Proposing Two Local Modeling Approaches for Discriminating PGI Sunite Lamb from Other Origins Using Stable Isotopes and Machine Learning
Abstract
:1. Introduction
2. Proposed Two Local Modeling Approaches
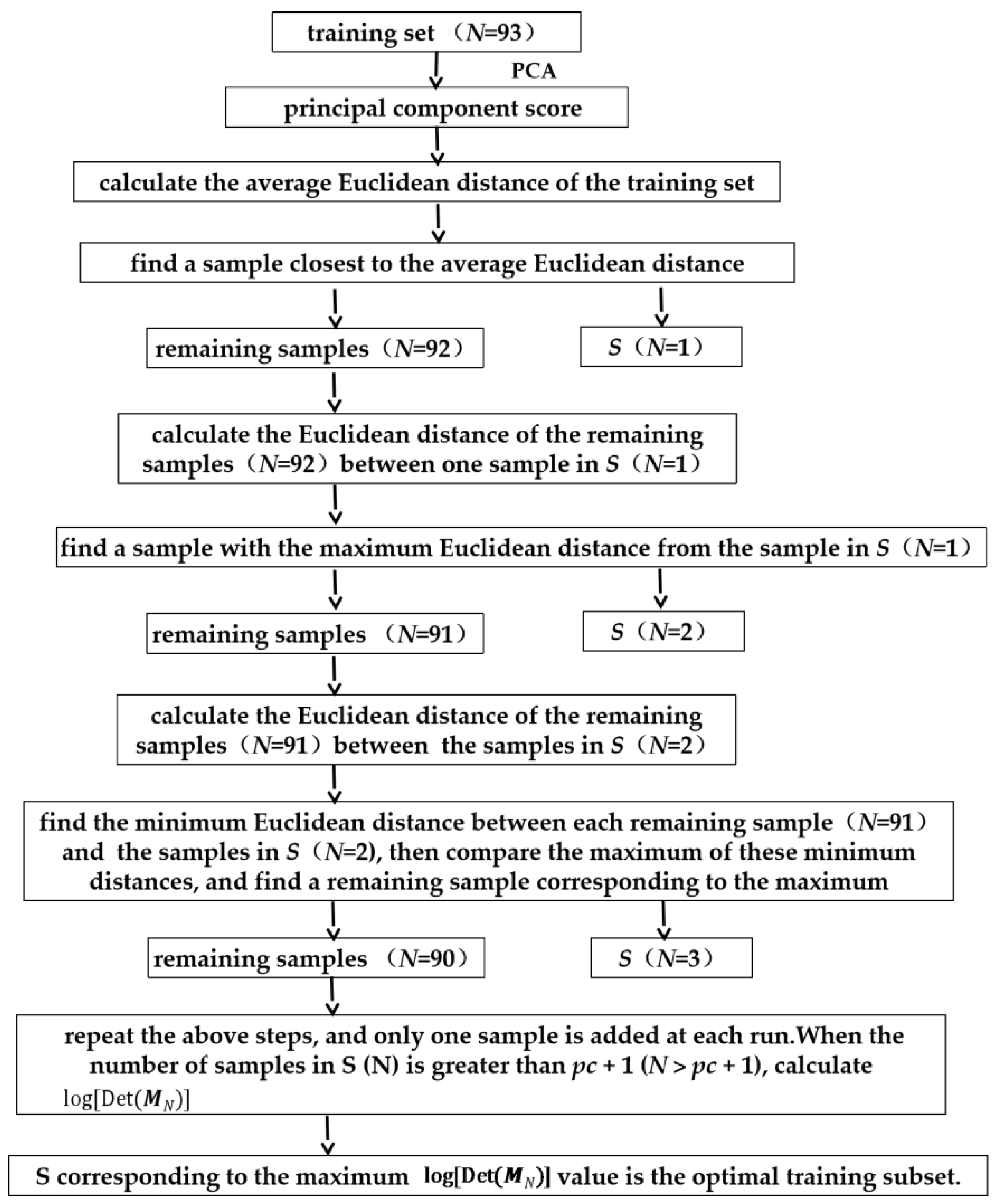
2.1. Adaptive Kennard–Stone (AKS)
2.2. An Approach of PCA–Full Distance (FD) Based on Data-Driven Soft Independent Modeling of Class Analogy (DD–SIMCA)
3. Predictive Performance of the Model
4. Materials and Methods
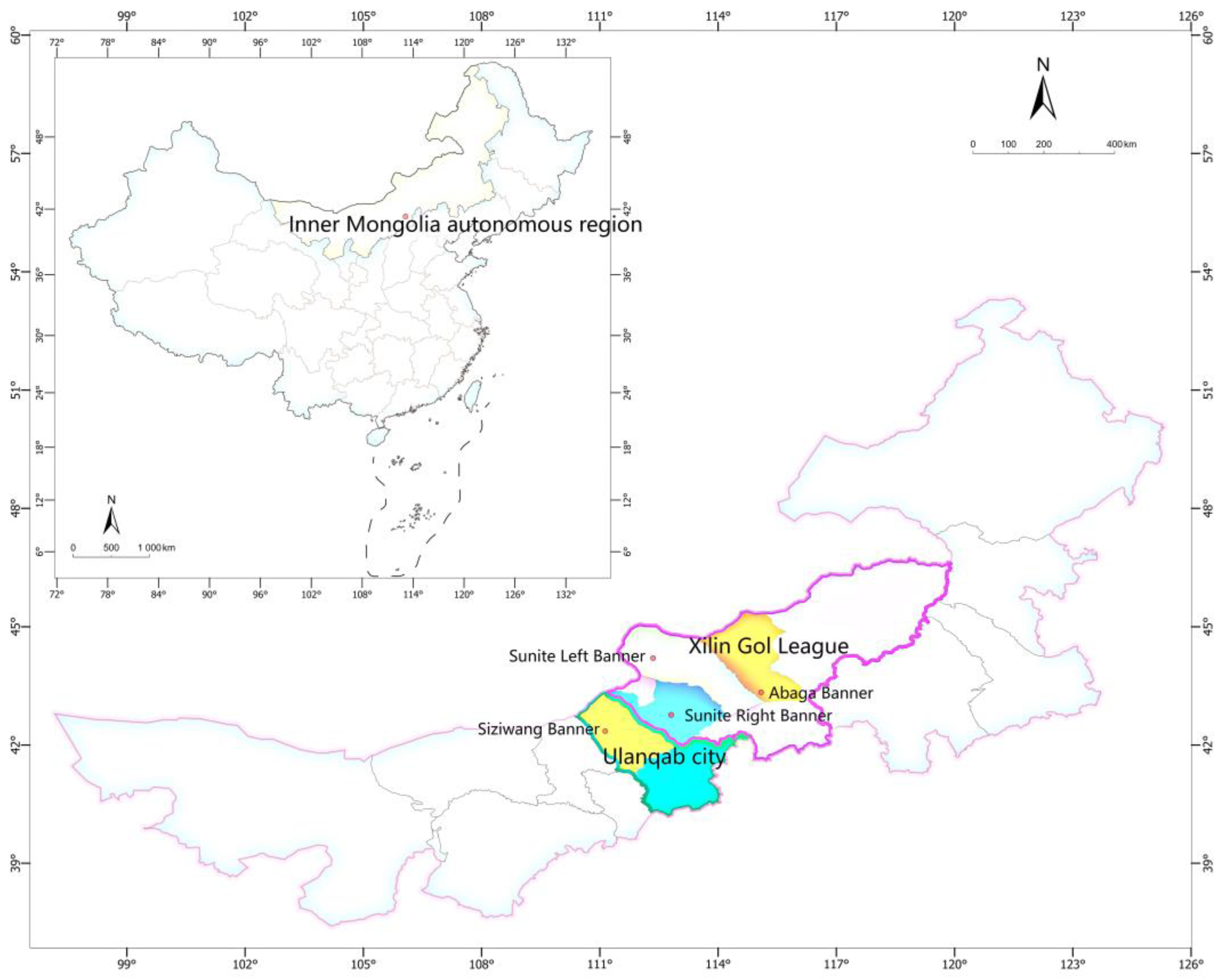
4.1. Materials
4.2. Stable Isotope Analysis
4.3. Statistical Analysis
5. Results and Discussions
5.1. Training Subset Obtained by Two Local Modeling Approaches
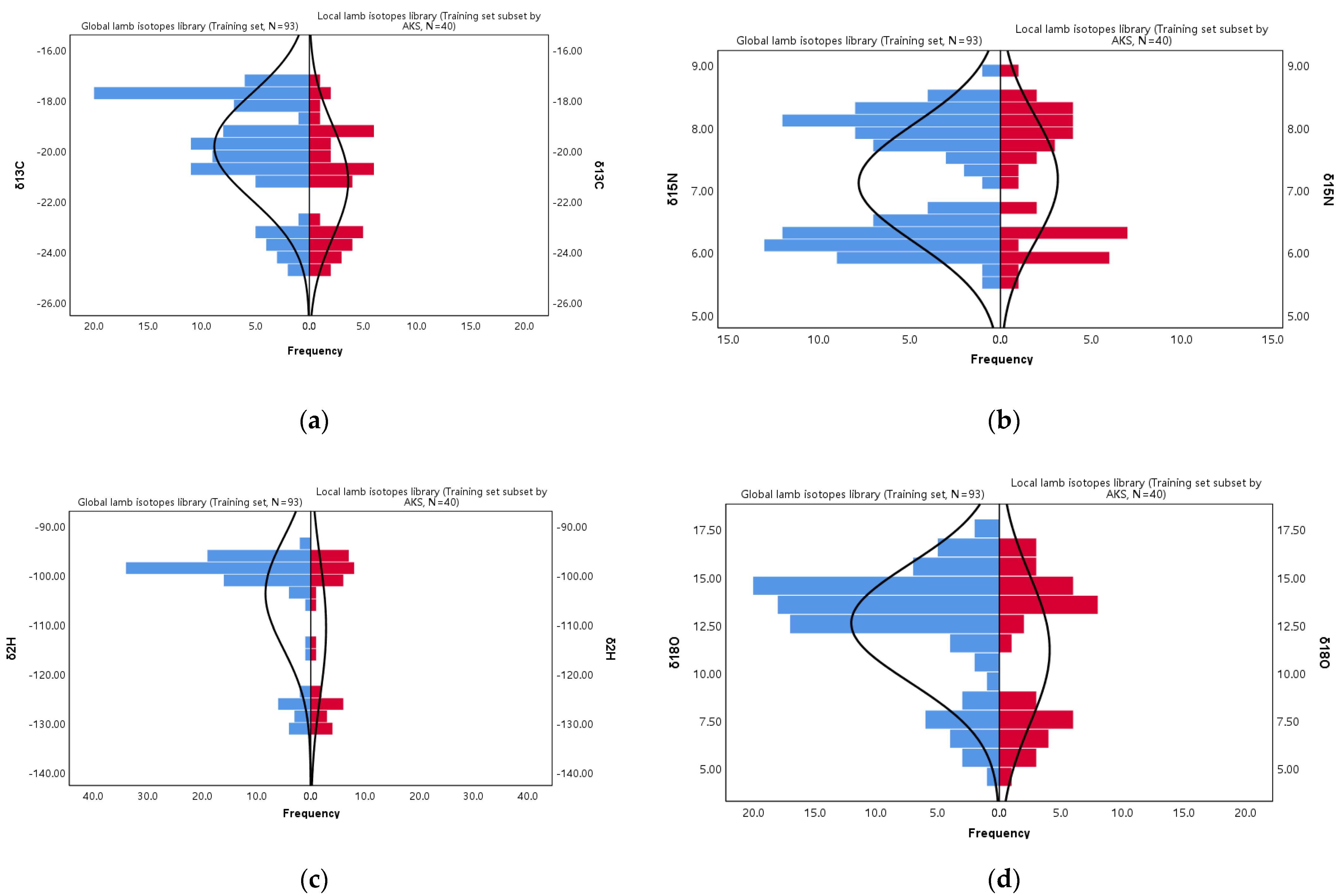
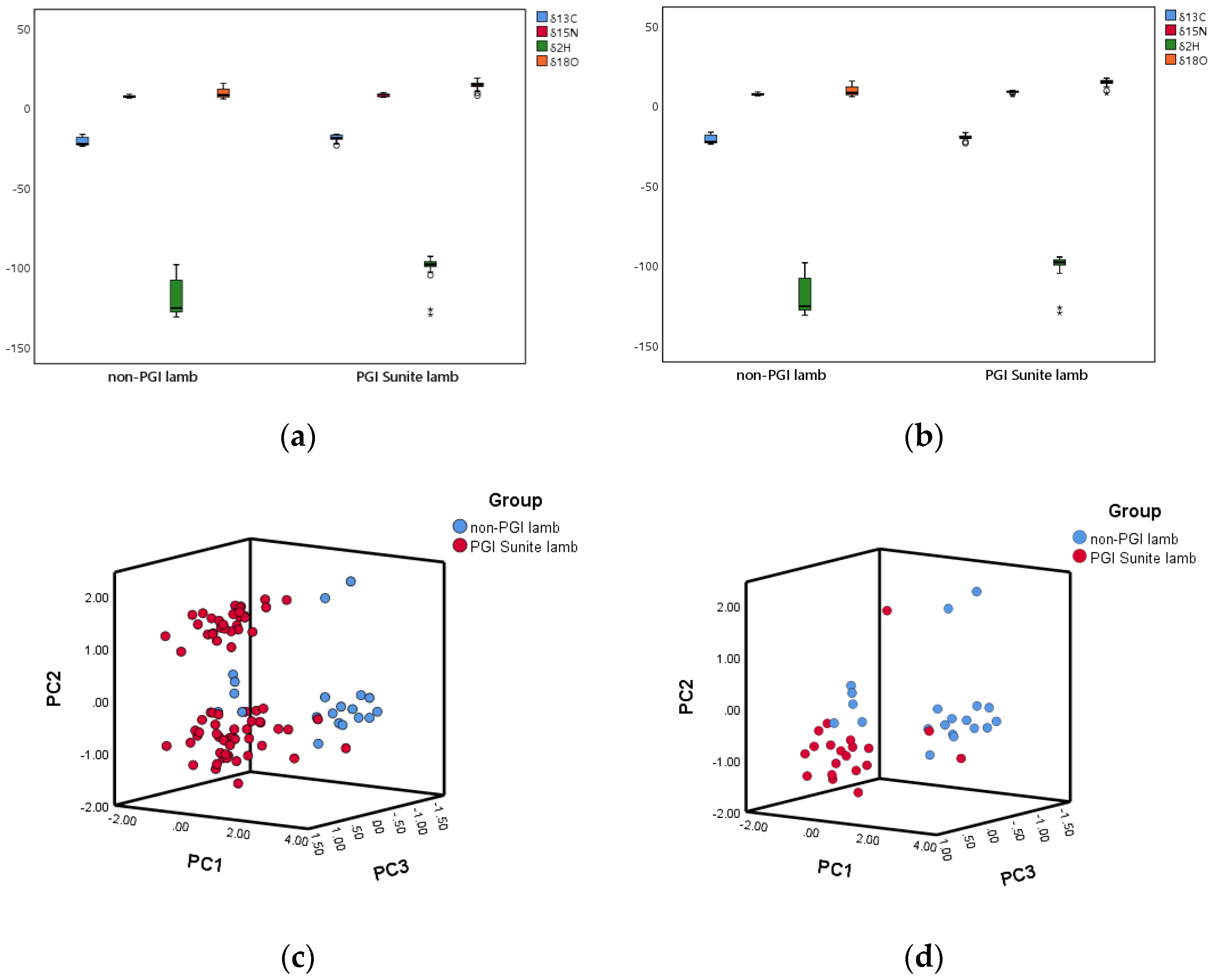
5.2. Main Characteristics of the Lamb Isotope Libraries
5.3. Predictive Performance
6. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Luo, Y.L.; Wang, B.H.; Jin, Z.M.; Liu, X.W.; Jin, Y. Effects of two feeding conditions on nutritional quality of Sunite sheep meat. Food Sci. 2016, 37, 227–231. [Google Scholar] [CrossRef]
- Su, R.; Luo, Y.L.; Wang, B.H.; Hou, Y.R.; Zhao, L.H.; Su, L.; Yao, D.; Qian, Y.; Jin, Y. Effects of physical exercise on meat quality characteristics of Sunite sheep. Small Rumin. Res. 2019, 183, 106023. [Google Scholar] [CrossRef]
- Nie, J.; Shao, S.Z.; Zhang, Y.Z.; Li, C.L.; Liu, Z.; Rogers, K.M.; Wu, M.C.; Lee, C.P.; Yuan, Y.W. Discriminating protected geographical indication Chinese Jinxiang garlic from other origins using stable isotopes and chemometrics. J. Food Compos. Anal. 2021, 99, 103856. [Google Scholar] [CrossRef]
- Zhou, G.; Feng, Y.M.; Li, Z.C.; Tao, L.Y.; Kong, W.S.; Xie, R.F.; Zhou, X. Fingerprinting and determination of hepatotoxic constituents in Polygoni Multiflori Radix Praeparata of different producing places by HPLC. J. Chromatogr. Sci. 2021, 1–10. [Google Scholar] [CrossRef] [PubMed]
