A Multistep DNA-Based Methodology for Accurate Authentication of Sturgeon Species
Abstract
:1. Introduction
2. Materials and Methods
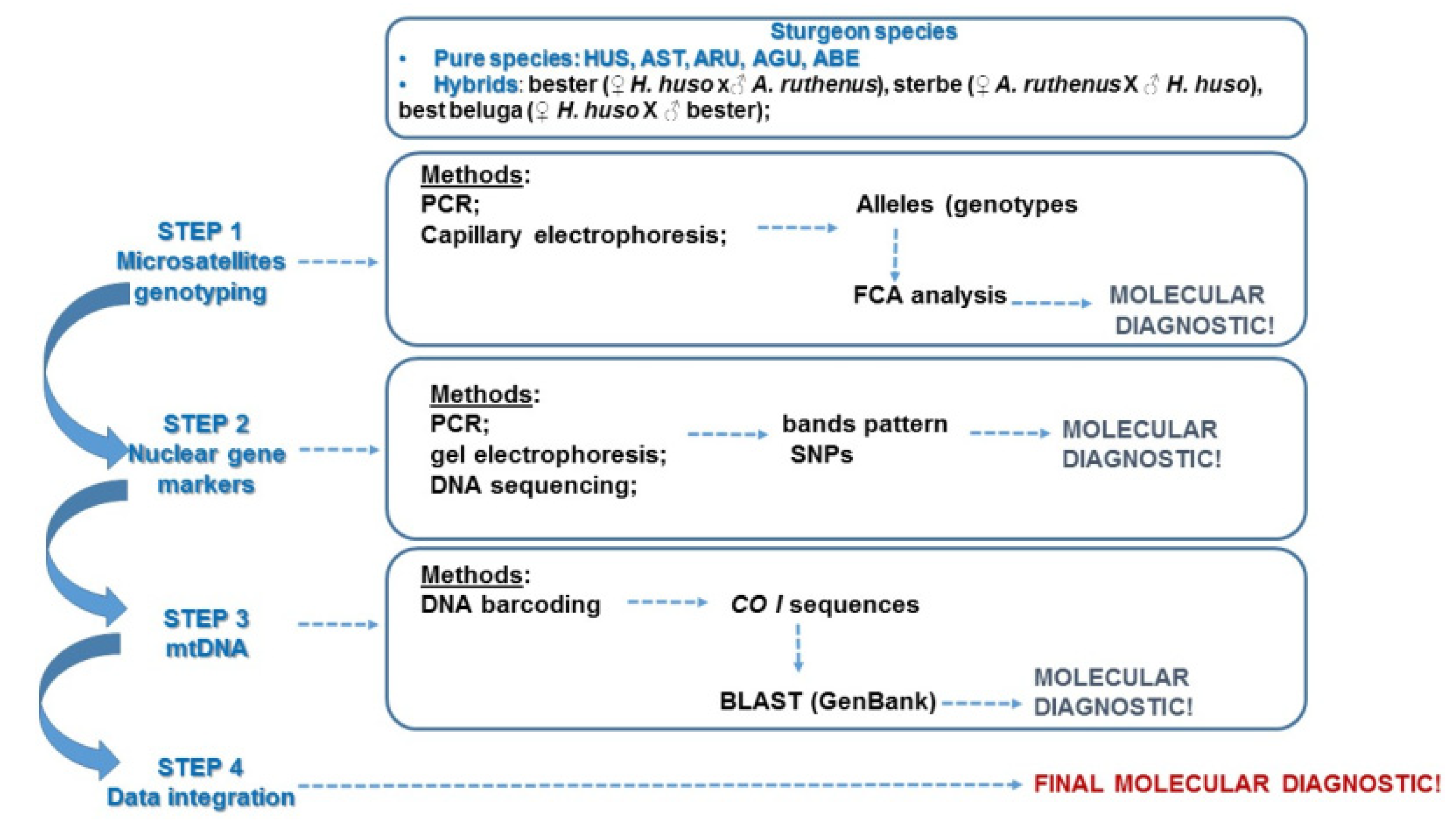
2.1. Schematic Overview of the Experimental Program
2.2. Sampling and DNA Extraction
2.3. Microsatellite Genotyping and Data Analysis
2.4. Nuclear Gene Marker Analysis
2.5. DNA Barcoding
3. Results
3.1. Step 1: Microsatellite Genotyping
- (i)
- On genotype data set of pure species.
