Differences between Kazak Cheeses Fermented by Single and Mixed Strains Using Untargeted Metabolomics
Abstract
:1. Introduction
2. Materials and Methods
2.1. Fermented Strains and Cheese Fermentation
2.2. Cheese Preparation
2.3. Measurement of Microbial Population
2.4. Determination of Physicochemical Properties
2.5. Determination of Free Amino Acids
2.6. Determination of Volatile Compounds
2.7. Determination of Nonvolatile Compounds
2.8. Statistical Analysis
3. Results and Discussion
3.1. Microbial Analysis of Cheese
3.2. Analysis of Physicochemical Properties of Cheese
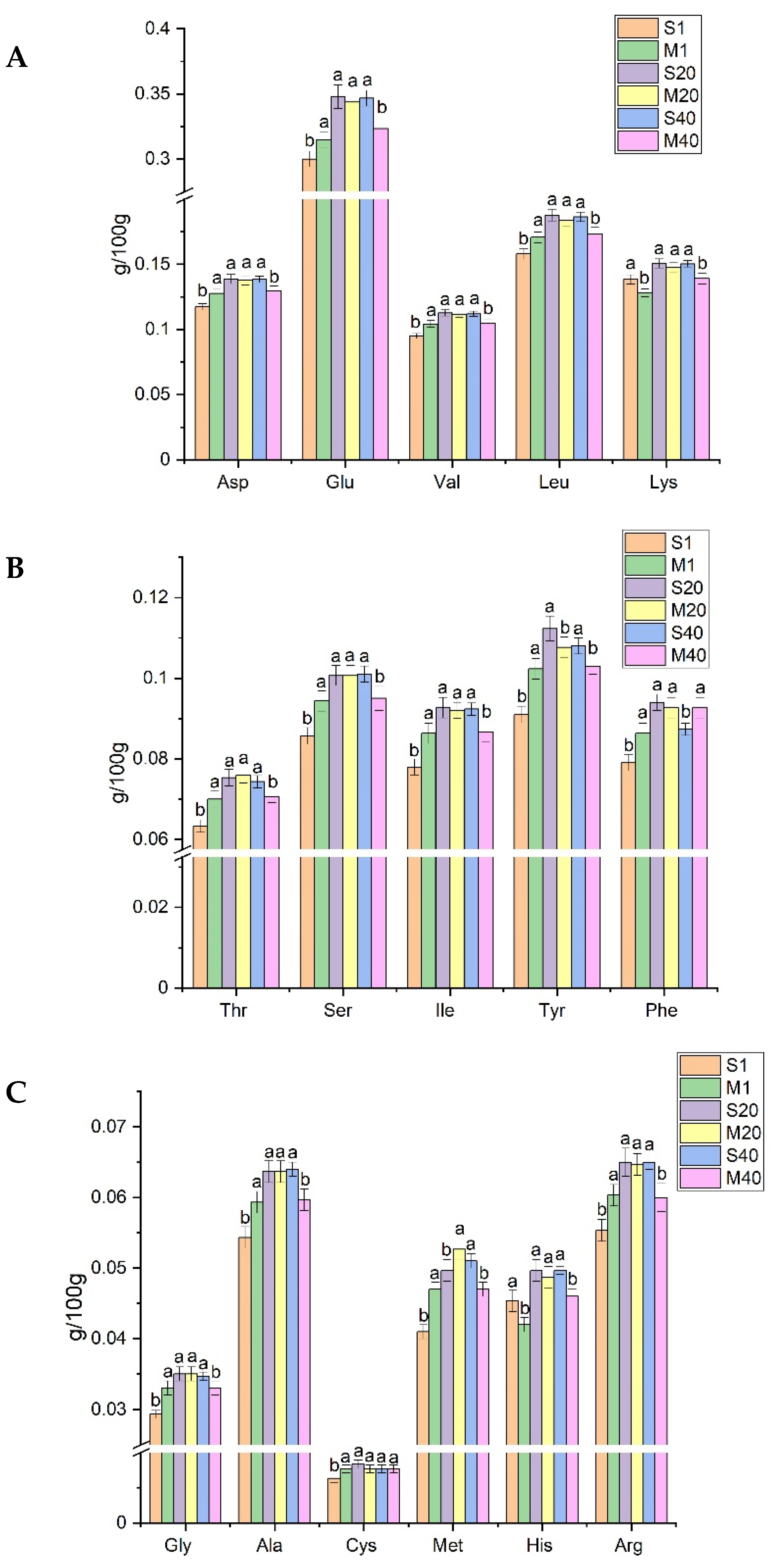
3.3. Analysis of FAA in Cheese
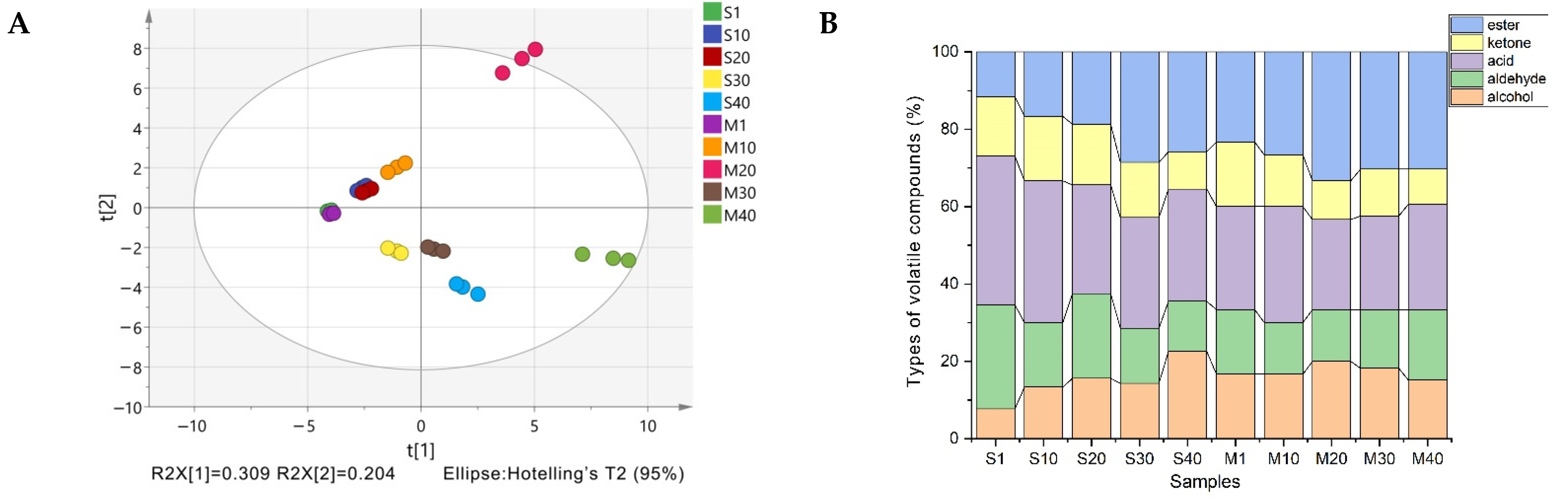
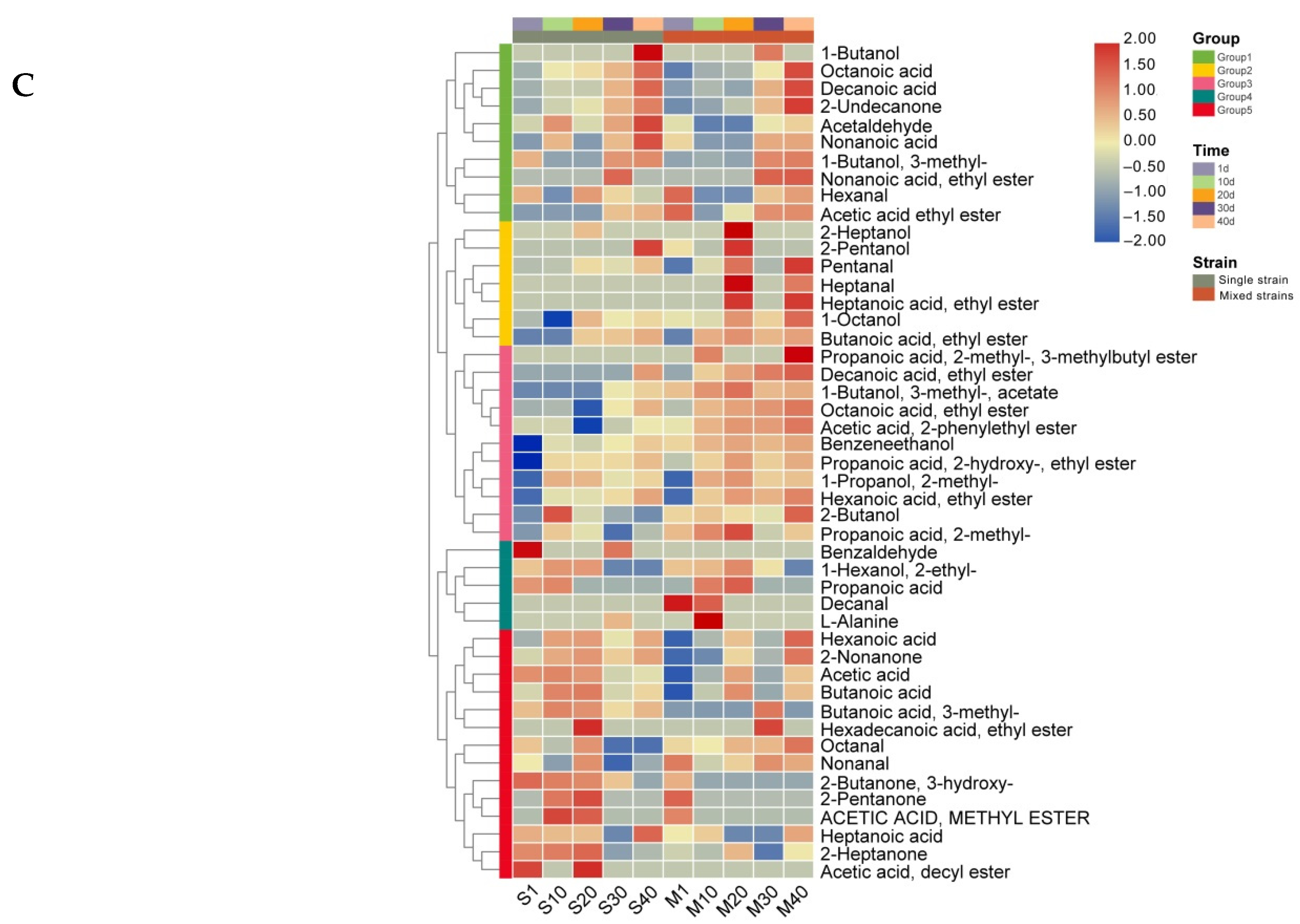
3.4. Analysis of Volatile Compounds in Cheese
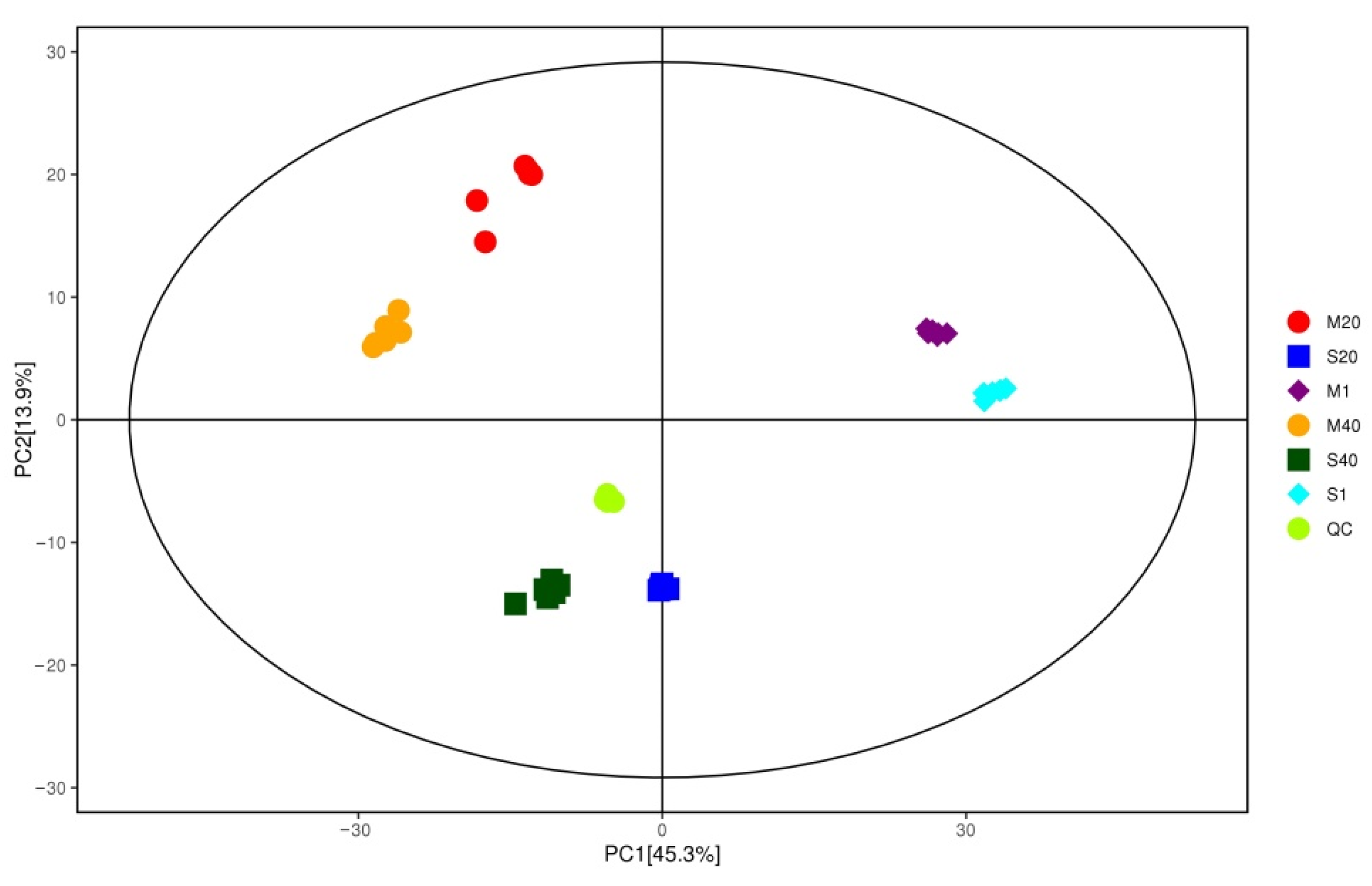
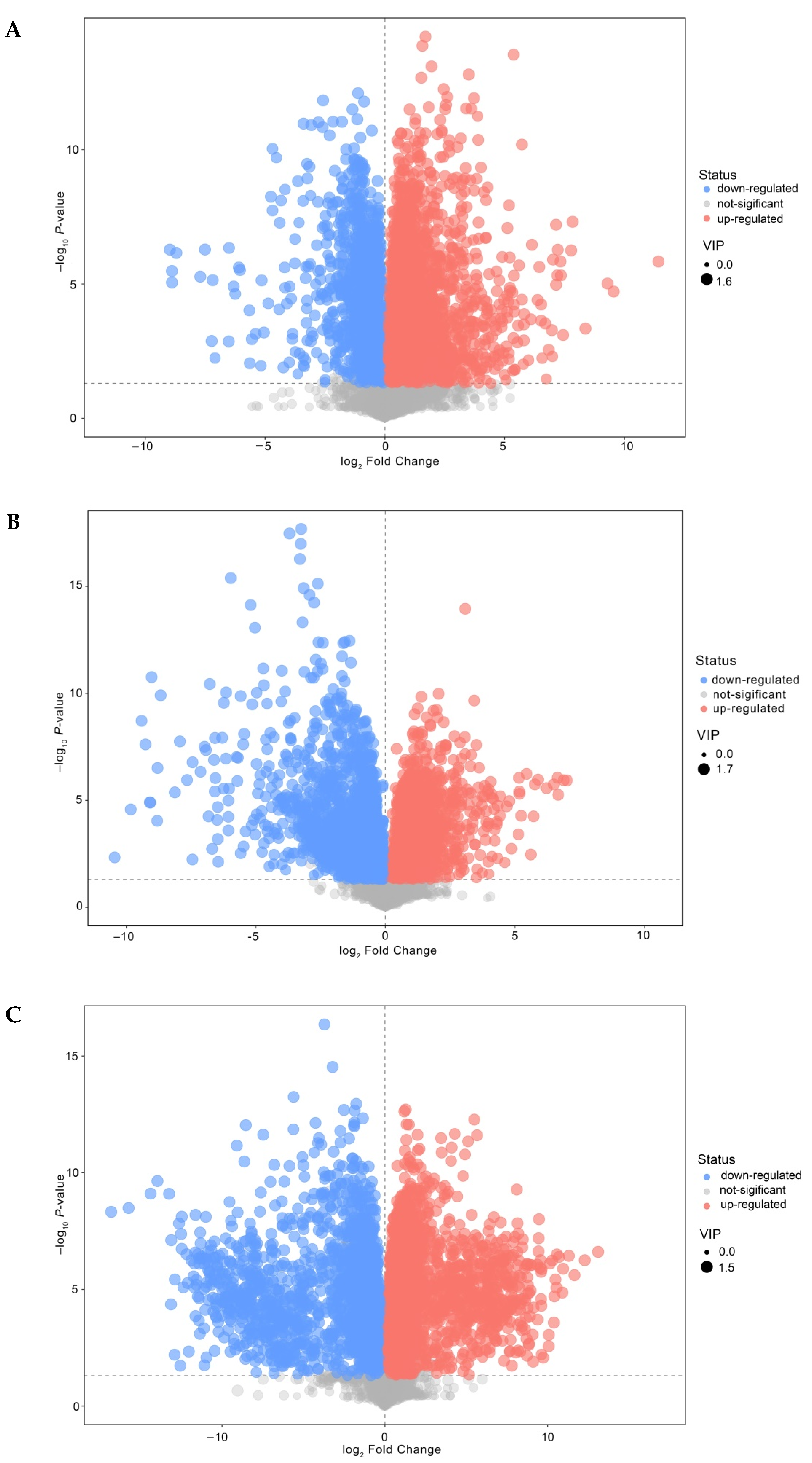
3.5. Analysis of Differential Nonvolatile Compounds in Cheese
3.6. Enrichment Analysis of Differential Metabolites
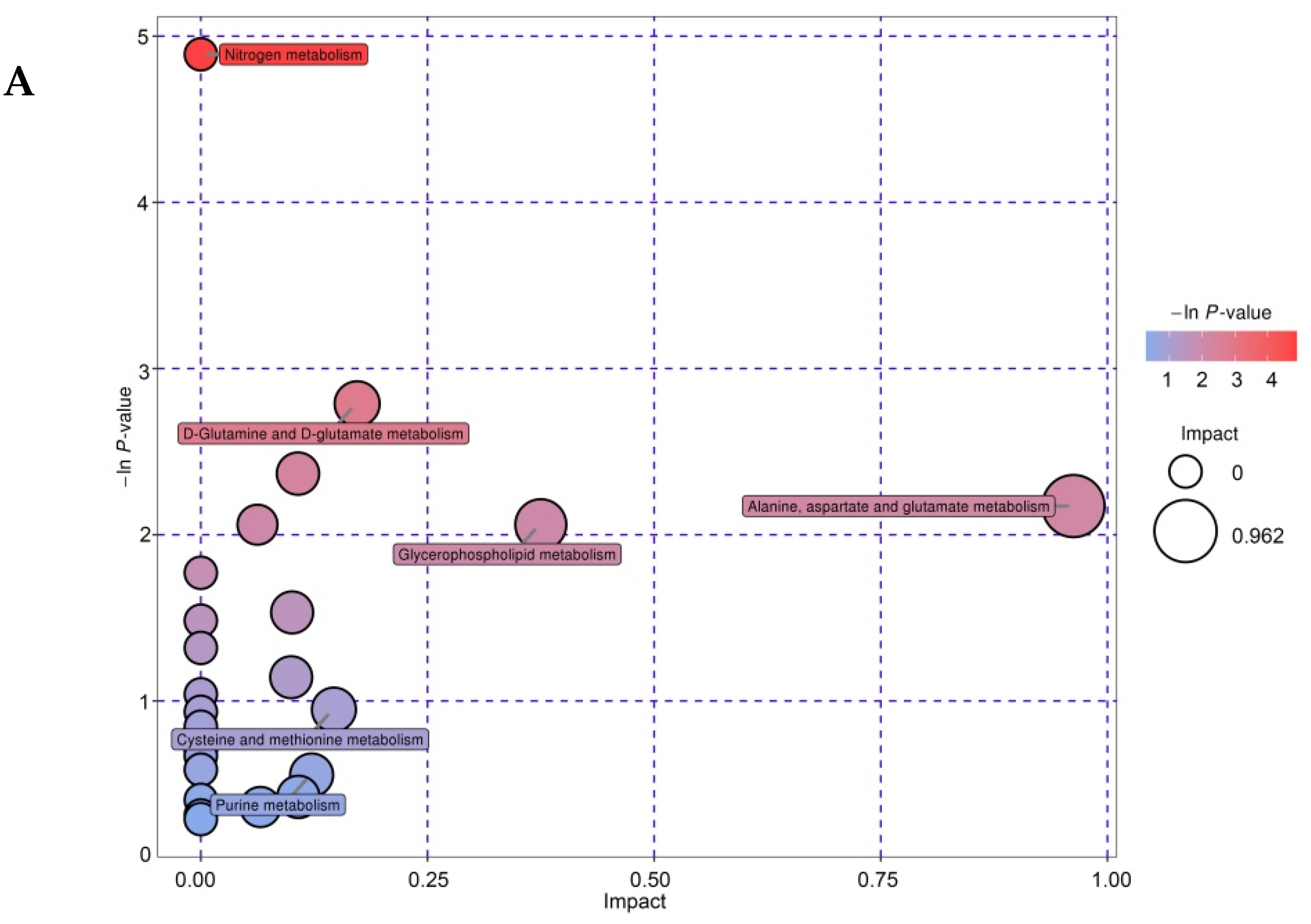
3.7. Metabolic Pathway Analysis of Differential Metabolites