- Sun, S.M.; Guo, B.L.; Wei, Y.M. Origin assignment by multi-element stable isotopes of lamb tissues. Food Chem. 2016, 213, 675–681. [Google Scholar] [CrossRef]
- Benincasa, C.; Lewis, J.; Sindona, G.; Tagarelli, A. The use of multi element profiling to differentiate between cow and buffalo milk. Food Chem. 2008, 110, 257–262. [Google Scholar] [CrossRef]
- Qie, M.J.; Zhang, B.; Li, Z.; Zhao, S.S.; Zhao, Y. Data fusion by ratio modulation of stable isotope, multi-element, and fatty acids to improve geographical traceability of lamb. Food Control. 2021, 120, 107549. [Google Scholar] [CrossRef]
- Zhao, R.T.; Su, M.C.; Zhao, Y.; Chen, G.; Chen, A.L.; Yang, S.M. Chemical analysis combined with multivariate statistical methods to determine the geographical origin of milk from four regions in China. Foods 2021, 10, 1119. [Google Scholar] [CrossRef]
- Xie, L.N.; Zhao, S.S.; Rogers, K.M.; Xia, Y.N.; Zhang, B.; Suo, R.; Zhao, Y. A case of milk traceability in small-scale districts-Inner Mongolia of China by nutritional and geographical parameters. Food Chem. 2020, 316, 126332. [Google Scholar] [CrossRef]
- Wang, J.S.; Xu, L.; Xu, Z.Z.; Wang, Y.; Niu, C.; Yang, S.M. Liquid chromatography quadrupole time-of-flight mass spectrometry and rapid evaporative ionization mass spectrometry were used to develop a lamb authentication method: A preliminary study. Foods 2020, 9, 1723. [Google Scholar] [CrossRef]
- Zhao, R.T.; Yang, S.M.; Zhao, Y. Research progress in traceability of agricultural products using stable isotope. J. Nucl. Agric. Sci. 2020, 34 (Suppl. S1), 120–128. [Google Scholar] [CrossRef]