- (ii)
- On a data set comprising the genotypes of hybrids bester, sterbe and best beluga together with their genitor species, A. ruthenus and H. huso.
- (iii)
- On the data set that consists in genotypes of pure species and two putative hybrids captured in the Danube River.
3.2. Step 2: Nuclear Gene Analysis
- (i)
- RP1.
- (ii)
- Vimentin (vim).
- (iii)
- Rhodopsin (Rh).
3.3. Step 3: mtDNA Analysis
3.4. Step 4: Data Integration
4. Discussion
- (i)
- A. stellatus—microsatellites supplemented with RP1 and mtDNA;
- (ii)
- H. huso—microsatellites supplemented with S7RPEx1, RP1 and mtDNA;
- (iii)
- A. ruthenus—microsatellites supplemented with vimentin and mtDNA;
- (iv)
- A. gueldenstaedtii—vimentin + mtDNA;
- (v)
- A. baerii –vimentin + mtDNA;
- (vi)
- bester (H. huso × A. ruthenus)—microsatellites+ vimentin + mtDNA;
- (vii)
- sterbe (A. ruthenus × H. huso)—microsatellites + mtDNA;
- (viii)
- best beluga (H. huso × bester)—microsatellites + mtDNA;
- (ix)
- H203—microsatellites+ RP1 + vimentin + mtDNA;
- (x)
- H204—microsatellites + RP1+ vimentin+ mtDNA.
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Bemis, W.E.; Findeis, E.K.; Grande, L. An overview of Acipenseriformes. Environ. Biol. Fish. 1997, 48, 25–71. [Google Scholar] [CrossRef]
- IUCN Red List of Threatened Species. Available online: https://www.iucnredlist.org/search?taxonomies=100315&searchType=species (accessed on 15 December 2021).
- Sturgeon. CITES. Available online: https://cites.org/eng/prog/sturgeon.php#:~:text=the%20CITES%20Animals%20Committee%20was,areas%2C%20primarily%20through%20illegal%20fishing (accessed on 15 December 2021).
- Billard, R.; Lecointre, G. Biology and conservation of sturgeon and paddlefish. Rev. Fish Biol. Fish. 2001, 10, 355–392. [Google Scholar] [CrossRef]
- Chandra, G.; Fopp-Bayat, D. Trends in aquaculture and conservation of sturgeons: A review of molecular and cytogenetic tools. Rev. Aquac. 2021, 13, 119–137. [Google Scholar] [CrossRef]
- Smederevac-Lalić, M.; Jarić, I.; Višnjić-Jeftić, Ž.; Skorić, S.; Cvijanović, G.; Gačić, Z.; Lenhardt, M. Management approaches and aquaculture of sturgeons in the lower Danube region countries. J. Appl. Ichthyol. 2011, 27, 94–100. [Google Scholar] [CrossRef]
- Holostenco, D.; Ciorpac, M.; Paraschiv, M.; Iani, M.; Hont, S.; Taflan, E.; Suciu, R.; Rasnoveanu, G. Overview of the Romanian Sturgeon Supportive Stocking Programme in the Lower Danube River System. Sci. Ann. Danub. Delta Inst. 2019, 24, 21–30. [Google Scholar]
- Arnold, M.L. Natural Hybridization and Evolution; Oxford University Press: New York, NY, USA, 1997; pp. 113–181. [Google Scholar]
- Ludwig, A.; Belfiore, N.M.; Pitra, C.; Svirsky, V.; Jenneckens, I. Genome duplication events and functional reduction of ploidy levels in sturgeon (Acipenser, Huso and Scaphirhynchus). Genetics 2001, 158, 1203–1215. [Google Scholar] [CrossRef] [PubMed]
- Fontana, F.; Congiu, L.; Mudrak, V.A.; Quattro, J.M.; Smith, T.I.; Ware, K.; Doroshov, S.I. Evidence of hexaploid karyotype in shortnose sturgeon. Genome 2008, 51, 113–119. [Google Scholar] [CrossRef]
- Boscari, E.; Barbisan, F.; Congiu, L. Inheritance pattern of microsatellite loci in the polyploid Adriatic sturgeon (Acipenser naccarii). Aquaculture 2011, 321, 223–229. [Google Scholar] [CrossRef]
