Metagenomic and Functional Characterization of Two Chilean Kefir Beverages Reveals a Dairy Beverage Containing Active Enzymes, Short-Chain Fatty Acids, Microbial β-Amyloids, and Bio-Film Inhibitors
Abstract
:1. Introduction
2. Materials and Methods
2.1. Materials
2.2. Culture Media
2.3. Production of Kefir Beverage and Whey
2.4. Characterization of Kefir Beverages
2.4.1. Nutritional Properties
2.4.2. Detection of Short-Chain Fatty Acids
2.4.3. Microbial Viability in Kefir Beverages
2.5. Metagenomic Sequencing
2.5.1. DNA Extraction
2.5.2. Shotgun Metagenomic Sequencing and Analysis of Kefir Beverages
2.5.3. Amplicon-Based 16S rDNA Metagenomic Sequencing
2.6. Enzymatic Activity in Kefir Beverages
2.7. Effect of Kefir Beverages on Biofilm Formation
2.8. Detection of β-Amyloids in Kefir Beverage
2.9. Statistical Analysis
2.10. Accession Numbers
3. Results
3.1. Nutritional and Microbiological Characterization of Kefir Beverages
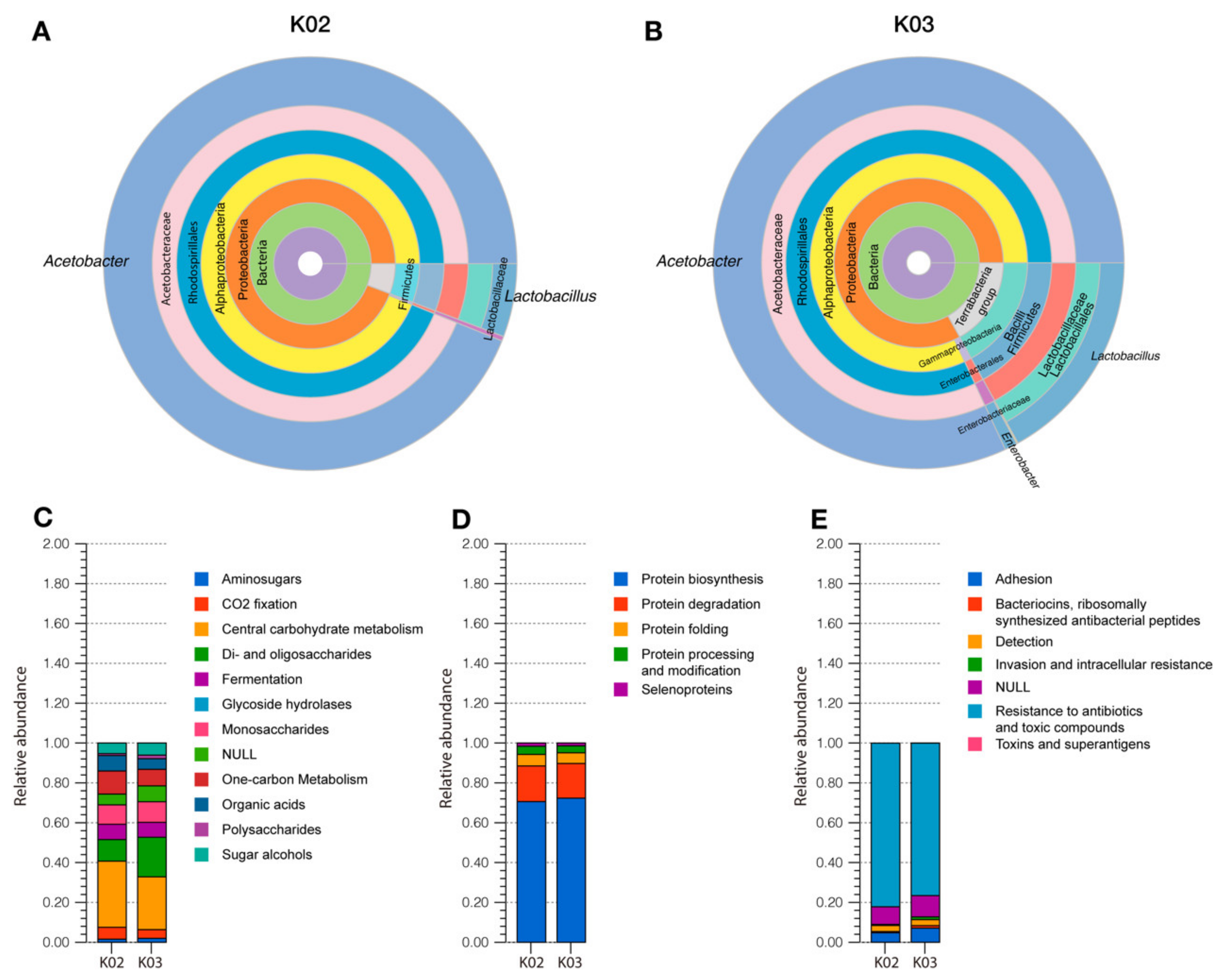
3.2. Taxonomic Analysis of Kefir Beverages
3.3. Functional Characterization of Kefir Beverages
3.3.1. Carbohydrate Metabolism
3.3.2. Protein Metabolism
3.3.3. Microbial Resistance to Antimicrobials: Heavy Metals and Antibiotics
3.3.4. Microbial Interaction in Kefir Beverages: Biofilm Formation and Host Interaction
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Farnworth, E.R. Kefir. J. Nutraceuticals Funct. Med Foods 1999, 1, 57–68. [Google Scholar] [CrossRef]
- Da Miguel, M.G.C.P.; Cardoso, P.G.; de Lago, L.A.; Schwan, R.F. Diversity of Bacteria Present in Milk Kefir Grains Using Culture-Dependent and Culture-Independent Methods. Food Res. Int. 2010, 43, 1523–1528. [Google Scholar] [CrossRef]
- Marsh, A.J.; O’Sullivan, O.; Hill, C.; Ross, R.P.; Cotter, P.D. Sequencing-Based Analysis of the Bacterial and Fungal Composition of Kefir Grains and Milks from Multiple Sources. PLoS ONE 2013, 8, e69371. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Guangsen, T.; Xiang, L.; Jiahu, G. Microbial Diversity and Volatile Metabolites of Kefir Prepared by Different Milk Types. CyTA J. Food 2021, 19, 399–407. [Google Scholar] [CrossRef]
- Leite, A.M.O.; Miguel, M.A.L.; Peixoto, R.S.; Ruas-Madiedo, P.; Paschoalin, V.M.F.; Mayo, B.; Delgado, S. Probiotic Potential of Selected Lactic Acid Bacteria Strains Isolated from Brazilian Kefir Grains. J. Dairy Sci. 2015, 98, 3622–3632. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Garrote, G.L.; Abraham, A.G.; De Antoni, G.L. Chemical and Microbiological Characterisation of Kefir Grains. J. Dairy Res. 2001, 68, 639–652. [Google Scholar] [CrossRef]
- Carasi, P.; Jacquot, C.; Romanin, D.E.; Elie, A.-M.; De Antoni, G.L.; Urdaci, M.C.; de los Serradell, M.A. Safety and Potential Beneficial Properties of Enterococcus Strains Isolated from Kefir. Int. Dairy J. 2014, 39, 193–200. [Google Scholar] [CrossRef]
- Sabir, F.; Beyatli, Y.; Cokmus, C.; Onal-Darilmaz, D. Assessment of Potential Probiotic Properties of Lactobacillus Spp., Lactococcus Spp. and Pediococcus Spp. Strains Isolated from Kefir. J. Food Sci. 2010, 75, M568–M573. [Google Scholar] [CrossRef]
- Simova, E.; Beshkova, D.; Angelov, A.; Hristozova, T.; Frengova, G.; Spasov, Z. Lactic Acid Bacteria and Yeasts in Kefir Grains and Kefir Made from Them. J. Ind. Microbiol. Biotechnol. 2002, 28, 1–6. [Google Scholar] [CrossRef]
- Vardjan, T.; Mohar Lorbeg, P.; Rogelj, I.; Čanžek Majhenič, A. Characterization and Stability of Lactobacilli and Yeast Microbiota in Kefir Grains. J. Dairy Sci. 2013, 96, 2729–2736. [Google Scholar] [CrossRef] [Green Version]
- TaŞ, T.K.Ö.K.; Ekinci, F.Y.; Guzel-Seydim, Z.B. Identification of Microbial Flora in Kefir Grains Produced in Turkey Using PCR. Int. J. Dairy Technol. 2012, 65, 126–131. [Google Scholar] [CrossRef]
- Garofalo, C.; Ferrocino, I.; Reale, A.; Sabbatini, R.; Milanović, V.; Alkić-Subašić, M.; Boscaino, F.; Aquilanti, L.; Pasquini, M.; Trombetta, M.F.; et al. Study of Kefir Drinks Produced by Backslopping Method Using Kefir Grains from Bosnia and Herzegovina: Microbial Dynamics and Volatilome Profile. Food Res. Int. 2020, 137, 109369. [Google Scholar] [CrossRef]