- Durante, C.; Lancellotti, L.; Manzini, D.; Rossi, M.C.; Sighinolfi, S.; Marchetti, A.; Tassi, L. 87Sr/86Sr ratio as traceability marker for Modena’s balsamic vinegars. LWT 2021, 147, 11571. [Google Scholar] [CrossRef]
- Zhao, Y.; LV, J.; Yang, S.M. Research progress and application of stable isotope technology in agricultural products tracing field. Qual. Saf. Agro-Products. 2015, 6, 35–40. [Google Scholar]
- Nie, J.; Shao, S.Z.; Xia, W.; Liu, Z.; Yu, C.C.; Li, R.; Wang, W.; Li, J.R.; Yuan, Y.W.; Rogers, K.M. Stable isotopes verify geographical origin of yak meat from Qinghai-Tibet plateau. Meat Sci. 2020, 165, 108113. [Google Scholar] [CrossRef] [PubMed]
- Erasmus, S.W.; Muller, M.; Butler, M.; Hoffman, L.C. The truth is in the isotopes: Authenticating regionally unique South African lamb. Food Chem. 2018, 239, 926–934. [Google Scholar] [CrossRef] [PubMed]
- Perini, M.; Camin, F.; Bontempo, L.; Rossmann, A.; Piasentier, E. Multielement (H, C, N, O, S) stable isotope characteristics of lamb meat from different Italian regions. Rapid Commun. Mass Spectrom. 2009, 23, 2573–2585. [Google Scholar] [CrossRef] [PubMed]
- Chung, I.M.; Kim, J.K.; Yang, Y.J.; An, Y.J.; Kim, S.Y.; Kwon, C.; Kim, S.H. A case study for geographical indication of organic milk in Korea using stable isotope ratios-based chemometric analysis. Food Control. 2020, 107, 106755. [Google Scholar] [CrossRef]
- Kang, X.M.; Zhao, Y.F.; Shang, D.R.; Zhai, Y.X.; Ning, J.S.; Ding, H.Y.; Sheng, X.F. Identification of the geographical origins of sea cucumbers in China: The application of stable isotope ratios and compositions of C., N., O and H. Food Control. 2020, 111, 107036. [Google Scholar] [CrossRef]
- Camin, F.; Bontempo, L.; Heinrich, K.; Horacek, M.; Kelly, S.D.; Schlicht, C.; Rossmann, A. Multi-element (H, C, N, S) stable isotope characteristics of lamb meat from different European regions. Anal. Bioanal Chem. 2007, 389, 309–320. [Google Scholar] [CrossRef] [Green Version]
