iTRAQ-Based Quantitative Proteomic Analysis of Antibacterial Mechanism of Milk-Derived Peptide BCp12 against Escherichia coli
Abstract
:1. Introduction
2. Materials and Methods
2.1. Materials and Chemicals
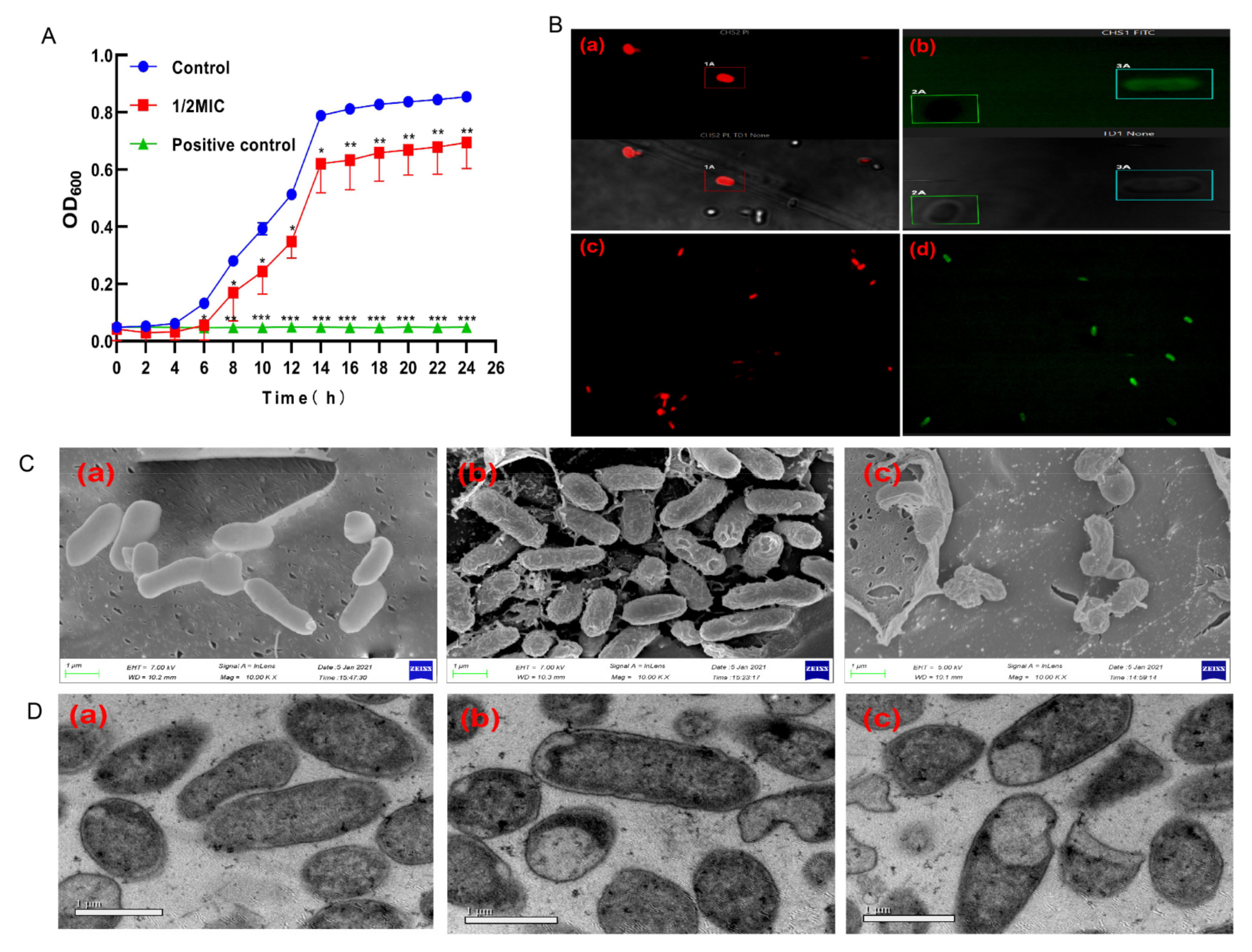
2.2. Growth Curves of E. coli
2.3. Confocal Laser Scanning Microscopy (CLSM) Assay
2.4. Determination of the Morphologies of E. coli
2.5. Transmission Electron Microscopy (TEM) Assay
2.6. Proteome Analysis of E. coli
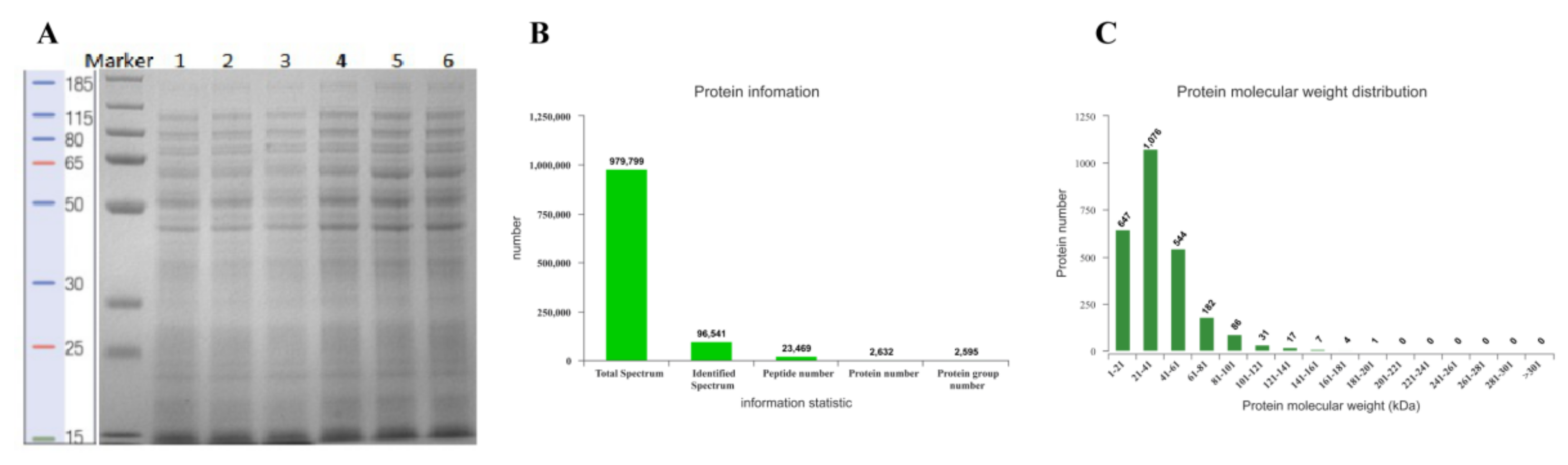
2.6.1. Total Protein Extraction
2.6.2. Protein Digestion and iTRAQ Labeling
2.6.3. High pH-RPLC Separation
2.6.4. LC-MS/MS Analysis
2.6.5. Protein Identification
2.7. Molecular Docking Analysis
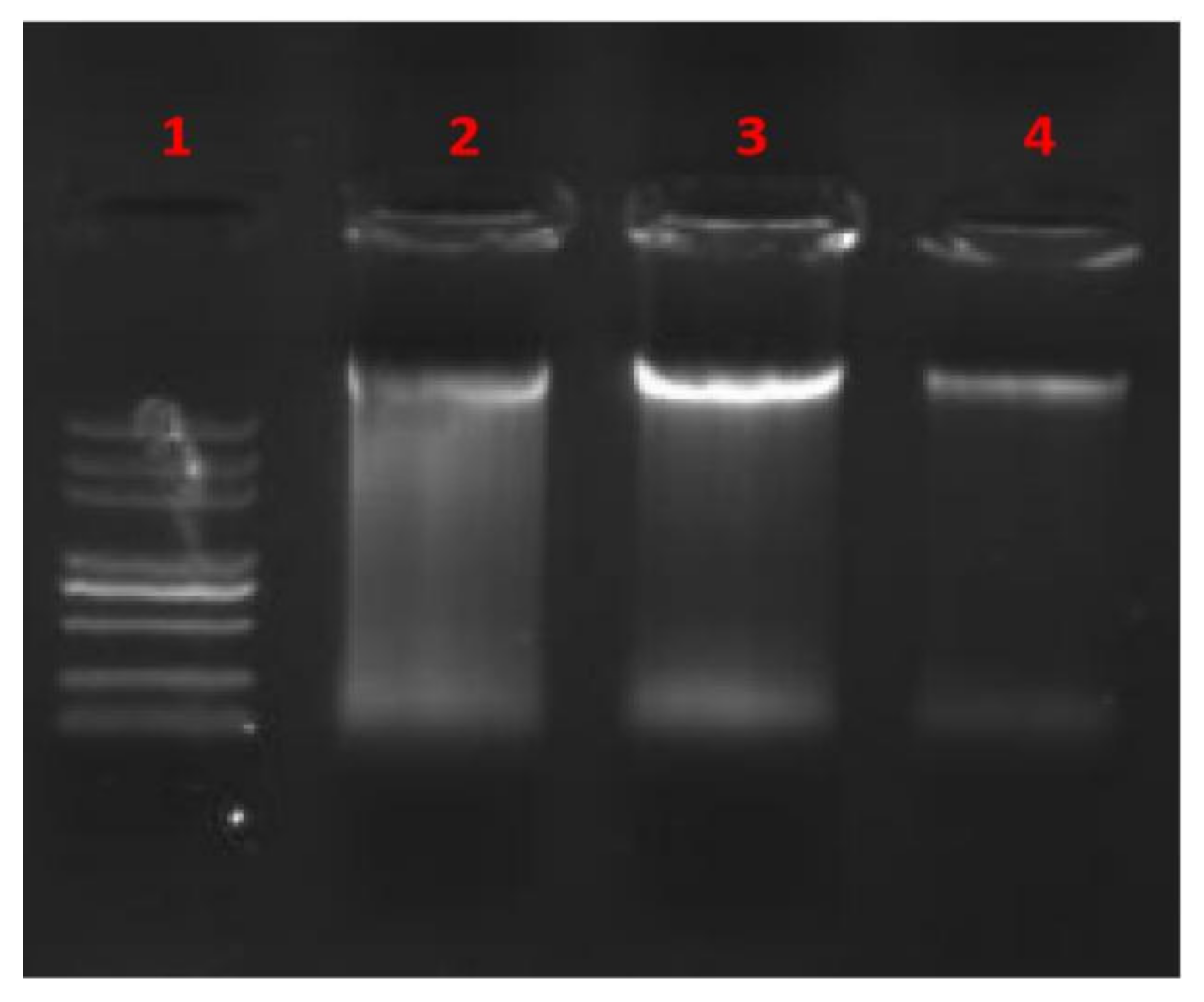
2.8. Gel Mobility Shift Assay
2.9. Statistical Analysis
3. Results
3.1. The Effect of BCp12 on E. coli Growth
3.2. Localization Analysis of BCp12 in E. coli Cells
3.3. Effect of BCp12 on E. coli Morphologies
3.4. The Effect of BCp12 on the Internal Structure of E. coli Cells (TEM Observation)
3.5. Proteome Analysis
3.5.1. Protein Extraction and Basic Identification Information
3.5.2. LC-MS/MS Analysis
3.5.3. Functional Annotation of Differentially Expressed Proteins
3.5.4. Expression Changes of Cell Membrane-Related Proteins
3.6. Molecular Docking Analysis
3.7. In Vitro Interaction between BCp12 and DNA
4. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Yi, L.; Luo, L.; Chen, J.; Sun, H.; Lv, X. Cell wall and DNA damage of Staphylococcus aureus by bacteriocin BM1157. LWT Food Sci. Technol. 2020, 134, 109842. [Google Scholar] [CrossRef]
- Ruegg, P.L. A 100-Year Review: Mastitis detection, management, and prevention. J. Dairy Sci. 2017, 100, 10381–10397. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cao, T.H.; Liu, G.H. Analysis on detection results of foodborne pathogens in meat and meat products in Tonghua city from 2014 to 2018. Chin. J. Public Heath Eng. 2020, 1, 39–41. [Google Scholar]
- Guo, Y.D.; Fan, L.; Tang, X.S. Investigation on microbial contamination status of takeaway catering in Yunnan province in 2018. J. Food Saf. Qual. 2019, 10, 6. [Google Scholar]