- Vasil’ev, V.P. Mechanisms of Polyploid Evolution in Fish: Polyploidy in Sturgeons. In Biology, Conservation and Sustainable Development of Sturgeons, 1st ed.; Fish and Fisheries, Series; Carmona, R., Domezain, A., Gallego, M.G., Hernando, J.A., Rodríguez, F., Ruiz-Rejón, M., Eds.; Springer: Amsterdam, The Netherlands, 2009; pp. 97–117. [Google Scholar]
- Ludwig, A.; Lippold, S.; Debus, L.; Reinartz, R. First evidence of hybridization between endangered sterlets (Acipenser ruthenus) and exotic Siberian sturgeons (Acipenser baerii) in the Danube River. Biol. Invasions 2009, 11, 753–760. [Google Scholar] [CrossRef] [Green Version]
- Doukakis, P.; Pikitch, E.K.; Rothschild, A.; DeSalle, R.; Amato, G.; Kolokotronis, S.O. Testing the effectiveness of an international conservation agreement: Marketplace forensic and CITES caviar trade regulation. PLoS ONE 2012, 7, e40907. [Google Scholar] [CrossRef] [Green Version]
- Burtsev, L.A. Bester in aquaculture. In Sturgeon Stocks and Caviar Trade Workshop; Birstein, A., Kaiser, A., Eds.; Pohlman, IUCN: Gland, Switzerland, 1997; pp. 35–432. [Google Scholar]
- Boscari, E.; Barmintseva, A.; Pujolar, M.J.; Doukakis, P.; Mugue, N.; Congiu, L. Species and hybrid identification of sturgeon caviar: A new molecular approach to detect illegal trade. Mol. Ecol. Res. 2014, 14, 489–498. [Google Scholar] [CrossRef] [PubMed]
- Ludwig, A.; Lieckfeldt, D.; Jahrl, J. Mislabeled and counterfeit sturgeon caviar from Bulgaria and Romania. J. Appl. Ichthyol. 2015, 31, 587–591. [Google Scholar] [CrossRef]
- Zhang, X.; Tinacci, L.; Xie, S.; Wang, J.; Ying, X.; Wen, J.; Armani, A. Caviar products sold on Chinese Business to customer (B2C) online platforms: Labelling assessment supported by molecular identification. Food Control 2022, 131, 108370. [Google Scholar] [CrossRef]
- Biodiversity Science. Available online: http://dna-barcoding.blogspot.ro/2015/07/fake-caviar.html (accessed on 22 January 2022).
- Bronzi, P.; Rosenthal, H. Present and future sturgeon and caviar production and marketing: A global market overview. J. Appl. Ichthyol. 2014, 30, 1536–1546. [Google Scholar] [CrossRef]
- Ludwig, A. Identification of Acipenseriformes species in trade. J. Appl. Ichthyol. 2008, 24 (Suppl. 1), 2–19. [Google Scholar] [CrossRef]
- Scarano, D.; Rao, R. DNA markers for food products authentication. Diversity 2014, 6, 579–596. [Google Scholar] [CrossRef]
- Ludwig, A.; Boner, M.; Boscari, E.; Congiu, L.; Gessner, J.; Jahrl, J.; Striebel, B. Identification of species and hybrids, source and geographical origin of sturgeon and paddlefish (Acipenseriformes spp.) specimens and products in trade. In Proceedings of the Addendum to Identification and Traceability of Sturgeons and Paddlefish (Acipenseriformes spp.), CITES—Thirty-First Meeting of the Animals Committee, Online, 31 May–22 June 2021. [Google Scholar]
- Pappalardo, A.M.; Petraccioli, A.; Capriglione, T.; Ferrito, V. From fish eggs to fish name: Caviar species discrimination by Coibar-RFLP, an efficient molecular approach to detect fraud in the caviar trade. Molecules 2019, 24, 2468. [Google Scholar] [CrossRef] [Green Version]
- NCBI. Available online: https://www.ncbi.nlm.nih.gov/nuccore/?term=mitochondrion+genome+Acipenseridae (accessed on 10 January 2022).
