Abstract
Salmonella enteritidis is a major causative agent of foodborne illnesses worldwide. As the traditional serotyping and quantification methods are labor-intensive, time-consuming, and expensive, faster and more convenient molecular diagnostic methods are needed. In this study, we developed and validated a rapid duplex TaqMan real-time polymerase chain reaction (PCR) for the accurate identification and quantification of S. enteritidis. The primers and TaqMan probes were designed based on the S. enteritidis-specific gene lygD and the Salmonella genus-specific gene invA. The melt curve and gel electrophoresis analysis showed that the designed primers had potent specificity for the amplification of lygD and invA. The duplex real-time PCR specifically identified S. enteritidis from a panel of 40 Salmonella strains that represented 29 serovars and 12 non-Salmonella organisms. The duplex real-time PCR assay detected four copies of S. enteritidis DNA per reaction. The intra- and inter- assays indicated a high degree of reproducibility. The real-time PCR could accurately detect and quantify S. enteritidis in chicken organs after Salmonella infection. Furthermore, the assay identified 100% of the S. enteritidis and Salmonella genus isolates from chicken egg samples with superior sensitivity after 6 h of pre-enrichment compared to the traditional culture method. Additionally, the most-probable-number (MPN) combined with qPCR and a shortened incubation time (MPN-qPCR-SIT) method was developed for the population determination of S. enteritidis and compared with various enumeration methods. Thus, we have established and validated a new duplex real-time PCR assay and MPN-qPCR-SIT method for the accurate detection and quantification of S. enteritidis, which could contribute to meeting the need for fast detection and identification in prevention and control measures for food safety.
1. Introduction
Salmonella is one of the most important food-borne pathogens, and can cause severe enteritis worldwide. Almost 1.3 billion cases of human salmonellosis occur and approximately 3.5 million patients die as a result of the disease annually [1]. Most of these cases result from the uptake of Salmonella-contaminated food such as pork, poultry, and eggs [2]. Human salmonellosis symptoms include gastroenteritis, fever, diarrhea, and serious systemic infections that may cause hospitalization [3].
S. enteritidis has the ability to survive in the egg white and efficiently contaminate eggs [4], which leads to significant economic and health burdens worldwide [5,6]. Raw or undercooked poultry meat and eggs are foods with a high risk of leading to human salmonellosis, and several outbreaks due to these foods have been reported [7]. Although more than 2600 Salmonella serovars exist, S. enteritidis is one of the most important agents resulting in severe infection [8,9], amounting to over 60% of human salmonellosis cases in Europe [10].
Traditional Salmonella serotyping is conducted according to the Kauffman–White scheme by using the specific antisera for the bacterial surface O and H antigens [11]. Despite its widespread use, the traditional culture-based method is relatively expensive, labor-intensive, and time-consuming, as it often involves several enrichment steps followed by biochemical or serological confirmation, taking 3–5 days [12,13]. Since an accurate surveillance method is important for controlling the spread of salmonellosis, developing a reliable method is critical for the identification of prevalent serovars isolated from contaminated foods.
Most of the foodborne outbreaks come from animal origin food, including beef meat, poultry, eggs, and milk products, which may be contaminated by multiple pathogens including Salmonella enterica [14]. Fresh-cut produce is at great risk of Salmonella contamination, and a one-step quantitative real-time polymerase chain reaction (qRT-PCR) assay has been used to detect Salmonella in fresh-cut vegetables [15]. A fluorescent biosensor with multiple fluorescent signal amplification based on a streptavidin biotin system was established to detect Salmonella in milk. The detection limit was ten times better than that of the conventional sandwich enzyme linked immunosorbent assay (ELISA) [16]. A sensitive immunosensor was successfully constructed based on a Fe3O4 graphene nanocomposite to detect Salmonella. The constructed immunosensor exhibited acceptable selectivity and reproducibility for detecting Salmonella in milk [17]; however, a convenient method for rapid, accurate, and quantitative identification of Salmonella is urgently needed.
Rapid molecular methods to identify Salmonella serovars, especially clinically important serovars, could promote routine surveillance and therefore, public health. Of these methods, polymerase chain reaction (PCR) has been widely studied because it has high throughput, and is rapid, facile, highly sensitive, and highly specific [18,19]. Unlike traditional PCR, real-time PCR methods have gained more attention recently because the results are monitored in real time. Therefore, no other analyses or assays, such as gel electrophoresis, are needed for conformation of the specific pathogens, and the data can be analyzed quantitatively. More importantly, real-time PCR assays provide high quality quantitative data for a specific pathogen in food products [20]. The most commonly targeted Salmonella gene is invA (invasion protein gene), which is necessary for virulence and encodes a membrane protein of the type III secretion system [21,22,23,24]. The invA gene has been widely used for the identification of the Salmonella genus and used as the internal control reference [25]. Previously we found that the Salmonella gene lygD shares 98–100% similarity in nucleotide sequences and only exists in S. enteritidis [26,27]. Thus, the lygD gene is a desirable candidate for the specific identification of S. enteritidis.
The most probable number (MPN) method is based on decimal dilutions and has been used for the quantification of bacterial contamination in low levels. In a ten-fold dilution MPN, the bacteria number increases in each well following the incubation. Eventually, bacteria-positive wells can be observed, even if at least one bacteria cell exists before incubation. The calculation of the number of bacteria was based on the standard MPN table [28]. This method has been widely applied for the quantification of low levels of Salmonella and Vibrio parahaemolyticus in samples [29,30,31]. The combination of qPCR and MPN methods has also been used for the quantification of Listeria monocytogenes and Vibrio parahaemolyticus, and the qPCR was used as a confirmation step following MPN assays by a shortened incubation time (SIT) to determine positive/negative results [32,33,34].
In the present study, we selected the lygD and invA genes for developing a fast and accurate TaqMan real-time PCR assay for the timely identification and quantification of S. enteritidis. We determined the specificity, sensitivity and its accuracy of the PCR method by comparing it with that of the traditional culture method. The developed method was applied for the identification of S. enteritidis in chicken organs or directly contaminated chicken egg samples.
2. Materials and Methods
2.1. Bacterial Strains
Forty strains of Salmonella (29 serovars) and 12 non-Salmonella organisms were used in this study. They were either commercially obtained, or previously isolated after routine monitoring (Table 1). The strains were used to test the sensitivity and specificity of the duplex real-time PCR method.

Table 1.
Different Salmonella serovars and other non-Salmonella bacteria used to determine the specificity of the established real-time PCR assay.
2.2. Bacterial Growth and Genomic DNA Extraction
Bacterial genomic DNA was prepared as previously described [27]. In brief, all bacteria used in this study were inoculated in either brain heart infusion broth (Becton, Dickinson and Company, Sparks, MD, USA) or Luria–Bertani (LB) broth (Oxoid, Basingstoke, Hampshire, UK) at 37 °C overnight in a shaker incubator. DNA extracts were prepared from 1.0 mL of overnight cultures of the test isolates using a TIANamp Bacterial DNA kit (TianGen Biotech Co. Ltd., Beijing, China). The DNA concentration was determined using a NanoDrop ND-1000 (Thermo Scientific, Wilmington, DE, USA) spectrophotometer. The number of genomic DNA copies present in the bacterial strains was determined based on the online URI Genomics & Sequencing Center copy number calculator for double-stranded DNA (http://cels.uri.edu/gsc/cndna.html, accessed on 9 June 2021). DNA extracts were stored at −20 °C until use.