- Garrote, G.L.; Abraham, A.G.; De Antoni, G.L. Microbial Interactions in Kefir: A Natural Probiotic Drink. In Biotechnology of Lactic Acid Bacteria: Novel Applications; Wiley-Blackwell: Hoboken, NJ, USA, 2010; 327p. [Google Scholar]
- Cheirsilp, B.; Shoji, H.; Shimizu, H.; Shioya, S. Interactions between Lactobacillus Kefiranofaciens and Saccharomyces Cerevisiae in Mixed Culture for Kefiran Production. J. Biosci. Bioeng. 2003, 96, 279–284. [Google Scholar] [CrossRef]
- Rosa, D.D.; Dias, M.M.S.; Grześkowiak, Ł.M.; Reis, S.A.; Conceição, L.L.; Peluzio, M.C.G. Milk Kefir: Nutritional, Microbiological and Health Benefits. Nutr. Res. Rev. 2017, 30, 82–96. [Google Scholar] [CrossRef]
- Farag, M.A.; Jomaa, S.A.; Abd El-Wahed, A.; El-Seedi, H.R. The Many Faces of Kefir Fermented Dairy Products: Quality Characteristics, Flavour Chemistry, Nutritional Value, Health Benefits, and Safety. Nutrients 2020, 12, 346. [Google Scholar] [CrossRef] [Green Version]
- Bellikci-Koyu, E.; Sarer-Yurekli, B.P.; Akyon, Y.; Aydin-Kose, F.; Karagozlu, C.; Ozgen, A.G.; Brinkmann, A.; Nitsche, A.; Ergunay, K.; Yilmaz, E.; et al. Effects of Regular Kefir Consumption on Gut Microbiota in Patients with Metabolic Syndrome: A Parallel-Group, Randomized, Controlled Study. Nutrients 2019, 11, 2089. [Google Scholar] [CrossRef] [Green Version]
- Hsu, Y.-J.; Huang, W.-C.; Lin, J.-S.; Chen, Y.-M.; Ho, S.-T.; Huang, C.-C.; Tung, Y.-T. Kefir Supplementation Modifies Gut Microbiota Composition, Reduces Physical Fatigue, and Improves Exercise Performance in Mice. Nutrients 2018, 10, 862. [Google Scholar] [CrossRef] [Green Version]
- Kim, D.-H.; Chon, J.-W.; Kim, H.; Seo, K.-H. Modulation of Intestinal Microbiota in Mice by Kefir Administration. Food Sci. Biotechnol. 2015, 24, 1397–1403. [Google Scholar] [CrossRef]
- Carasi, P.; Trejo, F.M.; Pérez, P.F.; De Antoni, G.L.; de los Serradell, M.A. Surface Proteins from Lactobacillus Kefir Antagonize In Vitro Cytotoxic Effect of Clostridium Difficile Toxins. Anaerobe 2012, 18, 135–142. [Google Scholar] [CrossRef]
- De Montijo-Prieto, S.; Moreno, E.; Bergillos-Meca, T.; Lasserrot, A.; Ruiz-López, M.-D.; Ruiz-Bravo, A.; Jiménez-Valera, M. A Lactobacillus Plantarum Strain Isolated from Kefir Protects against Intestinal Infection with Yersinia Enterocolitica O9 and Modulates Immunity in Mice. Res. Microbiol. 2015, 166, 626–632. [Google Scholar] [CrossRef]
- Urdaneta, E.; Barrenetxe, J.; Aranguren, P.; Irigoyen, A.; Marzo, F.; Ibáñez, F.C. Intestinal Beneficial Effects of Kefir-Supplemented Diet in Rats. Nutr. Res. 2007, 27, 653–658. [Google Scholar] [CrossRef]
- De Vrese, M.; Keller, B.; Barth, C.A. Enhancement of Intestinal Hydrolysis of Lactose by Microbial β-Galactosidase (EC 3.2.1.23) of Kefir. Br. J. Nutr. 1992, 67, 67–75. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hertzler, S.R.; Clancy, S.M. Kefir Improves Lactose Digestion and Tolerance in Adults with Lactose Maldigestion. J. Am. Diet. Assoc. 2003, 103, 582–587. [Google Scholar] [CrossRef] [PubMed]
- Chen, H.L.; Tung, Y.T.; Tsai, C.L.; Lai, C.W.; Lai, Z.L.; Tsai, H.C.; Lin, Y.L.; Wang, C.H.; Chen, C.M. Kefir Improves Fatty Liver Syndrome by Inhibiting the Lipogenesis Pathway in Leptin-Deficient Ob/ob Knockout Mice. Int. J. Obes. 2013, 38, 1172. [Google Scholar] [CrossRef] [PubMed]
- Chen, H.L.; Tsai, T.C.; Tsai, Y.C.; Liao, J.W.; Yen, C.C.; Chen, C.M. Kefir Peptides Prevent High-Fructose Corn Syrup-Induced Non-Alcoholic Fatty Liver Disease in a Murine Model by Modulation of Inflammation and the JAK2 Signaling Pathway. Nutr. Diabetes 2016, 6, e237. [Google Scholar] [CrossRef]
- Gao, J.; Gao, J.; Gu, F.; Ruan, H.; Chen, Q.; He, J.; He, G. Induction of Apoptosis of Gastric Cancer Cells SGC7901 in Vitro by a Cell-Free Fraction of Tibetan Kefir. Int. Dairy J. 2013, 30, 14–18. [Google Scholar] [CrossRef]
- Khoury, N.; El-Hayek, S.; Tarras, O.; El-Sabban, M.; El-Sibai, M.; Rizk, S. Kefir Exhibits Anti-proliferative and Pro-apoptotic Effects on Colon Adenocarcinoma Cells with No Significant Effects on Cell Migration and Invasion. Int. J. Oncol. 2014, 45, 2117–2127. [Google Scholar] [CrossRef]