- Zang, Y.; Jiang, J.; Fu, Z.Q. Surveillance of microbial contamination in processing of cooked-meat products in Nanchong from 2015 to 2017. J. Prev. Med. Inf. 2018, 34, 4. [Google Scholar]
- Mazdeh, F.Z.; Aftabdari, F.E.; Moradi-Khatoonabadi, Z.; Shaneshin, M.; Torabi, P.; Ardekani, M.S. Sodium benzoate and potassium sorbate preservatives in iranian doogh. Food Addit. Contam. Part B 2014, 7, 115119. [Google Scholar]
- Kashani, H.H.; Nikzad, H.; Mobaseri, S.; Hoseini, E.S. Synergism Effect of Nisin Peptide in Reducing Chemical Preservatives in Food Industry. Life Sci. J. 2012, 9, 496–501. [Google Scholar]
- Vaisocherová-Lísalová, H.; Víšová, I.; Ermini, M.L.; Špringer, T.; Chadtová Song, X.; Mrázek, J.; Lamačová, J.; Lynn, N.S.; Šedivák, P.; Homola, J. Low-fouling surface plasmon resonance biosensor for multi-step detection of foodborne bacterial pathogens in complex food samples. Biosens. Bioelectron. 2016, 80, 84–90. [Google Scholar] [CrossRef]
- Hao, G.; Hui, H.; Ma, D.Y. Studies on the inhibition of the synthesis of E. coli macromolecules by antimicrobial peptide BuforinⅡ derivatives. Microbiol. Bull. 2013, 40, 9. [Google Scholar]
- Pu, C.; Tang, W. Affinity and selectivity of anchovy antibacterial peptide for Staphylococcus aureus cell membrane lipid and its application in whole milk. Food Control 2017, 72, 153–163. [Google Scholar] [CrossRef]
- Ciumac, D.; Gong, H.; Hu, X.; Lu, J.R. Membrane targeting cationic antimicrobial peptides. J. Colloid Interface Sci. 2019, 537, 163–185. [Google Scholar] [CrossRef] [PubMed]
- Moraes, L.; Vieira, R.; Nakaie, C.R.; Miranda, A. Conformational and functional studies of gomesin analogues by cd, epr and fluorescence spectroscopies. BBA Biomembr. 2007, 1768, 52–58. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wood, S.J.; Park, Y.A.; Kanneganti, N.P.; Mukkisa, H.R.; Crisman, L.L.; Davis, S.E. Modified cysteine-deleted tachyplesin (cdt) analogs as linear antimicrobial peptides: Influence of chain length, positive charge, and hydrophobicity on antimicrobial and hemolytic activity. Int. J. Pept. Res. Ther. 2014, 20, 519–530. [Google Scholar] [CrossRef]