- Kasabov, N. Global, local and personalised modeling and pattern discovery in bioinformatics: An integrated approach. Pattern Recog Lett. 2007, 28, 673–685. [Google Scholar] [CrossRef]
- Guo, J.; Ni, L.; Zhang, L. The first Chinese Conference on Near Infrared Spectroscopy. Sampling Method for NIR Calibration Sample Based on Uniform-Design. In Proceedings of the Near Infrared Spectroscopy Technology in Modern China, Beijing, China, 18–21 October 2006. [Google Scholar]
- Centner, V.; de Noord, O.E.; Massart, D.L. Detection of nonlinearity in multivariate calibration. Anal. Chim. Acta 1998, 376, 153–168. [Google Scholar] [CrossRef]
- Chen, D.; Shao, X.G. The first Chinese Conference on Near Infrared Spectroscopy. Study On novel Strategies for Selecting Representative Samples of Near-Infrared Spectroscopy. In Proceedings of the Near Infrared Spectroscopy Technology in Modern China, Beijing, China, 18–21 October 2006. [Google Scholar]
- Leonardo, R.; Thorsten, B.; Karsten, S.; Antoine, S.; Jose, A.M.; Thomas, S. The spectrum-based learner: A new local approach for modeling soil vis-NIR spectra of complex datasets. Geoderma 2013, 195, 268–279. [Google Scholar] [CrossRef]
- Abhinav, G.; Hitesh, B.V.; Bhabani, S.D.; Aditya, K.C. Local modeling approaches for estimating soil properties in selected Indian soils using diffuse reflectance data over visible to near-infrared region. Geoderma 2018, 325, 59–71. [Google Scholar] [CrossRef]
- Palou, A.; Miró, A.; Blanco, M.; Larraz, R.; Gómez, J.F.; Martínez, T.; González, J.M.; Alcalà, M. Calibration sets selection strategy for the construction of robust PLS models for prediction of biodiesel/diesel blends physico-chemical properties using NIR spectroscopy. Spectrochim. Acta A 2017, 180, 119–126. [Google Scholar] [CrossRef] [PubMed]