- Prado, M.R.M.; Boller, C.; Zibetti, R.G.M.; de Souza, D.; Pedroso, L.L.; Soccol, C.R. Anti-Inflammatory and Angiogenic Activity of Polysaccharide Extract Obtained from Tibetan Kefir. Microvasc. Res. 2016, 108, 29–33. [Google Scholar] [CrossRef]
- Oryan, A.; Alemzadeh, E.; Eskandari, M.H. Kefir Accelerates Burn Wound Healing Through Inducing Fibroblast Cell Migration In Vitro and Modulating the Expression of IL-1β, TGF-β1, and bFGF Genes In Vivo. Probiotics Antimicrob. Proteins 2019, 11, 874–886. [Google Scholar] [CrossRef]
- Alves, E.; Gregório, J.; Baby, A.R.; Rijo, P.; Rodrigues, L.M.; Rosado, C. Homemade Kefir Consumption Improves Skin Condition—A Study Conducted in Healthy and Atopic Volunteers. Foods 2021, 10, 2794. [Google Scholar] [CrossRef]
- Zamberi, N.R.; Mohamad, N.E.; Yeap, S.K.; Ky, H.; Beh, B.K.; Liew, W.C.; Tan, S.W.; Ho, W.Y.; Boo, S.Y.; Chua, Y.H.; et al. 16S Metagenomic Microbial Composition Analysis of Kefir Grain Using MEGAN and BaseSpace. Food Biotechnol. 2016, 30, 219–230. [Google Scholar] [CrossRef]
- Ilıkkan, Ö.K.; Bağdat, E.Ş. Comparison of Bacterial and Fungal Biodiversity of Turkish Kefir Grains with High-Throughput Metagenomic Analysis. LWT 2021, 152, 112375. [Google Scholar] [CrossRef]
- Kazou, M.; Grafakou, A.; Tsakalidou, E.; Georgalaki, M. Zooming Into the Microbiota of Home-Made and Industrial Kefir Produced in Greece Using Classical Microbiological and Amplicon-Based Metagenomics Analyses. Front. Microbiol. 2021, 12, 64. [Google Scholar] [CrossRef]
- Yegin, Z.; Zafer Yurt, M.N.; Tasbasi, B.B.; Acar, E.E.; Altunbas, O.; Ucak, S.; Ozalp, V.C.; Sudagidan, M. Determination of Bacterial Community Structure of Turkish Kefir Beverages via Metagenomic Approach. Int. Dairy J. 2022, 129, 105337. [Google Scholar] [CrossRef]
- Kalamaki, M.S.; Angelidis, A.S. High-Throughput, Sequence-Based Analysis of the Microbiota of Greek Kefir Grains from Two Geographic Regions. Food Technol. Biotechnol. 2020, 58, 138–146. [Google Scholar] [CrossRef]
- Dertli, E.; Çon, A.H. Microbial Diversity of Traditional Kefir Grains and Their Role on Kefir Aroma. LWT Food Sci. Technol. 2017, 85, 151–157. [Google Scholar] [CrossRef]
- Zeng, X.; Wang, Y.; Jia, H.; Wang, Z.; Gao, Z.; Luo, Y.; Sheng, Q.; Yuan, Y.; Yue, T. Metagenomic Analysis of Microflora Structure and Functional Capacity in Probiotic Tibetan Kefir Grains. Food Res. Int. 2022, 151, 110849. [Google Scholar] [CrossRef]
- Melkonian, C.; Gottstein, W.; Blasche, S.; Kim, Y.; Abel-Kistrup, M.; Swiegers, H.; Saerens, S.; Edwards, N.; Patil, K.R.; Teusink, B.; et al. Finding Functional Differences Between Species in a Microbial Community: Case Studies in Wine Fermentation and Kefir Culture. Front. Microbiol. 2019, 10, 1347. [Google Scholar] [CrossRef] [Green Version]
- You, L.; Yang, C.; Jin, H.; Kwok, L.-Y.; Sun, Z.; Zhang, H. Metagenomic Features of Traditional Fermented Milk Products. LWT 2022, 155, 112945. [Google Scholar] [CrossRef]
- Nalbantoglu, U.; Cakar, A.; Dogan, H.; Abaci, N.; Ustek, D.; Sayood, K.; Can, H. Metagenomic Analysis of the Microbial Community in Kefir Grains. Food Microbiol. 2014, 41, 42–51. [Google Scholar] [CrossRef]
- Tenorio-Salgado, S.; Castelán-Sánchez, H.G.; Dávila-Ramos, S.; Huerta-Saquero, A.; Rodríguez-Morales, S.; Merino-Pérez, E.; Roa de la Fuente, L.F.; Solis-Pereira, S.E.; Pérez-Rueda, E.; Lizama-Uc, G. Metagenomic Analysis and Antimicrobial Activity of Two Fermented Milk Kefir Samples. Microbiologyopen 2021, 10, e1183. [Google Scholar] [CrossRef] [PubMed]
- Gao, J.; Gu, F.; He, J.; Xiao, J.; Chen, Q.; Ruan, H.; He, G. Metagenome Analysis of Bacterial Diversity in Tibetan Kefir Grains. Eur. Food Res. Technol. 2013, 236, 549–556. [Google Scholar] [CrossRef]
- Walsh, A.M.; Crispie, F.; Kilcawley, K.; O’Sullivan, O.; O’Sullivan, M.G.; Claesson, M.J.; Cotter, P.D.; Dutton, R.J. Microbial Succession and Flavor Production in the Fermented Dairy Beverage Kefir. mSystems 2016, 1, e00052-16. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Biçer, Y.; Telli, A.E.; Sönmez, G.; Turkal, G.; Telli, N.; Uçar, G. Comparison of Commercial and Traditional Kefir Microbiota Using Metagenomic Analysis. Int. J. Dairy Technol. 2021, 74, 528–534. [Google Scholar] [CrossRef]