- Turk, M.; Grimalt, J.; Gunde-Cimerman, N.; Plemenita, A. Salt-induced changes in lipid composition and membrane fluidity of halophilic yeast-like melanized fungi. Extremophiles 2004, 8, 53–61. [Google Scholar] [CrossRef]
- Li, Y.Q.; Han, Q.; Feng, J.L.; Tian, W.L.; Mo, H.Z. Antibacterial characteristics and mechanisms of ϵ-poly-lysine against Escherichia coli and Staphylococcus aureus. Food Control 2016, 59, 282–289. [Google Scholar]
- Tan, Z.; Shi, Y.; Xing, B.; Hou, Y.; Cui, J.; Jia, S. The antimicrobial effects and mechanism of ε-poly-lysine against Staphylococcus aureus. Bioresour. Bioprocess. 2019, 6, 11. [Google Scholar] [CrossRef]
- Scocchi, M.; Mardirossian, M.; Runti, G. Non-membrane permeabilizing modes of action of antimicrobial peptides on bacteria. Curr. Top. Med. Chem. 2016, 16, 70–88. [Google Scholar] [CrossRef]
- Maldonado-Barragan, A.; Cardenas, N.; Martinez, B.; Ruiz-Barba, J.L.; Garvicin, A. A novel class IId bacteriocin from Lactococcus garvieae that inhibits septum formation in L. garvieae strains. Appl. Environ. Microbiol. 2013, 79, 4336–4346. [Google Scholar] [CrossRef] [Green Version]
- Duan, F.; Li, X.; Cai, S.; Xin, G.; Liu, X.; Huang, W. Haloemodin as novel antibacterial agent inhibiting DNA gyrase and bacterial topoisomerase i. J. Med. Chem. 2014, 57, 3707–3714. [Google Scholar] [CrossRef]
- Mardirossian, M.; Perebaskine, N.; Benincasa, M.; Gambato, S.; Wilson, D.N. The Dolphin Proline-Rich Antimicrobial Peptide Tur1A Inhibits Protein Synthesis by Targeting the Bacterial Ribosome. Cell Chem. Biol. 2018, 25, 530–539. [Google Scholar] [CrossRef] [Green Version]
- Lü, A.; Hu, X.; Wang, Y.; Shen, X.; Li, X.; Zhu, A.; Tian, J.; Ming, Q.; Feng, Z. Itraq analysis of gill proteins from the zebrafish (Danio rerio) infected with Aeromonas hydrophila. Fish Shellfish Immun. 2014, 36, 229–239. [Google Scholar] [CrossRef]
- Miao, J.; Chen, F.; Duan, S.; Gao, X.; Chen, Y. Itraq-based quantitative proteomic analysis of the antimicrobial mechanism of peptide F1 against Escherichia coli. J. Agric. Food Chem. 2015, 63, 7190–7197. [Google Scholar] [CrossRef] [PubMed]