- Coppa, M.; Martin, B.; Hulin, S.; Guillemin, J.; Gauzentes, J.V.; Pecou, A.; Andueza, D. Prediction of indicators of cow diet composition and authentication of feeding specifications of Protected Designation of Origin cheese using mid-infrared spectroscopy on milk. J. Dairy Sci. 2021, 104, 112–125. [Google Scholar] [CrossRef] [PubMed]
- Rius, A.; Callao, M.P.; Ferré, J.; Rius, F.X. Assessing the validity of principal component regression models in different analytical conditions. Anal. Chim. Acta 1997, 337, 287–296. [Google Scholar] [CrossRef]
- De Maesschalck, R.; Jouan-Rimbaud, D.; Massart, D.L. The Mahalanobis distance. Chemom. Intell. Lab. Syst. 2000, 50, 1–18. [Google Scholar] [CrossRef]
- Pomerantsev, A.L. Acceptance areas for multivariate classification derived by projection methods. J. Chemometr. 2008, 22, 601–609. [Google Scholar] [CrossRef]
- Pomerantsev, A.L.; Rodionova, O. Concept and role of extreme objects in PCA/SIMCA. J. Chemometr. 2013, 28, 429–438. [Google Scholar] [CrossRef]
- Foody, G.M. Explaining the unsuitability of the kappa coefficient in the assessment and comparison of the accuracy of thematic maps obtained by image classification. Remote Sens. Environ. 2020, 239, 111630. [Google Scholar] [CrossRef]
- Zhao, S.S.; Zhang, H.B.; Zhang, B.; Xu, Z.Z.; Chen, A.L.; Zhao, Y. A rapid sample preparation method for the analysis of stable isotope ratios of beef samples from different countries. Rapid Commun. Mass Spectrom. 2020, 34, 8795. [Google Scholar] [CrossRef] [PubMed]
- Nietner, T.; Haughey, S.A.; Ogle, N.; Fauhl-Hassek, C.; Elliott, C.T. Determination of geographical origin of distillers dried grains and solubles using isotope ratio mass spectrometry. Food Res. Inter. 2014, 60, 146–153. [Google Scholar] [CrossRef]
