56 (0.258) | 0.88 (0.150) | 1.08 (0.085) | 1.33 (0.086) | 1.42 (0.075) | 1.44 (0.051) | NC |
| 2705 | 0.07 (0.055) | 0.12 (0.160) | 0.32 (0.149) | 0.39 (0.112) | 0.47 (0.041) | 0.60 (0.030) | 0.62 (0.040) | 0.66 (0.092) | 4 d |
| 2706 | 0.01 (0.017) | 0.02 (0.051) | 0.03 (0.058) | 0.10 (0.159) | 0.32 (0.375) | 0.51 (0.540) | 0.74 (0.564) | 0.64 (0.242) | 24 h |
| Lp. plantarum | |||||||||
| 2584 | 0.07 (0.032) | 0.19 (0.041) | 0.34 (0.015) | 0.42 (0.032) | 0.55 (0.02) | 0.64 (0.015) | 0.66 (0.005) | 0.66 (0.02) | 3 d |
| 2608 | 0.14 (0.005) | 0.30 (0.032) | 0.51 (0.051) | 0.64 (0.025) | 0.85 (0.130) | 0.91 (0.145) | 0.99 (0.137) | 1.05 (0.151) | 2 d |
| 2653 | 0.10 (0.028) | 0.30 (0.026) | 0.50 (0.026) | 0.78 (0.017) | 1.05 (0.101) | 1.25 (0.092) | 1.39 (0.083) | 1.48 (0.085) | 12 h |
| 2668 | 0.07 (0.043) | 0.06 (0.050) | 0.07 (0.032) | 0.37 (0.589) | 0.60 (0.335) | 1.32 (0.526) | 2.00 (0.104) | 2.05 (0.068) | 24 h |
| 2612 | 0.10 (0.020) | 0.23 (0.055) | 0.37 (0.098) | 0.50 (0.134) | 0.61 (0.185) | 0.67 (0.189) | 0.73 (0.172) | 0.77 (0.163) | 2 d |
| 2645 | 0.05 (0.005) | 0.18 (0.037) | 0.29 (0.028) | 0.31 (0.015) | 0.44 (0.025) | 0.44 (0.025) | 0.46 (0.026) | 0.47 (0.025) | 4 d |
| 2648 | 0.06 (0.032) | 0.14 (0.045) | 0.24 (0.05) | 0.43 (0.060) | 0.90 (0.066) | 1.19 (0.040) | 1.31 (0.041) | 1.36 (0.051) | 24 h |
| 2656 | 0.08 (0.015) | 0.23 (0.005) | 0.41 (0.020) | 0.52 (0.126) | 0.42 (0.040) | 0.47 (0.030) | 0.47 (0.040) | 0.51 (0.037) | 3 d |
| Lv. brevis | |||||||||
| 2596 | 0.05 (0.015) | 0.11 (0.017) | 0.24 (0.01) | 0.33 (0.047) | 0.67 (0.075) | 0.77 (0.051) | 0.88 (0.026) | 0.95 (0.015) | 2 d |
| 2624 | 0.07 (0.005) | 0.12 (0.005) | 0.16 (0.005) | 0.19 (0.015) | 0.15 (0.047) | 0.21 (0.069) | 0.21 (0.005) | 0.56 (0.597) | NC |
| 2664 | 0.05 (0.047) | 0.12 (0.205) | 0.30 (0.090) | 1.63 (0.272) | 1.98 (0.165) | 2.14 (0.120) | 2.20 (0.075) | 2.16 (0.066) | 6 h |
| 2665 | 0.06 (0.055) | 0.22 (0.015) | 0.39 (0.052) | 0.44 (0.015) | 0.45 (0.015) | 0.46 (0.037) | 0.48 (0.017) | 0.56 (0.085) | 3 d |
| 2757 | 0.09 (0.040) | 0.20 (0.020) | 0.23 (0.005) | 0.28 (0.005) | 0.28 (0.03) | 0.29 (0.017) | 0.31 (0.032) | 0.35 (0.017) | NC |
| 2760 | 0.06 (0.005) | 0.12 (0.005) | 0.15 (0.005) | 0.18 (0.015) | 0.15 (0.005) | 0.17 (0.005) | 0.23 (0.011) | 0.25 (0.015) | NC |
| 2763 | 0.06 (0.01) | 0.12 (0.01) | 0.14 (0.01) | 0.19 (0.015) | 0.13 (0.015) | 0.15 (0.015) | 0.21 (0.023) | 0.24 (0.040) | NC |
| 2688 | 0.1 (0.02) | 0.13 (0.051) | 0.16 (0.036) | 0.18 (0.040) | 0.09 (0.058) | 0.25 (0.046) | 0.26 (0.017) | 0.26 (0.034) | NC |
| W. paramesenteroides | |||||||||
| 2613 | 0.11 (0.028) | 0.20 (0.010) | 0.36 (0.010) | 0.43 (0.015) | 0.51 (0.020) | 0.53 (0.005) | 0.60 (0.075) | 0.61 (0.062) | NC |