- Zhao, Q.; Li, S.; Chen, C.H.; Huang, A. Preparation of antimicrobial peptides by hydrolysis of betel nut river buffalo casein with Hypericum perforatum protease. Food Ind. 2020, 41, 5. [Google Scholar]
- Zhao, Q.; Shi, Y.; Wang, X.; Huang, A. Characterization of a novel antimicrobial peptide from buffalo casein hydrolysate based on live bacteria adsorption. J. Dairy Sci. 2020, 103, 11116–11128. [Google Scholar] [CrossRef] [PubMed]
- Su, M.; Liu, F.; Luo, Z.; Wu, H.; Miao, Y. The antibacterial activity and mechanism of chlorogenic acid against foodborne pathogen Pseudomonas aeruginosa. Foodborne Pathog. Dis. 2019, 16, 823–830. [Google Scholar] [CrossRef] [PubMed]
- Wang, Q.; Miao, J.Y.; Feng, K. Antibacterial action of peptide F1 against colistin resistance E. coli shp45 (mcr-1). Food Funct. 2010, 11, 10231–10241. [Google Scholar] [CrossRef]
- Shu, J.L.; O’Brien-Simpson, N.M.; Pantarat, N.; Sulistio, A.; Qiao, G. Combating multidrug-resistant gram-negative bacteria with structurally nanoengineered antimicrobial peptide polymers. Nat. Microbiol. 2016, 1, 16162. [Google Scholar]
- Kang, J.; Jin, W.; Wang, J.; Sun, Y.; Wu, X.; Liu, L. Antibacterial and anti-biofilm activities of peppermint essential oil against Staphylococcus aureus. LWT 2019, 101, 639–645. [Google Scholar] [CrossRef]
- Jung, W.K.; Koo, H.C.; Kim, K.W.; Shin, S.; Kim, S.H.; Park, Y.H. Antibacterial activity and mechanism of action of the silver ion in Staphylococcus aureus and Escherichia coli. Appl. Environ. Microbiol. 2008, 74, 2171–2178. [Google Scholar] [CrossRef] [Green Version]
- Ma, W.; Zhang, D.; Li, G.; He, G.; Zhang, P. Antibacterial mechanism of daptomycin antibiotic against Staphylococcus aureus based on a quantitative bacterial proteome analysis. J. Proteom. 2017, 150, 242–251. [Google Scholar] [CrossRef] [Green Version]
- Bronwen, M.; Randall, B.; Becker, K.G.; Marjan, G.; Stuart, M. Itraq analysis of complex proteome alterations in 3xtgad Alzheimer’s mice: Understanding the interface between physiology and disease. PLoS ONE 2008, 7, e2750. [Google Scholar]
- Shi, X.; Wang, X.; Cheng, F.; Cao, H.; Liang, H.; Lu, J.; Kong, Q.; Bie, Z. Itraq-based quantitative proteomics analysis of cold stress-induced mechanisms in grafted watermelon seedlings. J. Proteom. 2019, 192, 311–320. [Google Scholar] [CrossRef] [PubMed]