- Zhao, Y.; Zhang, B.; Chen, G.; Chen, A.L.; Yang, S.M.; Ye, Z.H. Recent developments in application of stable isotope analysis on agro-product authenticity and traceability. Food Chem. 2014, 145, 300–305. [Google Scholar] [CrossRef] [PubMed]
- Yuan, H.C.; Li, C.Y.; Jian, Y.; Geng, M.M.; Xu, L.W.; Wang, J.R. Stable isotope technique in soil carbon cycling research of agricultural ecosystems. J. Isot. 2014, 27, 170–178. [Google Scholar] [CrossRef]
- Zhao, Y.; Zhang, B.; Chen, G.; Chen, A.L.; Yang, S.M.; Ye, Z.H. Tracing the geographic origin of beef in China on the basis of the combination of stable isotopes and multielement analysis. J. Agric. Food Chem. 2013, 61, 7055–7060. [Google Scholar] [CrossRef] [PubMed]
- Zhao, H.Y.; Wang, F.; Yang, Q.L. Origin traceability of peanut kernels based on multi-element fingerprinting combined with multivariate data analysis. J. Sci. Food. Agric. 2020, 100, 4040–4048. [Google Scholar] [CrossRef] [PubMed]
| Confusion Matrix | Predicted Class | ||
|---|---|---|---|
| Class 1 | Class 2 | ||
| Actual class | Class 1 | True positive (TP) | False negative (FN) |
| Class 2 | False positive (FP) | True negative (TN) | |
| (a) | ||||
| Parameter | Mean | Standard Deviation | Minimum | Maximum |
| Global lamb isotope library (Training set, n = 93) | ||||
| δ13C | −19.87 | 2.10 | −24.69 | −17.16 |
| δ15N | 7.10 | 0.95 | 5.58 | 8.88 |
| δ2H | −103.90 | 11.14 | −131.87 | −93.77 |
| δ18O | 12.55 | 3.08 | 4.92 | 17.98 |
| Local lamb isotope library (Training subset by AKS, n = 40) | ||||
| δ13C | −21.24 | 2.21 | −24.69 | −17.26 |
| δ15N | 7.16 | 1.01 | 5.58 | 8.88 |
| δ2H | −110.85 | 14.11 | −131.87 | −95.44 |
| δ18O | 11.15 | 3.90 | 4.92 | 16.56 |
| (b) | ||||