2.3. PCR Primer Pairs and TaqMan Probes
The duplex TaqMan real-time PCR was designed by targeting the S. enteritidis-specific lygD gene and the Salmonella genus-specific invA gene. The different primer/probe sets were designed based on sequence data available at the National Center for Biotechnology Information databases using Primer 3 software (http://bioinfo.ut.ee/primer3-0.4.0/, accessed on 16 May 2021; [38]) (Table 2). The specific probes and primers were synthesized by Takara (Dalian, China).

Table 2.
Primers and TaqMan probes for the two genes used in the duplex real-time PCR.
2.4. Conventional PCR
The conventional PCR assay was performed at a final volume of 20 μL including 10 μL of 2× Taq Master Mix (Vazyme, Nanjing, China), 0.5 μM of the lygD or invA F/R primers (Table 2), and the specified amount of DNA. The PCR amplification was carried out using a T100 Thermal Cycler (Bio-Rad, Hercules, CA, USA) with the following protocol: initial denaturation at 94 °C for 3 min, 30 cycles of 94 °C for 30 s, 60 °C for 30 s, 72 °C for 30 s, and a final extension of 72 °C for 10 min. The PCR products were run on 1% agarose gels.
2.5. SYBR-Based qRT-PCR
The melt curves for the lygD and invA products were collected by the SYBR-based qRT-PCR using serial concentrations of positive standard samples. The qRT-PCR was performed using ABI 7500 real-time instrument (Applied Biosystems, Carlsbad, CA, USA). The 20 μL PCR reactions contained 10 μL of 2× SYBR Green I Master Mix (Vazyme, Nanjing, China), 0.3 μM lygD or invA forward and reverse primers, nuclease-free water and the specified amount of DNA. The PCR profile consisted of initial denaturation at 95 °C for 5 min, followed by 40 cycles of 95 °C for 5 s, and 60 °C for 34 s. The melting curve analysis consisted of 1 cycle at 95 °C for 15 s and then 60 °C for 1 min, followed by a continuous increase of temperature to 95 °C at a rate of 0.5 °C/s. The fluorescence signal was monitored continuously and plotted against the temperature. The resulting PCR amplicons were directly visualized by 1% agarose gel electrophoresis.
2.6. Duplex TaqMan Real-Time PCR System
To identify S. enteritidis, the designed duplex PCR assay exploited the specific primers and probes for the lygD and invA. Real-time duplex PCR was conducted on an ABI 7500 instrument (Applied Biosystems, Foster, CA, USA) with the Premix Ex Taq Master kit (Takara). The reaction system (25 μL) contained DNA template (2.5 μL), 2× Premix Ex Taq Master (12.5 μL), 240 nM lygD forward and reverse primers (0.6 μL each), 100 nM probe (0.25 μL), 200 nM invA forward and reverse primers (0.5 μL each), 80 nM probe (0.2 μL), and ROX Reference Dye II (0.25 μL). The control tubes used the same mixture, without any DNA template. The real-time PCR reactions were performed for 30 s at 95 °C, followed by 40 cycles of denaturation at 95 °C for 5 s and annealing/extension at 60 °C for 34 s. The fluorescence was collected during the extension step of each cycle.
2.7. Specificity of the Duplex Real-Time PCR
The specificity of the two pairs of primers and probes in the duplex PCR method was determined from 40 different Salmonella strains and 12 other non-Salmonella strains using 105 copies of genomic DNA for each strain listed in Table 1.
2.8. Standard Curve and Detection Limit of the Duplex Real-Time PCR
The standard curve and detection limit of the duplex real-time PCR were evaluated using S. enteritidis C50041. Bacterial counts were verified based on 10-fold dilutions (101–108 dilutions) and a traditional plate counting assay. Genomic DNA was prepared from decimally diluted samples and amplified by the duplex real-time PCR. Ct values of each dilution were obtained and plotted against log10 colony-forming units (CFU). The linear ranges were assessed using the established standard curves.
2.9. Evaluation of the Reproducibility of the Method in Detecting S. enteritidis
Reproducibility for the identification and quantification of S. enteritidis was conducted on six standard samples of S. enteritidis. Different concentrations of S. enteritidis (4 × 101~4 × 106 copies/μL) were used as the templates for TaqMan real-time PCR assay. The intra-batch reproducibility experiment was conducted with three repetitions of the template in one TaqMan real-time assay. The inter-batch reproducibility experiment was conducted by measuring the same template three times by three operators independently. Finally, the coefficient of variation (CV) of the Ct values was determined based on the intra-assay or inter-assay results, so as to evaluate the reproducibility of the method.
2.10. Real-Time PCR for Quantification of S. enteritidis in Organs in a Chicken Model
Two-week-old specific-pathogen free (SPF) white Leghorn chickens were purchased from the poultry institute, at the Shandong academy of agricultural science. All chickens were housed in a room with controlled ventilation, light, and temperature. The procedures described in this study were approved by the Committee on the Ethics of Animal Experiments of Yangzhou University (Approval ID: SYXK (Su) 2017-0044). For oral infections, chickens were fasted overnight and subsequently inoculated orally with 1 × 106 CFU of bacteria in 0.2 mL phosphate-buffered saline (PBS). They were sacrificed three days post-infection, and the spleens and livers were collected to calculate the bacterial burden. The tissues were harvested in 2 mL pre-weighed tubes containing 0.1 mL PBS and weighed before homogenization using the Precellys 24 homogenizer (Rockville, MD, USA). The homogenate dilutions (100 μL each) were used for plate counting on LB agar. The data represent the number of CFU/mL of the organs. The 10−3 dilutions were incubated at 100 °C for 15 min and placed on ice immediately. The tubes were centrifuged for 5 min at 12,000 rpm at 4 °C. An aliquot of the supernatant (2 μL) was served as the template DNA in the duplex TaqMan real-time PCR.
2.11. Real-Time PCR for the Detection of S. enteritidis in Clinical Chicken Eggs
The sensitivity and accuracy of the real-time PCR in S. enteritidis detection in clinical dead egg samples was assessed using 70 samples from a chicken farm and compared to a traditional serotyping method. Samples from the chicken farm were collected as previously described [36,39]. Pre-PCR samples were prepared with a pre-enrichment step in buffered peptone water (BPW), followed by DNA extraction. In brief, 45 mL of BPW was added to the livers of the chicken eggs and incubated at 37 °C at 100 rpm for 6 h. The genomic DNA was extracted from one milliliter of each pre-enriched sample, and subsequently used for the real-time PCR.
2.12. Traditional Serotyping of Salmonella Isolates from Clinical Samples
The traditional serotyping method was conducted by incubating the sample enrichments as prepared above for an additional 18 h. Then, 0.1 mL of the broth culture was subcultured in 10 mL Rappaport–Vassiliadis (RV) enrichment broth (Difco, BD) at 42 °C for 24 h. One loopful of each RV broth culture was streaked on to xylose lysine tergitol 4 (Difco, BD) agar plates, and then incubated at 37 °C for 24–48 h. The presumptive Salmonella colony was picked from each plate and biochemically confirmed using an API-20E test kit (bioMérieux, Marcyl’Etoile, France). All Salmonella isolates from clinical contaminated samples were serotyped following the White–Kauffmann–Le Minor scheme based on agglutination with O- and H-antigen-specific sera (Tianrun Bio-Pharmaceutical, Ningbo, China). All samples were examined by conventional microbiological methods and compared with the duplex TaqMan real-time PCR in a blind manner.
2.13. Preparation of S. enteritidis Cell Suspension and Enumeration Methods
One colony of S. enteritidis on LB agar was expanded by growing cultures overnight at 37 °C. The S. enteritidis cells were harvested by centrifugation at 12,000 rpm for 2 min. The cell pellets were washed twice and resuspended in PBS. Serial dilutions of the cell suspension were used to obtain incrementally different bacterial concentrations. The populations of S. enteritidis were evaluated by different enumeration methods including traditional plating, traditional MPN, TaqMan real-time PCR, and the MPN-qPCR-SIT established in this study.