- Guo, W.; Zhou, X.; Xu, L.N.; Fu, X.J. Effects of cold storage on Microstructure and metabolites of grass carp meat. Food Sci. 2022, 1, 1–13. [Google Scholar]
- Bandyopadhyay, S.; Lee, M.; Sivaraman, J.; Chatterjee, C. Model membrane interaction and DNA-binding of antimicrobial peptide lasioglossin ii derived from bee venom. Biochem. Biophys. Res. Commun. 2013, 430, 1–6. [Google Scholar] [CrossRef]
- Boulos, L.; Prevost, M.; Barbeau, B.; Coallier, J.; Desjardins, R. LIVE/DEAD® BacLight™: Application of a new rapid staining method for direct enumeration of viable and total bacteria in drinking water. J. Microbiol. Methods 1999, 37, 77–86. [Google Scholar] [CrossRef]
- Shai, Y. Mode of action of membrane active antimicrobial peptides. Biopolymers 2002, 66, 236–248. [Google Scholar] [CrossRef]
- Chen, F.L. The Mechanism of Antibacterial Peptide Produced by Lactobacillus paracasei FX-6 against Staphylococcus aureus and Its Application. Master’s Thesis, South China Agricultural University, Guangzhou, China, 2016. [Google Scholar]
- Xiao, H.Q.; Li, Y.Z.; Lin, Q.L. Advances on multiple-targets action mechanism of antimicrobial peptides. J. Food Sci. Biotechnol. 2012, 21, 3–5. [Google Scholar]
- Brogden, K. Antimicrobial peptides: Pore formers or metabolic inhibitors in bacterial. Nat. Rev. Microbiol. 2005, 3, 238–250. [Google Scholar] [CrossRef]
- Chang, L.; Wang, J.; Tong, C.; Liu, X. Antibacterial mechanism of polyacrylonitrile fiber with organophosphorus groups against Escherichia coli. Fibers Polym. 2016, 17, 721–728. [Google Scholar] [CrossRef]
- Yang, S.; Li, J.; Aweya, J.; Yuan, Z.; Liu, G.M. Antimicrobial mechanism of Larimichthys crocea whey acidic protein-derived peptide (lcwap) against Staphylococcus aureus and its application in milk. Int. J. Food Microbiol. 2020, 335, 108891. [Google Scholar] [CrossRef]
- Maya, F.; Ganesan, N. Synergistic effect of low power ultrasonication on antimicrobial activity of cecropin p1 against E. coli in food systems. LWT 2018, 96, 175–181. [Google Scholar]