| Parameter | δ13C | δ15N | δ2H | δ18O |
| Global lamb isotope library (Training set, n = 93) | ||||
| PGI Sunite lamb | −19.31 ± 1.52 a | 7.28 ± 0.94 a | −99.48 ± 5.44 a | 13.65 ± 1.90 a |
| non-PGI lamb | −21.92 ± 2.65 b | 6.45 ± 0.70 b | −120.02 ± 11.84 b | 8.53 ± 3.28 b |
| Local lamb isotope library (Training subset by AKS, n = 40) | ||||
| PGI Sunite lamb | −20.57 ± 1.44 a | 7.88 ± 0.73 a | −101.68 ± 9.58 a | 13.78 ± 2.43 a |
| non-PGI lamb | −21.92 ± 2.65 b | 6.45 ± 0.70 b | −120.02 ± 11.84 b | 8.53 ± 3.28 b |
| (a) | Binary Discrimination Classes | ||||
| LDA | RF | SVM | BPNN | KNN | |
| Confusion matrix (No. of testing set samples) | |||||
| True positive | 17 | 17 | 17 | 18 | 17 |
| (tpi) | |||||
| False negative | 1 | 1 | 1 | 0 | 1 |
| (fni) | |||||
| True negative | 4 | 4 | 4 | 5 | 4 |
| (tni) | |||||
| False positive | 1 | 1 | 1 | 0 | 1 |
| (fpi) | |||||
| Performance evaluation | |||||
| Sensitivity | 0.9444 | 0.9444 | 0.9444 | 1.0000 | 0.9444 |
| Specificity | 0.8000 | 0.8000 | 0.8000 | 1.0000 | 0.8000 |
| Kappa | 0.7444 | 0.7444 | 0.7444 | 1.0000 | 0.7444 |
| Accuracy | 0.9130 | 0.9130 | 0.9130 | 1.0000 | 0.9130 |
| (b) | Binary Discrimination Classes | ||||
| LDA | RF | SVM | BPNN | KNN | |
| Confusion matrix (No. of testing set samples) | |||||
| True positive | 17 | 17 | 17 | 18 | 17 |
| (tpi) | |||||
| False negative | 1 | 1 | 1 | 0 | 1 |
| (fni) | |||||
| True negative | 4 | 5 | 4 | 5 | 4 |
| (tni) | |||||
| False positive | 1 | 0 | 1 | 0 | 1 |
| (fpi) | |||||
| Performance evaluation | |||||
| Sensitivity | 0.9444 | 0.9444 | 0.9444 | 1.0000 | 0.9444 |
| Specificity | 0.8000 | 1.0000 | 0.8000 | 1.0000 | 0.8000 |
| Kappa | 0.7444 | 0.8808 | 0.7444 | 1.0000 | 0.7444 |
| Accuracy | 0.9130 | 0.9565 | 0.9130 | 1.0000 | 0.9130 |