2.14. Traditional Plating and Traditional MPN Methods
Undiluted and serially diluted cell suspensions (100 µL of each sample) as prepared above were plated on LB agar, and subsequently incubated at 37 °C overnight. The traditional plating method was conducted by counting colonies for the determination of the S. enteritidis population. The conventional MPN method was conducted as previously described [34]. In brief, the assay was conducted using the 96-well sterile microtiter plates (Jet Biofil, Guangzhou, China). The 10 prepared samples containing different populations of Salmonella cells were serially diluted. A miniature of the three tube MPN assay was conducted by dispensing 200 µL of each diluent to three wells. The plates with the cell suspensions of S. enteritidis were cultured at 37 °C for 24 h. The negative and positive results of each well were evaluated visually via turbidity. The populations of S. enteritidis in each sample were determined based on the number of positive wells according to the standard MPN table [28].
2.15. TaqMan Real-Time PCR and MPN-qPCR-SIT Methods
To increase the DNA concentration of the Salmonella suspensions prepared above, the genomic DNA was extracted with a final elution volume of 20 µL from one milliliter of pure cultures. Extracted DNA (5 µL) was used as the template in the TaqMan real-time PCR system. The population of S. enteritidis was calculated based on the established standard curve. The MPN-qPCR-SIT method was prepared with the previously described conventional MPN procedure [34]. A short incubation time of 4 h was applied for the MPN plates. Genomic DNA from each well was harvested from the enriched cultures, and the negative or positive S. enteritidis levels were determined by the TaqMan real-time PCR.
2.16. Statistical Analysis
The significances of the differences of the bacterial loads in chicken organs, as determined by duplex real-time PCR and plate counting methods, were analyzed using the Student’s t-test with GraphPad Prism 5.0 (San Diego, CA, USA). Scatter plots were produced to present correlative relationships of the MPN-qPCR-SIT with the other methods. The MPN-qPCR-SIT and various enumeration methods were compared through linear regression and the coefficients from the scatter plots. A Bland–Altman plot was used to evaluate the 95% agreement boundaries of the different enumeration methods. The scatter plot, Bland–Altman plot, and regression trend lines were generated by using GraphPad Prism.
3. Results
3.1. Specificity Analysis of the Primers for the Amplification of lygD and invA
The melt curves of the SYBR-based qRT-PCR were analyzed to determine the specificity of the primers. The qRT-PCR assay was conducted with different concentrations of DNA, and the results were assessed by both melt curves and agarose gel electrophoresis. As seen in Figure S1A, the melt curve data showed single peaks of 79.58 °C for lygD and 80.56 °C for invA, suggesting that the primers had good specificity for the amplification of the both genes. In addition, a direct visualization of the products of the SYBR-based qRT-PCR showed that only one specific band was observed for either lygD or invA (Figure S1B). The conventional PCR results also showed that only one specific band corresponding to either lygD or invA of S. enteritidis was generated, respectively (Figure S1C).
3.2. Specificity of the Duplex TaqMan Real-Time PCR Assay
The specificity of the real-time PCR assay for the targets was determined using 40 strains of different Salmonella serovars and 12 other non-Salmonella bacterial strains. All Salmonella strains tested positive for invA. Both lygD and invA were amplified and detected in all S. enteritidis strains. None of the non-Salmonella strains produced signals for the lygD or invA targets. This indicated that lygD- and invA-based duplex TaqMan real-time PCR detects S. enteritidis specifically (Table 1).
3.3. Standard Curves and Sensitivity of the Developed Real-Time PCR
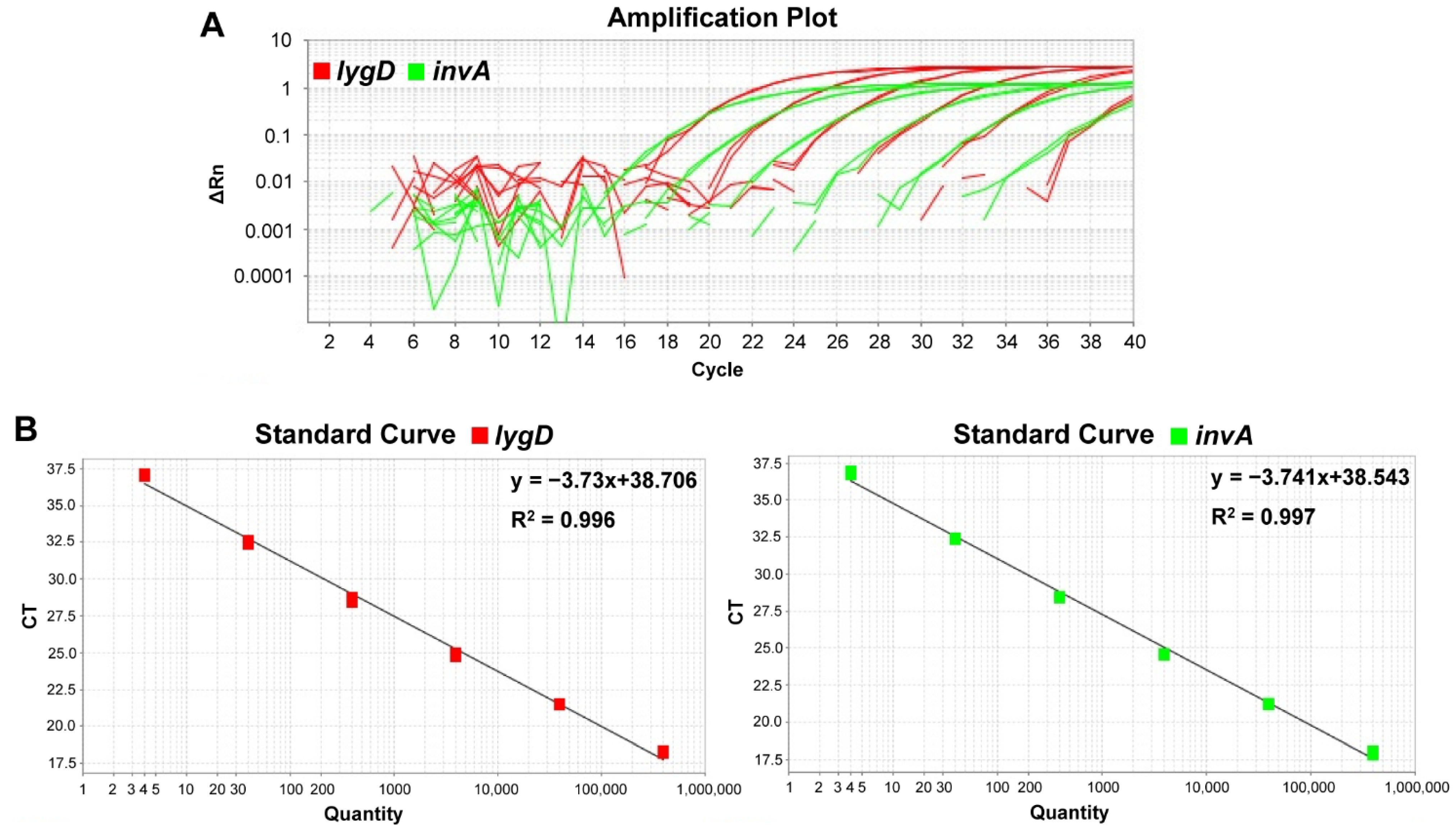
The detection limit of DNA concentration corresponding to the bacterial concentration was determined. Standard curves were generated by using the mean Ct values for various concentrations of S. enteritidis C50041 genomic DNA, ranging from 4–4 × 105 copies per reaction in the real-time PCR system. A good linearity of response (R2 = 0.996 and 0.997) for each reaction channel (Cy5 and BHQ-2 for lygD, FAM and TAMRA for invA) was observed between the Ct values and the amount of bacterial DNA. The results indicated that the duplex TaqMan real-time PCR could successfully detect as low as four copies per reaction of S. enteritidis DNA (Figure 1).