- Horwitz, W.; Latimer, G. Official Methods of Analysis of AOAC International, 18th ed.; AOAC International: Gaithersburg, MA, USA, 2006. [Google Scholar]
- Asarat, M.; Apostolopoulos, V.; Vasiljevic, T.; Donkor, O. Short-Chain Fatty Acids Produced by Synbiotic Mixtures in Skim Milk Differentially Regulate Proliferation and Cytokine Production in Peripheral Blood Mononuclear Cells. Int. J. Food Sci. Nutr. 2015, 66, 755–765. [Google Scholar] [CrossRef]
- Donkor, O.N.; Henriksson, A.; Vasiljevic, T.; Shah, N.P. Probiotic Strains as Starter Cultures Improve Angiotensin-Converting Enzyme Inhibitory Activity in Soy Yogurt. J. Food Sci. 2005, 70, m375–m381. [Google Scholar] [CrossRef]
- Schmieder, R.; Edwards, R. Quality Control and Preprocessing of Metagenomic Datasets. Bioinformatics 2011, 27, 863–864. [Google Scholar] [CrossRef] [Green Version]
- Truong, D.T.; Franzosa, E.A.; Tickle, T.L.; Scholz, M.; Weingart, G.; Pasolli, E.; Tett, A.; Huttenhower, C.; Segata, N. MetaPhlAn2 for Enhanced Metagenomic Taxonomic Profiling. Nat. Methods 2015, 12, 902. [Google Scholar] [CrossRef]
- Langmead, B.; Salzberg, S.L. Fast Gapped-Read Alignment with Bowtie 2. Nat. Methods 2012, 9, 357. [Google Scholar] [CrossRef] [Green Version]
- Nurk, S.; Meleshko, D.; Korobeynikov, A.; Pevzner, P.A. metaSPAdes: A New Versatile Metagenomic Assembler. Genome Res. 2017, 27, 824–834. [Google Scholar] [CrossRef] [Green Version]
- Lomsadze, A.; Zhu, W.; Borodovsky, M. Ab Initio Gene Identification in Metagenomic Sequences. Nucleic Acids Res. 2010, 38, e132. [Google Scholar] [CrossRef] [Green Version]
- Huerta-Cepas, J.; Forslund, K.; Coelho, L.P.; Bork, P.; von Mering, C.; Szklarczyk, D.; Jensen, L.J. Fast Genome-Wide Functional Annotation through Orthology Assignment by eggNOG-Mapper. Mol. Biol. Evol. 2017, 34, 2115–2122. [Google Scholar] [CrossRef] [Green Version]
- Buchfink, B.; Xie, C.; Huson, D.H. Fast and Sensitive Protein Alignment Using DIAMOND. Nat. Methods 2014, 12, 59. [Google Scholar] [CrossRef]
- Kanehisa, M.; Sato, Y.; Morishima, K. BlastKOALA and GhostKOALA: KEGG Tools for Functional Characterization of Genome and Metagenome Sequences. J. Mol. Biol. 2016, 428, 726–731. [Google Scholar] [CrossRef] [Green Version]
- Meyer, F.; Paarmann, D.; D’Souza, M.; Olson, R.; Glass, E.M.; Kubal, M.; Paczian, T.; Rodriguez, A.; Stevens, R.; Wilke, A.; et al. The Metagenomics RAST Server—A Public Resource for the Automatic Phylogenetic and Functional Analysis of Metagenomes. BMC Bioinform. 2008, 9, 386. [Google Scholar] [CrossRef] [Green Version]
- Caporaso, J.G.; Lauber, C.L.; Walters, W.A.; Berg-Lyons, D.; Lozupone, C.A.; Turnbaugh, P.J.; Fierer, N.; Knight, R. Global Patterns of 16S rRNA Diversity at a Depth of Millions of Sequences per Sample. Proc. Natl. Acad. Sci. USA 2011, 108, 4516–4522. [Google Scholar] [CrossRef] [Green Version]
- Caporaso, J.G.; Kuczynski, J.; Stombaugh, J.; Bittinger, K.; Bushman, F.D.; Costello, E.K.; Fierer, N.; Peña, A.G.; Goodrich, J.K.; Gordon, J.I.; et al. QIIME Allows Analysis of High-Throughput Community Sequencing Data. Nat. Methods 2010, 7, 335. [Google Scholar] [CrossRef] [Green Version]
- Edgar, R.C. Search and Clustering Orders of Magnitude Faster than BLAST. Bioinformatics 2010, 26, 2460–2461. [Google Scholar] [CrossRef] [Green Version]
- Huson, D.H.; Auch, A.F.; Qi, J.; Schuster, S.C. MEGAN Analysis of Metagenomic Data. Genome Res. 2007, 17, 377–386. [Google Scholar] [CrossRef] [Green Version]
- O’Toole, G.A.; Kolter, R. Initiation of Biofilm Formation in Pseudomonas Fluorescens WCS365 Proceeds via Multiple, Convergent Signalling Pathways: A Genetic Analysis. Mol. Microbiol. 1998, 28, 449–461. [Google Scholar] [CrossRef]
- McCrate, O.A.; Zhou, X.; Cegelski, L. Curcumin as an Amyloid-Indicator Dye in E. Coli. Chem. Commun. 2013, 49, 4193. [Google Scholar] [CrossRef] [Green Version]
- Bokranz, W.; Wang, X.; Tschäpe, H.; Römling, U. Expression of Cellulose and Curli Fimbriae by Escherichia Coli Isolated from the Gastrointestinal Tract. J. Med. Microbiol. 2005, 54, 1171–1182. [Google Scholar] [CrossRef]