| 2701 | 0.10 (0.040) | 0.13 (0.055) | 0.17 (0.041) | 0.19 (0.041) | 0.14 (0.050) | 0.18 (0.052) | 0.21 (0.020) | 0.21 (0.026) | NC |
| 2714 | 0.10 (0.030) | 0.19 (0.01) | 0.19 (0.107) | 0.36 (0.028) | 0.50 (0.017) | 0.56 (0.020) | 0.54 (0.028) | 0.55 (0.010) | NC |
| 2719 | 0.03 (0.010) | 0.10 (0.011) | 0.25 (0.047) | 0.57 (0.118) | 1.09 (0.049) | 1.19 (0.035) | 1.33 (0.026) | 1.40 (0.035) | 24 h |
| 2751 | 0.07 (0.005) | 0.19 (0.010) | 0.29 (0.010) | 0.37 (0.017) | 0.55 (0.020) | 0.57 (0.025) | 0.61 (0.038) | 0.62 (0.040) | 4 d |
| 2726 | 0.09 (0.032) | 0.20 (0.040) | 0.32 (0.015) | 0.42 (0.011) | 0.52 (0.020) | 0.54 (0.025) | 0.57 (0.025) | 0.64 (0.023) | 4 d |
| 2743 | 0.08 (0.041) | 0.24 (0.037) | 0.66 (0.036) | 0.90 (0.049) | 1.06 (0.050) | 1.19 (0.005) | 1.26 (0.020) | 1.33 (0.005) | 24 h |
| 2755 | 0.10 (0.023) | 0.20 (0.020) | 0.30 (0.011) | 0.39 (0.034) | 0.53 (0.026) | 0.53 (0.020) | 0.55 (0.015) | 0.56 (0.025) | NC |
| 2756 | 0.07 (0.026) | 0.25 (0.025) | 0.39 (0.005) | 0.48 (0.015) | 0.61 (0.011) | 0.62 (0.015) | 0.65 (0.036) | 0.66 (0.015) | 3 d |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Tsigkrimani, M.; Panagiotarea, K.; Paramithiotis, S.; Bosnea, L.; Pappa, E.; Drosinos, E.H.; Skandamis, P.N.; Mataragas, M. Microbial Ecology of Sheep Milk, Artisanal Feta, and Kefalograviera Cheeses. Part II: Technological, Safety, and Probiotic Attributes of Lactic Acid Bacteria Isolates. Foods 2022, 11, 459. https://doi.org/10.3390/foods11030459
Tsigkrimani M, Panagiotarea K, Paramithiotis S, Bosnea L, Pappa E, Drosinos EH, Skandamis PN, Mataragas M. Microbial Ecology of Sheep Milk, Artisanal Feta, and Kefalograviera Cheeses. Part II: Technological, Safety, and Probiotic Attributes of Lactic Acid Bacteria Isolates. Foods. 2022; 11(3):459. https://doi.org/10.3390/foods11030459
Chicago/Turabian StyleTsigkrimani, Markella, Konstantina Panagiotarea, Spiros Paramithiotis, Loulouda Bosnea, Eleni Pappa, Eleftherios H. Drosinos, Panagiotis N. Skandamis, and Marios Mataragas. 2022. "Microbial Ecology of Sheep Milk, Artisanal Feta, and Kefalograviera Cheeses. Part II: Technological, Safety, and Probiotic Attributes of Lactic Acid Bacteria Isolates" Foods 11, no. 3: 459. https://doi.org/10.3390/foods11030459
APA StyleTsigkrimani, M., Panagiotarea, K., Paramithiotis, S., Bosnea, L., Pappa, E., Drosinos, E. H., Skandamis, P. N., & Mataragas, M. (2022). Microbial Ecology of Sheep Milk, Artisanal Feta, and Kefalograviera Cheeses. Part II: Technological, Safety, and Probiotic Attributes of Lactic Acid Bacteria Isolates. Foods, 11(3), 459. https://doi.org/10.3390/foods11030459