Figure 1.
Sensitivity and standard curves of the developed duplex TaqMan real-time polymerase chain reaction (PCR) assay. The detection limit of the PCR assay was determined by testing various concentrations of Salmonella Enteritidis C50041 genomic DNA, ranging from 4–4 × 105 copies per reaction. The PCR system contained probes and primers specific for S. enteritidis and Salmonella spp. (A) Amplification plots of the duplex real-time PCR. X axis—PCR cycle numbers; y axis—fluorescence intensity. (B) Standard curves indicating the linearity for detecting lygD and invA by real-time PCR. There was a good linear correlation between the Ct values of lygD and invA and the logarithm of the DNA copy numbers over the whole range of DNA concentration. The Ct values were plotted against the corresponding Salmonella cell numbers.
3.4. Reproducibility of the TaqMan Real-Time PCR Assay
The duplex TaqMan real-time PCR assay produced very similar Ct values when tested on six samples of the target pathogen, S. enteritidis. The intra-assay CVs of the Ct values for the duplex real-time PCR ranged from 0.33% to 1.94% and 0.17% to 1.60% for lygD and invA, respectively. The inter-assay reproducibility experiments showed that the inter-assay CVs ranged from 0.80% to 2.22% and 0.54% to 2.54% for lygD and invA, respectively (Table 3). The results indicated a high degree of reproducibility of this assay.

Table 3.
Intra- and inter-assay reproducibility results of the developed TaqMan real-time PCR assay.
3.5. Quantification of S. enteritidis in a Chicken Model by Real-Time PCR Assay
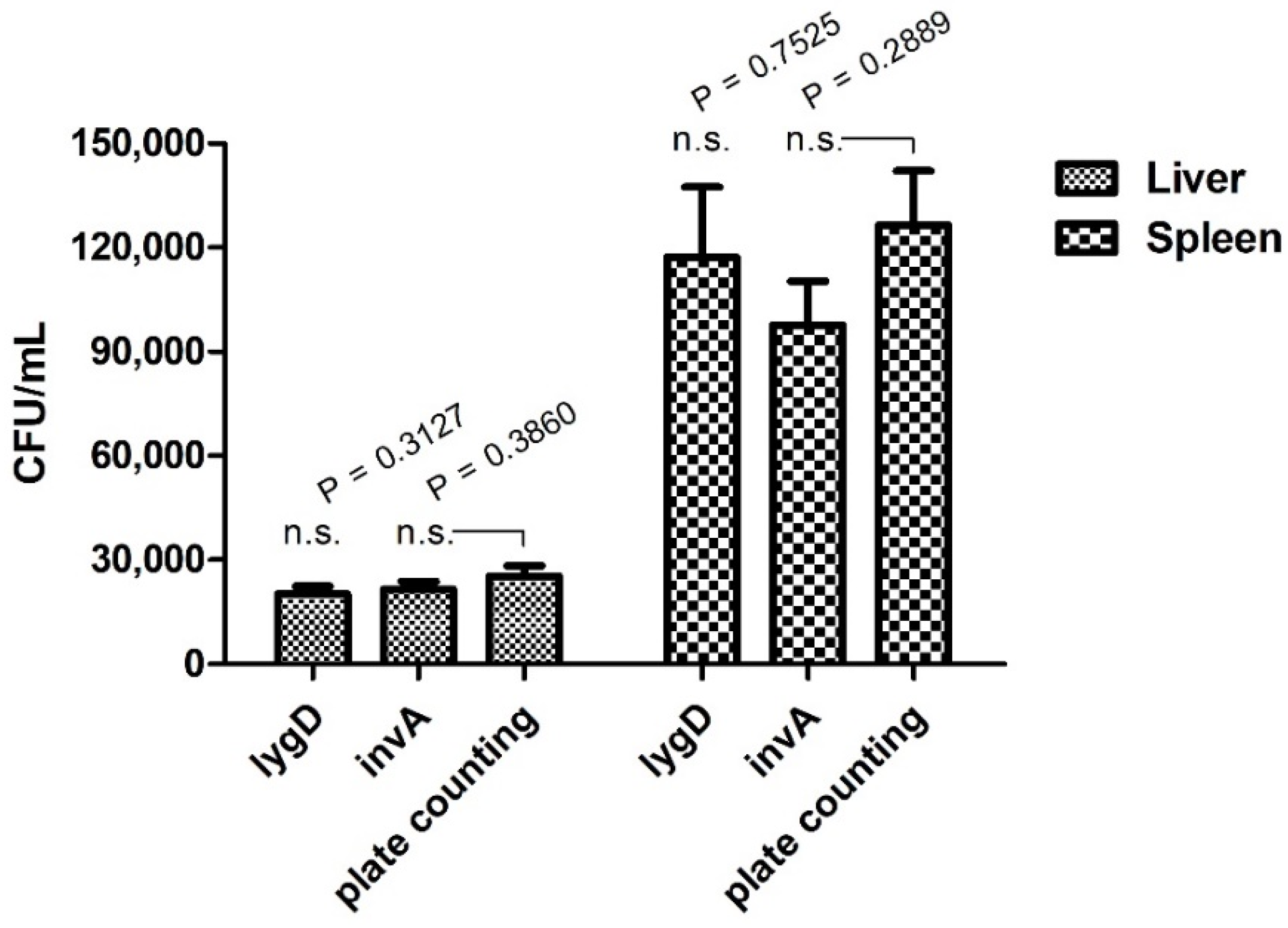
To evaluate the applicability and reliability of the assay in routine laboratories, a chicken infection model was applied. The bacterial burden in organs was determined by both the developed real-time PCR assay and plate counting methods. The results showed that lygD yielded Ct values of 32.36 ± 0.25 and 29.53 ± 0.40, 30 in 10−3 dilution of liver and spleen homogenates, respectively, while invA yielded Ct values of 32.08 ± 0.29 and 29.62 ± 0.30 in 10−3 dilution of liver and spleen homogenates, respectively. Of note, there was no significant difference in the quantification of Salmonella between the real-time PCR assay and the traditional plate counting method (Figure 2).
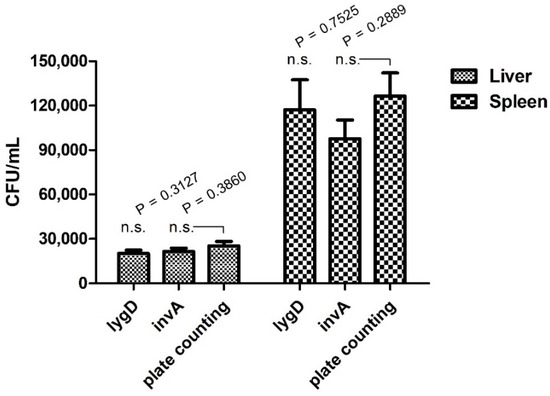
Figure 2.
Quantification analysis of the duplex real-time PCR for S. enteritidis in a chicken infection model. SPF white Leghorn chickens were infected with the indicated concentrations of bacteria in 0.2 mL phosphate-buffered saline orally. The spleens and livers were collected for analysis of bacterial burden. Organs were collected and homogenized. The serial 10-fold dilutions were plated on LB agar. The data represent the number of CFU/mL in 10−3 dilution of organs. n.s. indicates no significant difference.