| (c) | Binary Discrimination Classes | ||||
| LDA | RF | SVM | BPNN | KNN | |
| Confusion matrix (No. of testing set samples) | |||||
| True positive | 17 | 17 | 17 | 18 | 17 |
| (tpi) | |||||
| False negative | 1 | 1 | 1 | 0 | 1 |
| (fni) | |||||
| True negative | 4 | 4 | 4 | 5 | 5 |
| (tni) | |||||
| False positive | 1 | 1 | 1 | 0 | 0 |
| (fpi) | |||||
| Performance evaluation | |||||
| Sensitivity | 0.9444 | 0.9444 | 0.9444 | 1.0000 | 0.9444 |
| Specificity | 0.8000 | 0.8000 | 0.8000 | 1.0000 | 1.0000 |
| Kappa | 0.7444 | 0.7444 | 0.7444 | 1.0000 | 0.8808 |
| Accuracy | 0.9130 | 0.9130 | 0.9130 | 1.0000 | 0.9565 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhao, R.; Liu, X.; Wang, J.; Wang, Y.; Chen, A.-L.; Zhao, Y.; Yang, S. Proposing Two Local Modeling Approaches for Discriminating PGI Sunite Lamb from Other Origins Using Stable Isotopes and Machine Learning. Foods 2022, 11, 846. https://doi.org/10.3390/foods11060846
Zhao R, Liu X, Wang J, Wang Y, Chen A-L, Zhao Y, Yang S. Proposing Two Local Modeling Approaches for Discriminating PGI Sunite Lamb from Other Origins Using Stable Isotopes and Machine Learning. Foods. 2022; 11(6):846. https://doi.org/10.3390/foods11060846
Chicago/Turabian StyleZhao, Ruting, Xiaoxia Liu, Jishi Wang, Yanyun Wang, Ai-Liang Chen, Yan Zhao, and Shuming Yang. 2022. "Proposing Two Local Modeling Approaches for Discriminating PGI Sunite Lamb from Other Origins Using Stable Isotopes and Machine Learning" Foods 11, no. 6: 846. https://doi.org/10.3390/foods11060846
APA StyleZhao, R., Liu, X., Wang, J., Wang, Y., Chen, A.-L., Zhao, Y., & Yang, S. (2022). Proposing Two Local Modeling Approaches for Discriminating PGI Sunite Lamb from Other Origins Using Stable Isotopes and Machine Learning. Foods, 11(6), 846. https://doi.org/10.3390/foods11060846