- Lu, X.; Vora, H.; Khosla, C. Overproduction of free fatty acids in E. coli: Implications for biodiesel production. Metab. Eng. 2008, 10, 333–339. [Google Scholar] [CrossRef]
- Zhou, Y.S. The Effect of Reducing Power Producing Enzyme on E. coli Fatty Acid Synthesis. Ph.D. Thesis, Jiangnan University, Wuxi, China, 2013. [Google Scholar]
- Campbell, J.W.; Cronan, J.E. The enigmatic Escherichia coli fade gene is yafH. J. Bacteriol. 2002, 184, 3759. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Steen, E.J.; Kang, Y.; Bokinsky, G.; Hu, Z.; Schirmer, A.; McClure, A. Microbial production of fatty-acid-derived fuels and chemicals from plant biomass. Nature 2010, 463, 559–562. [Google Scholar] [CrossRef]
- Luo, L.; Yi, L.; Chen, J.; Liu, B.; Xin, L. Antibacterial mechanisms of bacteriocin BM1157 against Escherichia coli and Cronobacter sakazakii. Food Control. 2020, 123, 107730. [Google Scholar] [CrossRef]
- Patel, H.; Brunton, V.G. Loss of frma leads to increased cell-cell adhesion and impaired multi-cellular development of dictyostelium cells. Cell. Mol. Life Sci. 2009, 66, 145–155. [Google Scholar] [CrossRef] [PubMed]
- Mihaly, V.; Stephen, A.; Mandar, D.; Sreenath, N.; Cindy, N.; Galabina, Y. Alphafold protein structure database: Massively expanding the structural coverage of protein-sequence space with high-accuracy models. Nucleic Acids Res. 2021, 50, D439–D444. [Google Scholar]
- Callaway, E. DeepMind’s AI predicts structures for a vast trove of proteins. Nature 2021, 595, 635. [Google Scholar] [CrossRef]
- Han, Q.; Liu, F.; Wen, X.; Ni, Y. Kinetic, spectroscopic, and molecular docking studies on the inhibition of membrane-bound polyphenol oxidase from granny smith apples (malus domestica borkh). Food Chem. 2020, 338, 127928. [Google Scholar] [CrossRef]
- Xie, H.; Yang, X.; Ke, C.; Xu, H.; Guo, Y. The molecular mechanism of chemical synthesis of antimicrobial peptides in inhibiting pathogenic Escherichia coli F41. Mod. Food Sci. Technol. 2016, 1, 44–51. [Google Scholar]
- Yonezawa, A.; Kuwahara, J.; Fujii, N.; Sugiura, Y. Binding of tachyplesin i to DNA revealed by footprinting analysis: Significant contribution of secondary structure to DNA binding and implication for biological action. Biochemistry 1992, 31, 2998–3004. [Google Scholar] [CrossRef] [PubMed]