3.6. Application of the Duplex Real-Time PCR for Clinical Chicken Eggs
The established real-time PCR method was used to identify S. enteritidis in clinically contaminated egg samples from a chicken farm. A total of 70 egg samples were analyzed by the developed TaqMan real-time PCR method, and the results were compared with those obtained by traditional serotyping procedures. The prevalence (determined by real-time PCR) of the Salmonella genus and specifically, the S. enteritidis isolates were 42.86% (30/70) and 37.14% (26/70) for the TaqMan real-time PCR method and the traditional serotyping procedure respectively, after 6 h of non-selective pre-enrichment. Of the 70 samples tested, the traditional method detected 27 Salmonella isolates (38.57%, 27/70), including 23 strains of S. enteritidis (32.86%, 23/70) (Table 4). Of note, the PCR products of three S. enteritidis-positive isolates, not detected by the traditional method, were sequenced, and the results showed that all the PCR products were lygD- and invA-positive. Thus, the duplex TaqMan real-time PCR showed an improved clinical sensitivity and accuracy compared to the traditional serotyping method.

Table 4.
Identification of S. enteritidis and Salmonella genus from the contaminated chicken eggs.
3.7. Traditional Serotyping of Salmonella Isolates and Biochemical Identification
The isolation of Salmonella from the clinical chicken egg samples was also carried out according to the traditional standard method. The serotypes of Salmonella isolates from the chicken farm were determined using slide agglutination assays based on the specific O and H antisera. The results showed that the Salmonella isolates from 70 egg samples included 23 strains of S. enteritidis, one strain of S. Weltevreden, and three strains of S. London. Compared with the TaqMan real-time PCR results, three isolates (A8, C3 and F11) of S. enteritidis were not identified by the traditional method (Table 4).
3.8. Comparison of MPN-qPCR-SIT with Other Enumeration Methods
The developed MPN-qPCR-SIT method for population determination of S. enteritidis was compared to the traditional MPN, traditional plating, and TaqMan real-time PCR (Table 5). The results showed that the MPN-qPCR-SIT method and the traditional MPN presented the similar bacterial population. The average difference between them was 0.039 log MPN/mL and the greatest difference was 0.277 log MPN/mL. The MPN-qPCR-SIT also showed similar results to the traditional plating method. All bacterial populations based on the traditional plating assay fell in the 95% confidence interval of the developed MPN-qPCR-SIT. The MPN-qPCR-SIT showed a potent detection sensitivity compared to the traditional PCR and plating methods. The newly developed MPN-qPCR-SIT method could identify low levels of S. enteritidis bacterial cells that could not be detected by traditional plating and PCR.

Table 5.
Population determination of S. enteritidis (log MPN/mL or log CFU/mL) by different methods including the traditional plating, traditional MPN, TaqMan real-time PCR, and the MPN-qPCR-SIT.
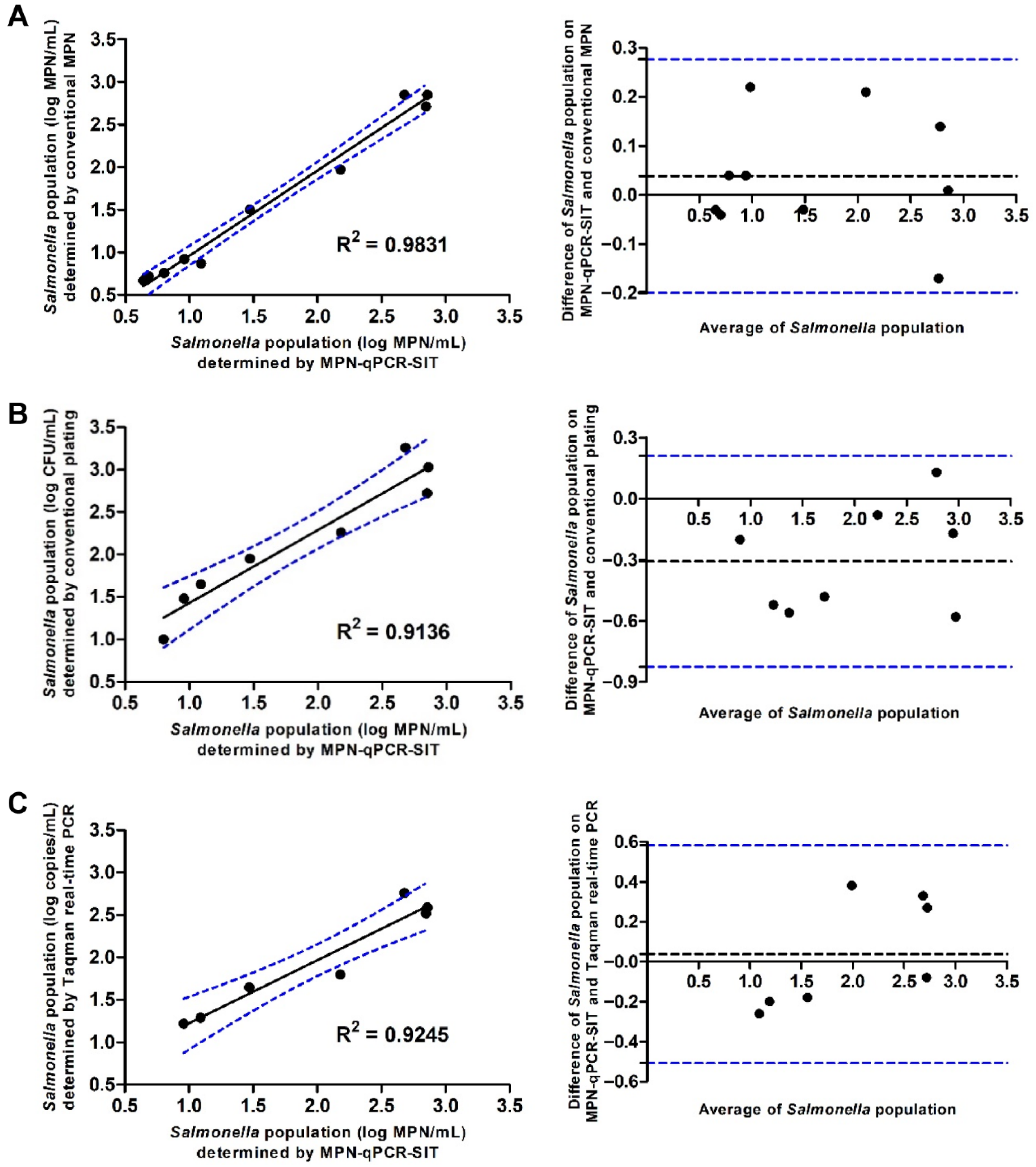
To determine the efficiency of the developed MPN-qPCR-SIT, correlation analysis was conducted on the S. enteritidis populations. The agreement between various methods was evaluated by the Bland–Altman method. The results showed that the MPN-qPCR-SIT presented a good correlation with the traditional MPN with an R2 = 0.9831. The 95% limits of agreement between the two methods ranged from −0.1988 to 0.2768 log MPN/mL, indicating that the MPN-qPCR-SIT had a similar sensitivity and effectiveness when compared to the traditional MPN method (Figure 3A). Linear correlations of the MPN-qPCR-SIT were also observed with the traditional plating assay (R2 = 0.9136, Figure 3B) and TaqMan real-time PCR (R2 = 0.9245, Figure 3C). The MPN-qPCR-SIT presented the 95% agreement boundaries of −0.8242 to 0.2092 and −0.5073 to 0.5816 with the traditional plating or TaqMan PCR, respectively.