- Baba, T.; Ara, T.; Hasegawa, M.; Takai, Y.; Okumura, Y.; Baba, M.; Datsenko, K.A.; Tomita, M.; Wanner, B.L.; Mori, H. Construction of Escherichia Coli K-12 in-Frame, Single-Gene Knockout Mutants: The Keio Collection. Mol. Syst. Biol. 2006, 2, 2006.0008. [Google Scholar] [CrossRef] [Green Version]
- De Filippis, F.; Troise, A.D.; Vitaglione, P.; Ercolini, D. Different Temperatures Select Distinctive Acetic Acid Bacteria Species and Promotes Organic Acids Production during Kombucha Tea Fermentation. Food Microbiol. 2018, 73, 11–16. [Google Scholar] [CrossRef]
- Soumahoro, S.; Ouattara, H.G.; Droux, M.; Nasser, W.; Niamke, S.L.; Reverchon, S. Acetic Acid Bacteria (AAB) Involved in Cocoa Fermentation from Ivory Coast: Species Diversity and Performance in Acetic Acid Production. J. Food Sci. Technol. 2020, 57, 1904–1916. [Google Scholar] [CrossRef]
- Ouoba, L.I.I.; Kando, C.; Parkouda, C.; Sawadogo-Lingani, H.; Diawara, B.; Sutherland, J.P. The Microbiology of Bandji, Palm Wine of Borassus akeassii from Burkina Faso: Identification and Genotypic Diversity of Yeasts, Lactic Acid and Acetic Acid Bacteria. J. Appl. Microbiol. 2012, 113, 1428–1441. [Google Scholar] [CrossRef]
- Li, L.; Wieme, A.; Spitaels, F.; Balzarini, T.; Nunes, O.C.; Manaia, C.M.; Van Landschoot, A.; De Vuyst, L.; Cleenwerck, I.; Vandamme, P. Acetobacter Sicerae Sp. Nov., Isolated from Cider and Kefir, and Identification of Species of the Genus Acetobacter by dnaK, groEL and rpoB Sequence Analysis. Int. J. Syst. Evol. Microbiol. 2014, 64, 2407–2415. [Google Scholar] [CrossRef]
- Wang, X.; Li, W.; Xu, M.; Tian, J.; Li, W. The Microbial Diversity and Biofilm-Forming Characteristic of Two Traditional Tibetan Kefir Grains. Foods 2022, 11, 12. [Google Scholar] [CrossRef] [PubMed]
- Goktas, H.; Dikmen, H.; Demirbas, F.; Sagdic, O.; Dertli, E. Characterisation of Probiotic Properties of Yeast Strains Isolated from Kefir Samples. Int. J. Dairy Technol. 2021, 74, 715–722. [Google Scholar] [CrossRef]
- Gut, A.M.; Vasiljevic, T.; Yeager, T.; Donkor, O.N. Characterization of Yeasts Isolated from Traditional Kefir Grains for Potential Probiotic Properties. J. Funct. Foods 2019, 58, 56–66. [Google Scholar] [CrossRef]
- Cho, Y.-J.; Kim, D.-H.; Jeong, D.; Seo, K.-H.; Jeong, H.S.; Lee, H.G.; Kim, H. Characterization of Yeasts Isolated from Kefir as a Probiotic and Its Synergic Interaction with the Wine Byproduct Grape Seed Flour/extract. LWT 2018, 90, 535–539. [Google Scholar] [CrossRef]
- Stadie, J.; Gulitz, A.; Ehrmann, M.A.; Vogel, R.F. Metabolic Activity and Symbiotic Interactions of Lactic Acid Bacteria and Yeasts Isolated from Water Kefir. Food Microbiol. 2013, 35, 92–98. [Google Scholar] [CrossRef] [PubMed]
- Farnworth, E.R. Kefir–A Complex Probiotic. Food Sci. Technol. Bull. Fu 2006, 2, 1–17. [Google Scholar] [CrossRef] [Green Version]
- Haghshenas, B.; Nami, Y.; Abdullah, N.; Radiah, D.; Rosli, R.; Barzegari, A.; Yari Khosroushahi, A. Potentially Probiotic Acetic Acid Bacteria Isolation and Identification from Traditional Dairies Microbiota. Int. J. Food Sci. Technol. 2015, 50, 1056–1064. [Google Scholar] [CrossRef]
- Haghshenas, B.; Nami, Y.; Abdullah, N.; Radiah, D.; Rosli, R.; Khosroushahi, A.Y. Anticancer Impacts of Potentially Probiotic Acetic Acid Bacteria Isolated from Traditional Dairy Microbiota. LWT Food Sci. Technol. 2015, 60, 690–697. [Google Scholar] [CrossRef]
- Hammes, W.P.; Vogel, R.F. The Genus Lactobacillus. In The Genera of Lactic Acid Bacteria; Wood, B.J.B., Holzapfel, W.H., Eds.; Springer: Boston, MA, USA, 1995; pp. 19–54. ISBN 978-1-4615-5817-0. [Google Scholar]
- Ramos, C.L.; Thorsen, L.; Schwan, R.F.; Jespersen, L. Strain-Specific Probiotics Properties of Lactobacillus Fermentum, Lactobacillus Plantarum and Lactobacillus Brevis Isolates from Brazilian Food Products. Food Microbiol. 2013, 36, 22–29. [Google Scholar] [CrossRef] [Green Version]
- Maragkoudakis, P.A.; Zoumpopoulou, G.; Miaris, C.; Kalantzopoulos, G.; Pot, B.; Tsakalidou, E. Probiotic Potential of Lactobacillus Strains Isolated from Dairy Products. Int. Dairy J. 2006, 16, 189–199. [Google Scholar] [CrossRef]