- Birstein, V.; Desalle, R.; Doukakis, P.; Hanner, R.; Ruban, G.; Wong, E. Testing taxonomic boundaries and the limit of DNA barcoding in the Siberian sturgeon, Acipenser baerii. Mitochondrial DNA 2009, 20, 110–118. [Google Scholar] [CrossRef]
- Popa, G.O.; Dudu, A.; Banaduc, D.; Curtean-Banaduc, A.; Barbalata, T.; Burcea, A.; Florescu, I.E.; Georgescu, S.E.; Costache, M. Use of DNA barcoding in the assignment of commercially valuable fish species from Romania. Aquat. Living Resour. 2017, 30, 20. [Google Scholar] [CrossRef]
- Havelka, M.; Fujimoto, T.; Hagihara, S.; Adachi, S.; Arai, K. Nuclear DNA markers for identification of Beluga and Sterlet sturgeons and their interspecific Bester hybrid. Sci. Rep. 2017, 7, 1694. [Google Scholar] [CrossRef] [Green Version]
- Boscari, E.; Barmintseva, A.E.; Zhang, S.; Yue, H.; Li, C.; Shedko, S.; Lieckfeldt, D.; Ludwig, A.; Wei, Q.W.; Mugue, N.S.; et al. Genetic identification of the caviar-producing Amur and Kaluga sturgeons revealed a high level of concealed hybridization. Food Control 2017, 82, 243–250. [Google Scholar] [CrossRef]
- Boscari, E.; Vitulo, N.; Ludwig, A.; Caruso, C.; Mugue, N.S.; Suciu, R.; Congiu, L. Fast genetic identification of the Beluga sturgeon and its sought-after caviar to stem illegal trade. Food Control 2017, 75, 145–152. [Google Scholar] [CrossRef]
- Taggart, J.B.; Hynes, R.A.; Prodöuhl, P.A.; Ferguson, A. A simplified protocol for routine total DNA isolation from salmonid fishes. Fish Biol. 1992, 40, 963–965. [Google Scholar] [CrossRef]
- May, B.; Krueger, C.C.; Kincaid, H.L. Genetic variation at microsatellite loci in sturgeon: Primer sequence homology in Acipenser and Scaphirhynchus. Can. J. Fish. Aquat. Sci. 1997, 54, 1542–1547. [Google Scholar] [CrossRef]
- King, T.L.; Lubinski, B.A.; Spidle, A.P. Microsatellite DNA variation in Atlantic sturgeon (Acipenser oxyrinchus oxyrinchus) and cross-species amplification in the Acipenseridae. Conserv. Genet. 2001, 2, 103–119. [Google Scholar] [CrossRef]
- Henderson-Arzapalo, A.; King, T.L. Novel microsatellite markers for Atlantic sturgeon (Acipenser oxyrinchus) population delineation and broodstock management. Mol. Ecol. Notes 2002, 2, 437–439. [Google Scholar] [CrossRef]
- Forlani, A.; Fontana, F.; Congiu, L. Isolation of microsatellite loci from the endemic and endangered Adriatic sturgeon (Acipenser naccarii). Conserv. Genet. 2008, 9, 461–463. [Google Scholar] [CrossRef]
- Dudu, A.; Suciu, R.; Paraschiv, M.; Georgescu, S.E.; Costache, M.; Berrebi, P. Nuclear markers of Danube sturgeon hybridization. Int. J. Mol. Sci. 2011, 12, 6796–6809. [Google Scholar] [CrossRef] [Green Version]
- Web de GENETIX, Institut des Sciences de l’Evolution. Available online: https://kimura.univ-montp2.fr/genetix/ (accessed on 11 December 2021).