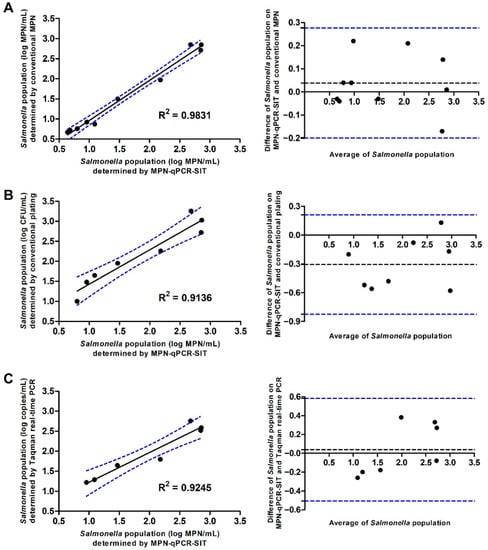
Figure 3.
Scatter plots and Bland–Altman plots to present correlative relationships to evaluate the 95% agreement boundaries by comparison of the MPN-qPCR-SIT with other methods (y-axis) including (A) traditional MPN, (B) traditional plating, and (C) TaqMan real-time PCR for the quantification of S. enteritidis in bacterial cell suspensions.
4. Discussion
Salmonella causes over one million foodborne infection cases in the US annually, and its accurate and rapid detection continues to be of important interest for both clinical diagnosis and food safety surveillance [5,40]. Compared to traditional serotyping, PCR methods are advantageous because of their simplicity and speed when many samples are to be tested for a few selected serotypes [41]. To identify S. enteritidis isolates directly from poultry eggs, a duplex TaqMan real-time PCR was developed and validated using two pairs of primers and TaqMan probes targeting the lygD and invA genes, which have been proven to be S. enteritidis-specific and Salmonella genus-specific, respectively [26,27]. SYBR-based qRT-PCR and conventional PCR assays showed that the designed primers had good specificity for the amplification of lygD and invA. The TaqMan real-time PCR results confirmed the specificity of the lygD and invA primer-probe sets. The present method directed toward S. enteritidis showed high selectivity and accuracy.
The logarithm of the Salmonella DNA copy numbers correlated excellently with the lygD and invA Ct values in the developed PCR assay. The correlation was linear in the entire range of DNA concentrations (4–4 × 105 copies), showing that this PCR method could be used to quantify the Salmonella numbers present in samples. The detection limit of the duplex real-time PCR assay was 4 copies per reaction for S. enteritidis, which was comparable to previous studies [42], indicating that the real-time PCR method developed in this study had sufficient sensitivity to be used for diagnostic purposes and as an alternative to the traditional methods. The reproducibility of the duplex TaqMan real-time PCR assay was evaluated by testing different concentrations of the positive standard samples. The results indicated satisfactory intra- and inter-reproducibility with the CVs lower than 3%, showing better reproducibility than a previous study, where the CVs were lower than 5% [43]. Additionally, the duplex TaqMan real-time PCR provided a double check for S. enteritidis identification by detecting the two genes of lygD and invA simultaneously.
The current developed duplex real-time PCR method for identifying S. enteritidis is highly accurate and could be performed directly on 6-h pre-enriched clinical samples. The TaqMan real-time PCR method is more sensitive and saves time compared with the traditional PCR method for the identification of Salmonella in clinical samples [44]; however, the 6 h of non-selective pre-enrichment is necessary for the accurate identification of S. enteritidis isolates from the clinical samples. Thus, the pre-enrichment step should be further optimized in the future study. The PCR results of chicken egg samples demonstrated 100% sensitivity, confirming the reliability and validity of this PCR method for detecting S. enteritidis. The entire identification time was approximately 7 h, compared to 5–6 days for the conventional culture and serotyping method [12]. Thus, the PCR method can be a reliable and alternative rapid method for screening and quantification of S. enteritidis and the Salmonella genus in such clinical samples. Besides, the developed method in this study is not limited by the sample size. As its high accuracy and efficiency, the duplex real-time PCR could be used as an effective tool to timely identify S. enteritidis and Salmonella genus, especially in high-throughput screening situations [45].
The developed real-time PCR identified three more S. enteritidis isolates, indicating that it is more sensitive and accurate for the detection of Salmonella from clinical egg samples compared to the traditional method. Other studies also showed that qPCR is more sensitive for the detection of Salmonella from cattle lymph nodes samples, as compared to the culture method [18]. The present real-time PCR generated more accurate data with increased detection sensitivity for the prevalence of Salmonella in chicken farms. Thus, the application of the present method in food control systems, for example, at slaughterhouses, could be helpful in generating more accurate data for future surveillance of Salmonella serotypes of veterinary and clinical relevance.
A new enumeration method of MPN-qPCR-SIT was developed based on the established TaqMan real-time PCR. The coefficient of the MPN-qPCR-SIT presented a high level of correlation compared to the traditional MPN. Traditional MPN generally needs 1–2 days for the plates or tubes incubation [29,30], while the MPN-qPCR-SIT needs only 4 h for incubating MPN plates. Thus, the MPN-qPCR-SIT method needs a much shorter time (less than 5 h totally) than the traditional MPN. These results showed that the MPN-qPCR-SIT is an alternative method that is fast and reliable for the quantification of S. enteritidis. In future studies, integrating multiplex qPCR into the MPN-qPCR-SIT would be a highly useful detection technology which could identify and quantify multiple targets simultaneously within a single reaction, such as different Salmonella serovars or even other pathogens.
5. Conclusions
In summary, we developed an accurate duplex TaqMan real-time PCR method and the new MPN-qPCR-SIT enumeration method for the identification and quantification of S. enteritidis. The method has potent selectivity, excellent sensitivity, good reproducibility and can be easily performed. This new real-time PCR method was well applicable for the identification of S. enteritidis in chicken organs and clinical chicken eggs with 100% specificity in a short time, which was superior than the traditional method. Additionally, the MPN-qPCR-SIT method presented a potent detection sensitivity and showed stronger advantages than the traditional MPN method for the population determination of S. enteritidis. This would be useful especially in the timely detection and quantification of low levels of Salmonella contamination in food production systems. The validated new duplex TaqMan real-time PCR assay could be used as a convenient tool for monitoring the Salmonella contamination in the chicken production chain and strengthen public health through effective screening of S. enteritidis in clinical samples.
Supplementary Materials
The following are available online at: https://www.mdpi.com/article/10.3390/foods11050742/s1, Figure S1: SYBR-based qRT-PCR and conventional PCR assays using ten-fold serially diluted DNA of S. enteritidis C50041.
Author Contributions
Conceptualization, Z.P. and X.J.; methodology, D.X.; software, D.X. and Y.Z.; validation, L.S. and B.L.; formal Analysis, D.X.; investigation, B.L.; resources, B.L., C.M. and X.C.; data curation, D.X. and R.P.; writing—original draft preparation, D.X.; writing—review and editing, Z.P. and X.J.; visualization, D.X.; supervision, Z.P. and X.J.; project administration, D.X., Z.P. and X.J. All authors have read and agreed to the published version of the manuscript.
Funding
This research was funded by the National Natural Science Foundation of China, grant numbers 31920103015, 31972685 and 32102669, the Natural Science Foundation of Jiangsu Province, grant number BK20210802, the China Postdoctoral Science Foundation, grant number 2020M681748, the Postdoctoral Science Foundation of Jiangsu Province, grant number 2021K095A, and the Priority Academic Program Development of Jiangsu Higher Education Institutions (PAPD).
Institutional Review Board Statement
The animal study protocol was approved by the Ethics Committee of Yangzhou University (protocol code SYXK (Su) 2017-0044) on 20 September 2021.
Informed Consent Statement
Not applicable.