- Bao, Y.; Zhang, Y.; Zhang, Y.; Liu, Y.; Wang, S.; Dong, X.; Wang, Y.; Zhang, H. Screening of Potential Probiotic Properties of Lactobacillus Fermentum Isolated from Traditional Dairy Products. Food Control 2010, 21, 695–701. [Google Scholar] [CrossRef]
- Kumar, A.; Kumar, D. Characterization of Lactobacillus Isolated from Dairy Samples for Probiotic Properties. Anaerobe 2015, 33, 117–123. [Google Scholar] [CrossRef]
- Fernandez-Pacheco, P.; Arévalo-Villena, M.; Bevilacqua, A.; Corbo, M.R.; Briones Pérez, A. Probiotic Characteristics in Saccharomyces Cerevisiae Strains: Properties for Application in Food Industries. LWT 2018, 97, 332–340. [Google Scholar] [CrossRef]
- Azhar, M.A.; Munaim, M.S.A. Identification and Evaluation of Probiotic Potential in Yeast Strains Found in Kefir Drink Samples from Malaysia. Int. J. Food Eng. 2019, 15, 20180347. [Google Scholar] [CrossRef]
- Agarbati, A.; Canonico, L.; Marini, E.; Zannini, E.; Ciani, M.; Comitini, F. Potential Probiotic Yeasts Sourced from Natural Environmental and Spontaneous Processed Foods. Foods 2020, 9, 287. [Google Scholar] [CrossRef] [Green Version]
- Silva, T.; Reto, M.; Sol, M.; Peito, A.; Peres, C.M.; Peres, C.; Malcata, F.X. Characterization of Yeasts from Portuguese Brined Olives, with a Focus on Their Potentially Probiotic Behavior. LWT Food Sci. Technol. 2011, 44, 1349–1354. [Google Scholar] [CrossRef]
- Zajšek, K.; Kolar, M.; Goršek, A. Characterisation of the Exopolysaccharide Kefiran Produced by Lactic Acid Bacteria Entrapped within Natural Kefir Grains. Int. J. Dairy Technol. 2011, 64, 544–548. [Google Scholar] [CrossRef]
- Swallow, D.M. Genetics of Lactase Persistence and Lactose Intolerance. Annu. Rev. Genet. 2003, 37, 197–219. [Google Scholar] [CrossRef]
- Ríos-Covián, D.; Ruas-Madiedo, P.; Margolles, A.; Gueimonde, M.; De los Reyes-Gavilán, C.G.; Salazar, N. Intestinal Short Chain Fatty Acids and Their Link with Diet and Human Health. Front. Microbiol. 2016, 7, 185. [Google Scholar] [CrossRef] [Green Version]
- Mamlouk, D.; Gullo, M. Acetic Acid Bacteria: Physiology and Carbon Sources Oxidation. Indian J. Microbiol. 2013, 53, 377–384. [Google Scholar] [CrossRef] [Green Version]
- Güzel-Seydim, Z.B.; Seydim, A.C.; Greene, A.K.; Bodine, A.B. Determination of Organic Acids and Volatile Flavor Substances in Kefir during Fermentation. J. Food Compos. Anal. 2000, 13, 35–43. [Google Scholar] [CrossRef]
- Adler, P.; Frey, L.J.; Berger, A.; Bolten, C.J.; Hansen, C.E.; Wittmann, C. The Key to Acetate: Metabolic Fluxes of Acetic Acid Bacteria under Cocoa Pulp Fermentation-Simulating Conditions. Appl. Environ. Microbiol. 2014, 80, 4702–4716. [Google Scholar] [CrossRef] [Green Version]
- Halstead, F.D.; Rauf, M.; Moiemen, N.S.; Bamford, A.; Wearn, C.M.; Fraise, A.P.; Lund, P.A.; Oppenheim, B.A.; Webber, M.A. The Antibacterial Activity of Acetic Acid against Biofilm-Producing Pathogens of Relevance to Burns Patients. PLoS ONE 2015, 10, e0136190. [Google Scholar] [CrossRef] [Green Version]
- Vieira, C.P.; Rosario, A.I.L.S.; Lelis, C.A.; Rekowsky, B.S.S.; Carvalho, A.P.A.; Rosário, D.K.A.; Elias, T.A.; Costa, M.P.; Foguel, D.; Conte-Junior, C.A. Bioactive Compounds from Kefir and Their Potential Benefits on Health: A Systematic Review and Meta-Analysis. Oxid. Med. Cell. Longev. 2021, 2021, 1–34. [Google Scholar] [CrossRef] [PubMed]
- Johansen, P.; Jespersen, L. Impact of Quorum Sensing on the Quality of Fermented Foods. Curr. Opin. Food Sci. 2017, 13, 16–25. [Google Scholar] [CrossRef]
- Jonas, K.; Tomenius, H.; Kader, A.; Normark, S.; Römling, U.; Belova, L.M.; Melefors, O. Roles of Curli, Cellulose and BapA in Salmonella Biofilm Morphology Studied by Atomic Force Microscopy. BMC Microbiol. 2007, 7, 70. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Prigent-Combaret, C.; Prensier, G.; Le Thi, T.T.; Vidal, O.; Lejeune, P.; Dorel, C. Developmental Pathway for Biofilm Formation in Curli-Producing Escherichia Coli Strains: Role of Flagella, Curli and Colanic Acid. Environ. Microbiol. 2000, 2, 450–464. [Google Scholar] [CrossRef]