- Chow, S.; Hazama, K. Universal PCR primers for S7 ribosomal protein gene introns in fish. Mol. Ecol. 1998, 7, 1255–1256. [Google Scholar]
- Rehbein, H. Difference of fish species by PCR-based DNA analysis of nuclear genes. Eur. Food Res. Technol. 2013, 236, 979–990. [Google Scholar] [CrossRef]
- Pendas, A.M.; Moran, P.; Martinez, J.L.; Garcia-Vazquez, E. Applications of 5S rDNA in Atlantic salmon, brown trout, and in Atlantic salmon brown trout hybrid identification. Mol. Ecol. 1995, 4, 275–276. [Google Scholar] [CrossRef] [PubMed]
- Sun, M.; Liang, C.; Gao, H.; Lin, C.; Deng, M. Detection of parvalbumin, a common fish allergen gene in food, by real-time polymerase chain reaction. J. AOAC Int. 2009, 92, 234–240. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hall, T.A. BioEdit: A user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucleic Acids Symp. Ser. 1999, 41, 95–98. [Google Scholar]
- Altschul, S.F.; Madden, T.L.; Schäffer, A.A.; Zhang, J.; Zhang, Z.; Miller, W.; Lipman, D.J. Gapped BLAST and PSI-BLAST: A new generation of protein database search programs. Nucleic Acids Res. 1997, 25, 3389–3402. [Google Scholar] [CrossRef] [Green Version]
- Larkin, M.A.; Blackshields, G.; Brown, N.P.; Chenna, R.; McGettigan, P.A.; McWilliam, H.; Valentin, F.; Wallace, I.M.; Wilm, A.; Lopez, R.; et al. Clustal W and Clustal X version 2. 0. Bioinformatics 2007, 23, 2947–2948. [Google Scholar] [CrossRef] [Green Version]
- Untergasser, A.; Cutcutache, I.; Koressaar, T.; Ye, J.; Faircloth, B.C.; Remm, M.; Rozen, S.G. Primer3–new capabilities and interfaces. Nucl. Acids Res. 2012, 40, e115. [Google Scholar] [CrossRef] [Green Version]
- Jenneckens, I.; Meyer, J.N.; Hörstgen-Schwark, G.; May, B.; Debus, L.; Wedekind, H.; Ludwig, A. A fixed allele at microsatellite locus LS-39 exhibiting species-specificity for the black caviar producer Acipenser stellatus. J. Appl. Ichthyol. 2001, 17, 39–42. [Google Scholar] [CrossRef]







| Common Name | Scientific Name/Genitor Species (in Case of Hybrids) | Abbreviation | No of Individuals | Origin | Sample Type | |
|---|---|---|---|---|---|---|
| Pure species | Beluga sturgeon | H. huso | HUS | 58 | Aquaculture | Fin, eggs |
| Stellate sturgeon | A. stellatus | STE | 40 | Aquaculture | Fin, eggs | |
| Russian sturgeon | A. gueldenstaedtii | AGU | 15 | Aquaculture | Fin, eggs | |
| Siberian sturgeon | A. baerii | ABE | 15 | Aquaculture | Fin | |
| Sterlet | A. ruthenus | RUT | 40 | Danube River | Fin, meat | |
| Hybrids | Bester | H. huso × A. ruthenus | − | 18 | Aquaculture | Fin, eggs |
| Sterbe | A. ruthenus × H. huso | − | 10 | Aquaculture | Fin | |
| Best beluga | Bester × H. huso | − | 12 | Aquaculture | Fin, eggs | |
| Unknown | − | − | 2 | Danube River | Fin |