Data Availability Statement
The data that support the findings of this study are available from the corresponding author upon reasonable request.
Acknowledgments
The authors would like to thank Manhong Ye of Yangzhou University for her valuable technical assistance.
Conflicts of Interest
The authors declare no conflict of interest.
References
- De Masi, L.; Yue, M.; Hu, C.; Rakov, A.V.; Rankin, S.C.; Schifferli, D.M. Cooperation of adhesin alleles in Salmonella-host tropism. mSphere 2017, 2, e00066-17. [Google Scholar] [CrossRef] [PubMed]
- Ehuwa, O.; Jaiswal, A.K.; Jaiswal, S. Salmonella, food safety and food handling practices. Foods 2021, 10, 907. [Google Scholar] [CrossRef] [PubMed]
- Gunn, J.S.; Marshall, J.M.; Baker, S.; Dongol, S.; Charles, R.C.; Ryan, E.T. Salmonella chronic carriage: Epidemiology, diagnosis, and gallbladder persistence. Trends Microbiol. 2014, 22, 648–655. [Google Scholar] [CrossRef]
- Raspoet, R.; Eeckhaut, V.; Vermeulen, K.; De Smet, L.; Wen, Y.; Nishino, K.; Haesebrouck, F.; Ducatelle, R.; Devreese, B.; Van Immerseel, F. The Salmonella Enteritidis TolC outer membrane channel is essential for egg white survival. Poult. Sci. 2019, 98, 2281–2289. [Google Scholar] [CrossRef] [PubMed]
- Chen, J.; Zhang, L.; Paoli, G.C.; Shi, C.; Tu, S.I.; Shi, X. A real-time PCR method for the detection of Salmonella enterica from food using a target sequence identified by comparative genomic analysis. Int. J. Food Microbiol. 2010, 137, 168–174. [Google Scholar] [CrossRef] [PubMed]
- Hugas, M.; Beloeil, P. Controlling Salmonella along the food chain in the European Union-progress over the last ten years. Eurosurveillance 2014, 19, 20804. [Google Scholar] [CrossRef]
- Kosa, K.M.; Cates, S.C.; Bradley, S.; Chambers, E., IV; Godwin, S. Consumer-reported handling of raw poultry products at home: Results from a national survey. J. Food Prot. 2015, 78, 180–186. [Google Scholar] [CrossRef]
- Nesbitt, A.; Ravel, A.; Murray, R.; McCormick, R.; Savelli, C.; Finley, R.; Parmley, J.; Agunos, A.; Majowicz, S.E.; Gilmour, M.; et al. Integrated surveillance and potential sources of Salmonella enteritidis in human cases in Canada from 2003 to 2009. Epidemiol. Infect. 2012, 140, 1757–1772. [Google Scholar] [CrossRef]
- Ding, J.; Lin, Q.; Zhang, J.; Young, G.M.; Jiang, C.; Zhong, Y.; Zhang, J. Rapid identification of pathogens by using surface-enhanced Raman spectroscopy and multi-scale convolutional neural network. Anal. Bioanal. Chem. 2021, 413, 3801–3811. [Google Scholar] [CrossRef]
- Kagambèga, A.; Lienemann, T.; Aulu, L.; Traoré, A.S.; Barro, N.; Siitonen, A.; Haukka, K. Prevalence and characterization of Salmonella enterica from the feces of cattle, poultry, swine and hedgehogs in Burkina Faso and their comparison to human Salmonella isolates. BMC Microbiol. 2013, 13, 253. [Google Scholar] [CrossRef]
- Grimont, P.A.D.; Weill, F.X. Antigenic Formulae of the Salmonella serovars, 9th ed.; WHO Collaborating Center for Reference and Research on Salmonella; Institut Pasteur: Paris, France, 2007. [Google Scholar]
- Soria, M.C.; Soria, M.A.; Bueno, D.J. A comparative study of culture methods and PCR assay for Salmonella detection in poultry drinking water. Poult. Sci. 2013, 92, 225–232. [Google Scholar] [CrossRef] [PubMed]
- Kasturi, K.N. A real-time PCR for rapid identification of Salmonella enterica Gaminara serovar. J. Microbiol. Methods 2020, 169, 105729. [Google Scholar] [CrossRef] [PubMed]
- Boukharouba, A.; González, A.; García-Ferrús, M.; Ferrús, M.A.; Botella, S. Simultaneous detection of four main foodborne pathogens in ready-to-eat food by using a simple and rapid multiplex PCR (mPCR) assay. Int. J. Environ. Res. Public Health 2022, 19, 1031. [Google Scholar] [CrossRef] [PubMed]
- Miao, Y.J.; Xiong, G.T.; Bai, M.Y.; Ge, Y.; Wu, Z.F. Detection of live Salmonella enterica in fresh-cut vegetables by a TaqMan-based one-step reverse transcription real-time PCR. Lett. Appl. Microbiol. 2018, 66, 447–454. [Google Scholar] [CrossRef]
- Ding, S.; Hu, H.; Yue, X.; Feng, K.; Gao, X.; Dong, Q.; Yang, M.; Tamer, U.; Huang, G.; Zhang, J. A fluorescent biosensor based on quantum dot-labeled streptavidin and poly-l-lysine for the rapid detection of Salmonella in milk. J. Dairy Sci. 2022, in press. [Google Scholar] [CrossRef]
- Feng, K.; Li, T.; Ye, C.; Gao, X.; Yue, X.; Ding, S.; Dong, Q.; Yang, M.; Huang, G.; Zhang, J. A novel electrochemical immunosensor based on Fe3O4@ graphene nanocomposite modified glassy carbon electrode for rapid detection of Salmonella in milk. J. Dairy Sci. 2022, 105, 2108–2118. [Google Scholar] [CrossRef]
- Bai, J.; Trinetta, V.; Shi, X.; Noll, L.W.; Magossi, G.; Zheng, W.; Porter, E.P.; Cernicchiaro, N.; Renter, D.G.; Nagaraja, T.G. A multiplex real-time PCR assay, based on invA and pagC genes, for the detection and quantification of Salmonella enterica from cattle lymph nodes. J. Microbiol. Methods 2018, 148, 110–116. [Google Scholar] [CrossRef] [PubMed]
- Ríos-Castillo, A.G.; Ripolles-Avila, C.; Rodríguez-Jerez, J.J. Detection of Salmonella Typhimurium and Listeria monocytogenes biofilm cells exposed to different drying and pre-enrichment times using conventional and rapid methods. Int. J. Food Microbiol. 2020, 324, 108611. [Google Scholar] [CrossRef]
- Park, S.H.; Ricke, S.C. Development of multiplex PCR assay for simultaneous detection of Salmonella genus, Salmonella subspecies I, Salm. Enteritidis, Salm. Heidelberg and Salm. Typhimurium. J. Appl. Microbiol. 2015, 118, 152–160. [Google Scholar] [CrossRef]
- Worrall, L.J.; Vuckovic, M.; Strynadka, N.C. Crystal structure of the C-terminal domain of the Salmonella type III secretion system export apparatus protein InvA. Protein Sci. 2010, 19, 1091–1096. [Google Scholar] [CrossRef] [PubMed]
- Barletta, F.; Mercado, E.H.; Lluque, A.; Ruiz, J.; Cleary, T.G.; Ochoa, T.J. Multiplex real-time PCR for detection of Campylobacter, Salmonella, and Shigella. J. Clin. Microbiol. 2013, 51, 2822–2829. [Google Scholar] [CrossRef] [PubMed]