- Larsen, P.; Nielsen, J.L.; Dueholm, M.S.; Wetzel, R.; Otzen, D.; Nielsen, P.H. Amyloid Adhesins Are Abundant in Natural Biofilms. Environ. Microbiol. 2007, 9, 3077–3090. [Google Scholar] [CrossRef]
- Ambalam, P.; Kondepudi, K.K.; Nilsson, I.; Wadström, T.; Ljungh, Å. Bile Stimulates Cell Surface Hydrophobicity, Congo Red Binding and Biofilm Formation of Lactobacillus strains. FEMS Microbiol. Lett. 2012, 333, 10–19. [Google Scholar] [CrossRef] [Green Version]
- Chiti, F.; Dobson, C.M. Protein Misfolding, Functional Amyloid, and Human Disease. Annu. Rev. Biochem. 2006, 75, 333–366. [Google Scholar] [CrossRef] [Green Version]
- Alonso, R.; Pisa, D.; Fernández-Fernández, A.M.; Carrasco, L. Infection of Fungi and Bacteria in Brain Tissue From Elderly Persons and Patients With Alzheimer’s Disease. Front. Aging Neurosci. 2018, 10, 159. [Google Scholar] [CrossRef] [Green Version]
- Higashi, T.; Nishii, R.; Kagawa, S.; Kishibe, Y.; Takahashi, M.; Okina, T.; Suzuki, N.; Hasegawa, H.; Nagahama, Y.; Ishizu, K.; et al. 18F-FPYBF-2, a New F-18-Labelled Amyloid Imaging PET Tracer: First Experience in 61 Volunteers and 55 Patients with Dementia. Ann. Nucl. Med. 2018, 32, 206–216. [Google Scholar] [CrossRef] [Green Version]
- Brothers, H.M.; Gosztyla, M.L.; Robinson, S.R. The Physiological Roles of Amyloid-β Peptide Hint at New Ways to Treat Alzheimer’s Disease. Front. Aging Neurosci. 2018, 10, 118. [Google Scholar] [CrossRef]
- Eimer, W.A.; Vijaya Kumar, D.K.; Navalpur Shanmugam, N.K.; Rodriguez, A.S.; Mitchell, T.; Washicosky, K.J.; György, B.; Breakefield, X.O.; Tanzi, R.E.; Moir, R.D. Alzheimer’s Disease-Associated β-Amyloid Is Rapidly Seeded by Herpesviridae to Protect against Brain Infection. Neuron 2018, 99, 56–63.e3. [Google Scholar] [CrossRef] [Green Version]
- De Lorenzi, E.; Chiari, M.; Colombo, R.; Cretich, M.; Sola, L.; Vanna, R.; Gagni, P.; Bisceglia, F.; Morasso, C.; Lin, J.S.; et al. Evidence That the Human Innate Immune Peptide LL-37 May Be a Binding Partner of Amyloid-β and Inhibitor of Fibril Assembly. J. Alzheimer’s Dis. 2017, 59, 1213–1226. [Google Scholar] [CrossRef] [Green Version]
- Frost, G.R.; Li, Y.-M. The Role of Astrocytes in Amyloid Production and Alzheimer’s Disease. Open Biol. 2017, 7, 170228. [Google Scholar] [CrossRef] [Green Version]
- Blanco, L.P.; Evans, M.L.; Smith, D.R.; Badtke, M.P.; Chapman, M.R. Diversity, Biogenesis and Function of Microbial Amyloids. Trends Microbiol. 2012, 20, 66–73. [Google Scholar] [CrossRef] [Green Version]
- Oppong, G.O.; Rapsinski, G.J.; Newman, T.N.; Nishimori, J.H.; Biesecker, S.G.; Tükel, Ç. Epithelial Cells Augment Barrier Function via Activation of the Toll-Like Receptor 2/Phosphatidylinositol 3-Kinase Pathway upon Recognition of Salmonella enterica Serovar Typhimurium Curli Fibrils in the Gut. Infect. Immun. 2013, 81, 478. [Google Scholar] [CrossRef] [Green Version]
- Oppong, G.O.; Rapsinski, G.J.; Tursi, S.A.; Biesecker, S.G.; Klein-Szanto, A.J.P.; Goulian, M.; McCauley, C.; Healy, C.; Wilson, R.P.; Tükel, C. Biofilm-Associated Bacterial Amyloids Dampen Inflammation in the Gut: Oral Treatment with Curli Fibres Reduces the Severity of Hapten-Induced Colitis in Mice. NPJ Biofilms Microbiomes 2015, 1, 15019. [Google Scholar] [CrossRef] [Green Version]
- Chen, S.G.; Stribinskis, V.; Rane, M.J.; Demuth, D.R.; Gozal, E.; Roberts, A.M.; Jagadapillai, R.; Liu, R.; Choe, K.; Shivakumar, B.; et al. Exposure to the Functional Bacterial Amyloid Protein Curli Enhances Alpha-Synuclein Aggregation in Aged Fischer 344 Rats and Caenorhabditis Elegans. Sci. Rep. 2016, 6, 1–10. [Google Scholar] [CrossRef]
- Bian, Z.; Yan, Z.-Q.; Hansson, G.K.; Thorén, P.; Normark, S. Activation of Inducible Nitric Oxide Synthase/Nitric Oxide by Curli Fibers Leads to a Fall in Blood Pressure during Systemic Escherichia Coli Infection in Mice. J. Infect. Dis. 2001, 183, 612–619. [Google Scholar] [CrossRef] [Green Version]
- Brindhadevi, K.; LewisOscar, F.; Mylonakis, E.; Shanmugam, S.; Verma, T.N.; Pugazhendhi, A. Biofilm and Quorum Sensing Mediated Pathogenicity in Pseudomonas Aeruginosa. Process Biochem. 2020, 96, 49–57. [Google Scholar] [CrossRef]