- Li, H.; Shen, X.F.; Zhou, X.E. Antibacterial mechanism of high-mobility group nucleosomal-binding domain 2 on the Gram-negative bacteria Escherichia coli. J. Zhejiang Univ. Sci. B 2017, 18, 410–420. [Google Scholar] [CrossRef] [PubMed] [Green Version]



| Target Protein | Docking Score (kcal/mol) | Hydrogen Bond Binding Site | Hydrophobic Interaction Binding Site |
|---|---|---|---|
| 3-ketoacyl-CoA thiolase (fadA) | −6.3 | ala-174,pro-133 | tyr-178, phe-394, val-67, gln-69, pro-133, pro-41, val-136 and val-171 |
| acyl-CoA dehydrogenase (fadE) | −6.9 | arg-483, asn-429, gly-428, ser-191, thr-185, Glu-270, pro-220 and arg-416 | ille-423, pro-419, leu-261 and phe-216 |
| S-(hydroxymethyl)glutathione dehydrogenase (frmA) | −7 | tyr-88, ile-287 and gln-294 | phe-45, val-291, val-289, ile-296, trp-309, phe-314, leu-105 and ile-287 |
| Acyltransferase (aas) | −7.5 | phe-344, arg-10, ser-78 and phe-341 | met-85, pro-86, VAL-89, Leu-93, leu-97, phe-146, leu-143, pro-340 and phe-341 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Yang, K.; Shi, Y.; Li, Y.; Wei, G.; Zhao, Q.; Huang, A. iTRAQ-Based Quantitative Proteomic Analysis of Antibacterial Mechanism of Milk-Derived Peptide BCp12 against Escherichia coli. Foods 2022, 11, 672. https://doi.org/10.3390/foods11050672
Yang K, Shi Y, Li Y, Wei G, Zhao Q, Huang A. iTRAQ-Based Quantitative Proteomic Analysis of Antibacterial Mechanism of Milk-Derived Peptide BCp12 against Escherichia coli. Foods. 2022; 11(5):672. https://doi.org/10.3390/foods11050672
Chicago/Turabian StyleYang, Kun, Yanan Shi, Yufang Li, Guangqiang Wei, Qiong Zhao, and Aixiang Huang. 2022. "iTRAQ-Based Quantitative Proteomic Analysis of Antibacterial Mechanism of Milk-Derived Peptide BCp12 against Escherichia coli" Foods 11, no. 5: 672. https://doi.org/10.3390/foods11050672
APA StyleYang, K., Shi, Y., Li, Y., Wei, G., Zhao, Q., & Huang, A. (2022). iTRAQ-Based Quantitative Proteomic Analysis of Antibacterial Mechanism of Milk-Derived Peptide BCp12 against Escherichia coli. Foods, 11(5), 672. https://doi.org/10.3390/foods11050672