- Garrido, A.; Chapela, M.J.; Román, B.; Fajardo, P.; Vieites, J.M.; Cabado, A.G. In-house validation of a multiplex real-time PCR method for simultaneous detection of Salmonella spp., Escherichia coli O157 and Listeria monocytogenes. Int. J. Food Microbiol. 2013, 164, 92–98. [Google Scholar] [CrossRef]
- Zhang, Q.; Chen, T.; Yang, S.; Wang, X.; Guo, H. Response surface methodology to design a selective enrichment broth for rapid detection of Salmonella spp. by SYBR Green Ι real-time PCR. Appl. Microbiol. Biotechnol. 2013, 97, 4149–4158. [Google Scholar] [CrossRef] [PubMed]
- Ramtahal, M.A.; Somboro, A.M.; Amoako, D.G.; Abia, A.L.K.; Perrett, K.; Bester, L.A.; Essack, S.Y. Molecular epidemiology of Salmonella enterica in poultry in South Africa using the farm-to-fork approach. Int. J. Microbiol. 2022, 2022, 5121273. [Google Scholar] [CrossRef] [PubMed]
- Zhu, C.; Yue, M.; Rankin, S.; Weill, F.X.; Frey, J.; Schifferli, D.M. One-step identification of five prominent chicken Salmonella serovars and biotypes. J. Clin. Microbiol. 2015, 53, 3881–3883. [Google Scholar] [CrossRef]
- Xiong, D.; Song, L.; Tao, J.; Zheng, H.; Zhou, Z.; Geng, S.; Pan, Z.; Jiao, X. An efficient multiplex PCR-based assay as a novel tool for accurate inter-serovar discrimination of Salmonella Enteritidis, S. Pullorum/Gallinarum and S. Dublin. Front. Microbiol. 2017, 8, 420. [Google Scholar] [CrossRef]
- Sutton, S. The most probable number method and its uses in enumeration, qualification, and validation. J. Valid. Technol. 2010, 16, 35–38. [Google Scholar]
- Rodgers, C.; Parveen, S.; Chigbu, P.; Jacobs, J.; Rhodes, M.; Harter-Dennis, J. Prevalence of Vibrio parahaemolyticus, and Vibrio vulnificus in blue crabs (Callinectes sapidus), seawater and sediments of the Maryland Coastal Bays. J. Appl. Microbiol. 2014, 117, 1198–1209. [Google Scholar] [CrossRef]
- Pauly, N.; Wichmann-Schauer, H.; Ballhausen, B.; Torres Reyes, N.; Fetsch, A.; Tenhagen, B.A. Detection and quantification of methicillin-resistant Staphylococcus aureus in fresh broiler meat at retail in Germany. Int. J. Food Microbiol. 2019, 292, 8–12. [Google Scholar] [CrossRef]
- Rao, M.; Tamber, S. Microbiological analysis of frozen profiteroles and mini chocolate eclairs implicated in a national salmonellosis outbreak. Food Microbiol. 2021, 100, 103871. [Google Scholar] [CrossRef]
- Jones, J.L.; Lüdeke, C.H.; Bowers, J.C.; DeRosia-Banick, K.; Carey, D.H.; Hastback, W. Abundance of Vibrio cholerae, V. vulnificus, and V. parahaemolyticus in oysters (Crassostrea virginica) and clams (Mercenaria mercenaria) from Long Island Sound. Appl. Environ. Microbiol. 2014, 80, 7667–7672. [Google Scholar] [CrossRef]
- Russo, P.; Botticella, G.; Capozzi, V.; Massa, S.; Spano, G.; Beneduce, L. A fast, reliable, and sensitive method for detection and quantification of Listeria monocytogenes and Escherichia coli O157:H7 in ready-to-eat fresh-cut products by MPN-qPCR. Biomed Res. Int. 2014, 2014, 608296. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Kim, S.A.; Park, S.H.; Lee, S.I.; Ricke, S.C. Development of a rapid method to quantify Salmonella Typhimurium using a combination of MPN with qPCR and a shortened time incubation. Food Microbiol. 2017, 65, 7–18. [Google Scholar] [CrossRef] [PubMed]
- Wigley, P.; Hulme, S.; Powers, C.; Beal, R.; Smith, A.; Barrow, P. Oral infection with the Salmonella enterica serovar Gallinarum 9R attenuated live vaccine as a model to characterize immunity to fowl typhoid in the chicken. BMC Vet. Res. 2005, 1, 2. [Google Scholar] [CrossRef]
- Cai, Y.; Tao, J.; Jiao, Y.; Fei, X.; Zhou, L.; Wang, Y.; Zheng, H.; Pan, Z.; Jiao, X. Phenotypic characteristics and genotypic correlation between Salmonella isolates from a slaughterhouse and retail markets in Yangzhou, China. Int. J. Food Microbiol. 2016, 222, 56–64. [Google Scholar] [CrossRef]
- Li, Y.; Cai, Y.; Tao, J.; Kang, X.; Jiao, Y.; Guo, R.; Wang, G.; Pan, Z.; Jiao, X. Salmonella isolated from the slaughterhouses and correlation with pork contamination in free market. Food Control 2016, 59, 591–600. [Google Scholar] [CrossRef]
- Untergasser, A.; Cutcutache, I.; Koressaar, T.; Ye, J.; Faircloth, B.C.; Remm, M.; Rozen, S.G. Primer3-new capabilities and interfaces. Nucleic Acids Res. 2012, 40, e115. [Google Scholar] [CrossRef] [PubMed]
- Zhou, Z.; Li, J.; Zheng, H.; Jin, X.; Shen, Y.; Lei, T.; Sun, X.; Pan, Z.; Jiao, X. Diversity of Salmonella isolates and their distribution in a pig slaughterhouse in Huaian, China. Food Control 2017, 78, 238–246. [Google Scholar] [CrossRef]
- Costard, S.; Pouzou, J.G.; Belk, K.E.; Morley, P.S.; Schmidt, J.W.; Wheeler, T.L.; Arthur, T.M.; Zagmutt, F.J. No change in risk for antibiotic-resistant salmonellosis from beef, United States, 2002–2010. Emerg. Infect. Dis. 2020, 26, 2108–2117. [Google Scholar] [CrossRef]
- Parichehr, M.; Mohammad, K.; Abbas, D.; Mehdi, K. Developing a multiplex real-time PCR with a new pre-enrichment to simultaneously detect four foodborne bacteria in milk. Future Microbiol. 2019, 14, 885–898. [Google Scholar] [CrossRef]
- Siala, M.; Barbana, A.; Smaoui, S.; Hachicha, S.; Marouane, C.; Kammoun, S.; Gdoura, R.; Messadi-Akrout, F. Screening and detecting Salmonella in different food matrices in Southern Tunisia using a combined enrichment/real-time PCR method: Correlation with conventional culture method. Front. Microbiol. 2017, 8, 2416. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Q.; Yang, F.; Gao, J.; Zhang, W.; Xu, X. Development of multiplex TaqMan qPCR for simultaneous detection and differentiation of eight common swine viral and bacterial pathogens. Braz. J. Microbiol. 2021, 53, 359–368. [Google Scholar] [CrossRef] [PubMed]
- Zhou, Y.Y.; Kang, X.L.; Meng, C.; Xiong, D.; Xu, Y.; Geng, S.Z.; Pan, Z.M.; Jiao, X.A. Multiple PCR assay based on the cigR gene for detection of Salmonella spp. and Salmonella Pullorum/Gallinarum identification. Poult. Sci. 2020, 99, 5991–5998. [Google Scholar] [CrossRef] [PubMed]
- Kralik, P.; Ricchi, M. A basic guide to real time PCR in microbial diagnostics: Definitions, parameters, and everything. Front. Microbiol. 2017, 8, 108. [Google Scholar] [CrossRef] [PubMed]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).