- Surgers, L.; Boyd, A.; Girard, P.-M.; Arlet, G.; Decré, D. Biofilm Formation by ESBL-Producing Strains of Escherichia Coli and Klebsiella Pneumoniae. Int. J. Med. Microbiol. 2019, 309, 13–18. [Google Scholar] [CrossRef]
- Uruén, C.; Chopo-Escuin, G.; Tommassen, J.; Mainar-Jaime, R.C.; Arenas, J. Biofilms as Promoters of Bacterial Antibiotic Resistance and Tolerance. Antibiotics 2021, 10, 3. [Google Scholar] [CrossRef] [PubMed]
- Kim, N.-N.; Kim, W.J.; Kang, S.-S. Anti-Biofilm Effect of Crude Bacteriocin Derived from Lactobacillus Brevis DF01 on Escherichia Coli and Salmonella Typhimurium. Food Control 2019, 98, 274–280. [Google Scholar] [CrossRef]
- Shangguan, W.; Xie, T.; Zhang, R.; Lu, C.; Han, X.; Zhong, Q. Anti-Biofilm Potential of Kefir-Derived Lactobacillus paracasei L10 against Vibrio Parahaemolyticus. Lett. Appl. Microbiol. 2021, 73, 750–758. [Google Scholar] [CrossRef] [PubMed]




| Characterization | Parameter | K02 | K03 |
|---|---|---|---|
| Nutritional | Lipid (% w/w) | 3.41 ± 0.10 a | 3.15 ± 0.08 b |
| Protein (% w/w) | 3.06 ± 0.06 a | 3.07 ± 0.08 a | |
| Dietary fiber (% w/w) | 1.71 ± 0.24 a | 1.28 ± 0.18 a | |
| Carbohydrates (% w/w) | 1.95 ± 0.07 a | 2.61 ± 0.16 b | |
| Energy (kcal/100 g) | 50.76 a | 51.07 a | |
| Acetic acid (g/L) | 2.71 ± 1.01 a | 1.78 ± 0.32 b | |
| Butyric acid (g/L) | 0.0 ± 0.0 a | 0.0 ± 0.0 a | |
| Lactic acid (g/L) | 6.96 ± 0.55 a | 6.45 ± 0.60 a | |
| Propionic acid (g/L) | 0.59 ± 0.20 a | 0.57 ± 0.15 a | |
| Microbiological | Total microorganism (CFU/mL) | 3 × 109 ± 3 × 108 a | 3 × 109 ± 1 × 108 a |
| AAB (CFU/mL) | 2 × 109 ± 7 × 107 a | 3 × 109 ± 1 × 108 a | |
| LAB (CFU/mL) | 1 × 108 ± 3 × 107 a | 1 × 108 ± 4 × 107 a | |
| Enterobacteria | ND | ND | |
| Yeast (CFU/mL) | 3 × 108 ± 2 × 108 a | 2 × 108 ± 6 × 107 a |
| Enzymatic Activity | K02 | K02W | K03 | K03W |
|---|---|---|---|---|
| Beta-galactosidase | + | − | + | − |
| Arginine dehydrolase | − | − | − | − |
| Lysine decarboxylase | − | − | − | − |
| Ornithine decarboxylase | − | − | − | − |
| Citrate utilization | − | − | − | − |
| H2S production | − | − | − | − |
| Urea hydrolysis | − | − | − | − |
| Deaminase | − | − | − | − |
| Indole production | − | − | − | − |
| Acetoin production | + | + | + | + |
| Gelatinase | − | − | − | − |
| Fermentation/Oxidation of glucose | + | + | + | + |
| Fermentation/Oxidation of mannitol | − | + | − | + |
| Fermentation/Oxidation of inositol | − | + | − | + |
| Fermentation/Oxidation of sorbitol | − | + | − | + |
| Fermentation/Oxidation of rhamnose | − | + | − | + |
| Fermentation/Oxidation of sucrose | − | + | − | + |
| Fermentation/Oxidation of melibiose | − | + | − | + |
| Fermentation/Oxidation of amygdalin | − | + | − | + |
| Fermentation/Oxidation of arabinose | − | + | − | + |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ibacache-Quiroga, C.; González-Pizarro, K.; Charifeh, M.; Canales, C.; Díaz-Viciedo, R.; Schmachtenberg, O.; Dinamarca, M.A. Metagenomic and Functional Characterization of Two Chilean Kefir Beverages Reveals a Dairy Beverage Containing Active Enzymes, Short-Chain Fatty Acids, Microbial β-Amyloids, and Bio-Film Inhibitors. Foods 2022, 11, 900. https://doi.org/10.3390/foods11070900
Ibacache-Quiroga C, González-Pizarro K, Charifeh M, Canales C, Díaz-Viciedo R, Schmachtenberg O, Dinamarca MA. Metagenomic and Functional Characterization of Two Chilean Kefir Beverages Reveals a Dairy Beverage Containing Active Enzymes, Short-Chain Fatty Acids, Microbial β-Amyloids, and Bio-Film Inhibitors. Foods. 2022; 11(7):900. https://doi.org/10.3390/foods11070900
Chicago/Turabian StyleIbacache-Quiroga, Claudia, Karoll González-Pizarro, Mariam Charifeh, Christian Canales, Rodrigo Díaz-Viciedo, Oliver Schmachtenberg, and M. Alejandro Dinamarca. 2022. "Metagenomic and Functional Characterization of Two Chilean Kefir Beverages Reveals a Dairy Beverage Containing Active Enzymes, Short-Chain Fatty Acids, Microbial β-Amyloids, and Bio-Film Inhibitors" Foods 11, no. 7: 900. https://doi.org/10.3390/foods11070900