| Locus | Primer Sequence from 5′ to 3′ | Annealing Temperature (°C) | Annealing Time (s) | Reference |
|---|---|---|---|---|
| Afu19 | F: * CATCTTAGCCGTCTGTGGTAC R: CAGGTCCCTAATACAATGGC | 55 | 30 | [32] |
| Afu34 | F: * TACATACCTTCTGCAACG R: GATCCCTTCTGTTATCAAC | 55 | 30 | [32] |
| Afu54 | F: * CTCTAGTCTTTGTTGATTACAG R: CAAAGGACTTGAAACTAGG | 55 | 30 | [32] |
| Afu39 | F: * TTCTGAAGTTCACACATTG R: ATGGAGCATTATTGGAAGG | 55 | 30 | [32] |
| Aox27 | F: * AATAACAATAACGGCAGAACCT R: TGTGTTGCTCAAGACAGTATGA | 60 | 45 | [33] |
| AoxD234 | F: * AACTGGCTTTGTGATTGATCC R: TGAAGCAAAGGGTATTATTTGAG | 52 | 30 | [34] |
| AnacC11 | F: * AAATTTCCATTGGGGTGT R: CTTCGTTTTGAGAACCCG | 50 | 45 | [35] |
| Anac E4 | F: * TCAGCTACAGGGTTCTGGG R: GTTGTTACTCATTGGAACTC | 55 | 45 | [35] |
| Locus | Primer Name/Sequence from 5′ to 3′ | Annealing Temperature (°C) | Reference |
|---|---|---|---|
| S7RPEx1 | S7RPEx1 F: TGGCCTCTTCCTTGGCCGTC S7RPEx1 R: AACTCGTCTGGCTTTTCGCC | 49 | [38] |
| RP1 | Rut_Bae F: TTACATTAATTACCTGTGTTAAGATAG RP1_Locus A R: ATCCAAGTACAAGCTTGAACA | 49 | [16] |
| Vimentin | Bae154B7F: TCCAGGGTTTCCTACACCAGCCAAT Bae154B7R: CCACCCTCGCTTTTCGTTGGTTTG | 59 | [16] |
| Rhodopsin | Rh1F: GTYACCMTBGARCACAAGGAARTC Rh4R: TCRAYYCCRCAYGAGCAYTGCAT | 53 | [39] |
| 5S rRNA | 5S rRNA F: TACGCCCGATCTCGTCCGATC 5S rRNA R: CAGGCTGGTATGGCCGTAAGC | No amplification | [40] |
| Parvalbumin | Parv F: CAGGACAAGAGTGGCTTCAT Parv R: GAAGTTCTGCAGGAACAGCTT | No amplification | [41] |
| Marker | Species | Nucleotide Position | Marker | Species | Nucleotide Position | Marker | Species | Nucleotide Position | ||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| RP1 | 50 | 147 | Vim | 60 | Rh | 13 | 262 | 317 | 374 | |||
| ARU | A | G | AST | C | AGU | G | G | G | G | |||
| ABE | T | T | ABE | C | ARU | A | G | G | A | |||
| HUS | C/A | AST | G | A | G | A | ||||||
| HUS | A | G | C | G | ||||||||
| Category | Species | Species Matched by BLAST | Matched Accession Number | % Identity |
|---|---|---|---|---|
| Pure species | A. stellatus | A. stellatus | JQ623906 | 100% |
| A. nudiventris | KC500105 | 100% | ||
| A. ruthenus | A. ruthenus | MG648396 | 100% | |
| MG648371 | 100% | |||
| MG648364 | 100% | |||
| OV754669 | 100% | |||
| LR824036 | 100% | |||
| A. gueldenstaedtii | A. gueldenstaedtii | KJ789859 | 100% | |
| FJ392605 | 100% | |||
| A. naccarii | MK078265 | 100% | ||
| A. persicus | MW713795 | 100% | ||
| MK213065 | 100% | |||
| A. sinensis | EU719645 | 100% | ||
| A. gueldenstaedtii × A. baerii | KJ321189 | 100% | ||
| A. baerii | A. baerii | KP833617 | 100% | |
| MW856904 | 100% | |||
| MW856903 | 100% | |||
| A. gueldenstaedtii | MT410937 | 100% | ||
| KM286425 | ||||
| A. persicus | FJ809722 | 100% | ||
| A. gueldenstaedtii × A. baerii | OM049247 | 100% | ||
| A. baerii × A. schrenckii | KC578843 | 100% | ||
| H. huso | H. huso | AY442351 | 100% | |
| Hybrids | Bester | H. huso | AY442351 | 100% |
| Sterbe | A. ruthenus | MG648396 | 100% | |
| MG648371 | 100% | |||
| MG648364 | 100% | |||
| OV754669 | 100% | |||
| LR824036 | 100% | |||
| Best beluga | H. huso | AY442351 | 100% | |
| H203 | H. huso | AY442351 | 100% | |
| H204 | A. ruthenus | OV754669 | 100% | |
| LR824036 | 100% |
| Microsatellites | Nuclear Gene Markers | DNA Barcoding | |||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| D | S7RPEx1 | RP1 | Vimentin | Rhodopsin | D | ||||||||||
| BP | SNPs | D | BP | SNPs | D | BP | SNPs | D | BP | SNPs | D | ||||
| Pure species | A. stellatus | YES | 700/900 | − | NO | − | − | YES | 373 | − | NO | NS | + | ? | YES |
| H. huso | YES | 700/700 | − | YES | − | − | YES | 373 | − | NO | NS | + | ? | YES | |
| A. ruthenus | YES | 700/900 | − | NO | 169 | + | ? | − | − | YES | NS | + | ? | YES | |
| A. gueldenstaedtii | NO | 700/900 | − | NO | 169 | + | ? | 200/450/700/900 | − | YES | NS | + | ? | YES | |
| A. baerii | NO | 700/900 | − | NO | 169 | + | ? | 373 | − | YES | NS | − | X | YES | |
| Hybrids | Bester | Partially Identifies genitor species: HUS and RUT | 700/900 | − | NO | 169 | − | NO | − | − | YES | NS | − | X | YES Maternal species HUS |
| Sterbe | Partially identifies genitor species: HUS and RUT | 700/900 | − | NO | 169 | − | NO | 373 | − | NO | NS | − | X | YES Maternal species RUT | |
| Best beluga | YES | 700/700 | − | NO | 169 | − | NO | 373 | − | NO | NS | − | NO | YES Maternal species HUS | |
| H203 | Partially identifies the possible genitors: HUS, ARU, AGU + ABE | 700/900 | − | NO | 169 | − | YES * | 373 | − | YES ** | NS | − | NO | YES Maternal species HUS | |
| H204 | Partially identifies one genitor: AGU/ABE Other possible genitor: ARU/HUS | 700/900 | − | NO | 169/169? | − | YES *** | − | − | YES **** | NS | − | NO | YES Maternal species ARU | |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Dudu, A.; Samu, M.; Maereanu, M.; Georgescu, S.E. A Multistep DNA-Based Methodology for Accurate Authentication of Sturgeon Species. Foods 2022, 11, 1007. https://doi.org/10.3390/foods11071007
Dudu A, Samu M, Maereanu M, Georgescu SE. A Multistep DNA-Based Methodology for Accurate Authentication of Sturgeon Species. Foods. 2022; 11(7):1007. https://doi.org/10.3390/foods11071007
Chicago/Turabian StyleDudu, Andreea, Maria Samu, Marilena Maereanu, and Sergiu Emil Georgescu. 2022. "A Multistep DNA-Based Methodology for Accurate Authentication of Sturgeon Species" Foods 11, no. 7: 1007. https://doi.org/10.3390/foods11071007
APA StyleDudu, A., Samu, M., Maereanu, M., & Georgescu, S. E. (2022). A Multistep DNA-Based Methodology for Accurate Authentication of Sturgeon Species. Foods, 11(7), 1007. https://doi.org/10.3390/foods11071007