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Xu, Z.; Chen, J.; Shi, X.; Wang, B.; Zheng, X.; Zheng, X. Characteristic physicochemical indexes and flavor compounds in Xinjiang Kazak cheese during ripening. Food Biosci. 2020, 35, 100586. [Google Scholar] [CrossRef]
- Li, J.; Huang, Q.; Zheng, X.; Ge, Z.; Lin, K.; Zhang, D.; Chen, Y.; Wang, B.; Shi, X. Investigation of the Lactic Acid Bacteria in Kazak Cheese and Their Contributions to Cheese Fermentation. Front. Microbiol. 2020, 11, 228. [Google Scholar] [CrossRef] [PubMed]
- Zheng, X.; Liu, F.; Shi, X.; Wang, B.; Li, K.; Li, B.; Zhuge, B. Dynamic correlations between microbiota succession and flavor development involved in the ripening of Kazak artisanal cheese. Food Res. Int. 2018, 105, 733–742. [Google Scholar] [CrossRef] [PubMed]
- Li, S.; Li, Y.; Du, Z.; Li, B.; Liu, Y.; Gao, Y.; Zhang, Y.; Zhang, K.; Wang, Q.; Lu, S.; et al. Impact of NSLAB on Kazakh cheese flavor. Food Res. Int. 2021, 144, 110315. [Google Scholar] [CrossRef]
- Peralta, G.H.; Wolf, I.V.; Perotti, M.C.; Bergamini, C.V.; Hynes, E.R. Formation of volatile compounds, peptidolysis and carbohydrate fermentation by mesophilic lactobacilli and streptoccocci cultures in a cheese extract. Dairy Sci. Technol. 2016, 96, 1–19. [Google Scholar] [CrossRef] [Green Version]
- Leeuwendaal, N.; Stanton, C.; O’Toole, P.W.; Beresford, T.P. The potential of non-starter lactic acid bacteria from Cheddar cheese to colonise the gut. J. Funct. Foods 2021, 83, 104425. [Google Scholar] [CrossRef]
- Liu, N.; Qin, L.; Mazhar, M.; Miao, S. Integrative transcriptomic-proteomic analysis revealed the flavor formation mechanism and antioxidant activity in rice-acid inoculated with Lactobacillus paracasei and Kluyveromyces marxianus. J. Proteom. 2021, 238, 104158. [Google Scholar] [CrossRef]
- Bartkiene, E.; Zavistanaviciute, P.; Lele, V.; Ruzauskas, M.; Bartkevics, V.; Bernatoniene, J.; Gallo, P.; Tenore, G.C.; Santini, A. Lactobacillus plantarum LUHS135 and paracasei LUHS244 as functional starter cultures for the food fermentation industry: Characterisation, mycotoxin-reducing properties, optimisation of biomass growth and sustainable encapsulation by using dairy by-products. LWT-Food Sci. Technol. 2018, 93, 649–658. [Google Scholar]
- McSweeney, P.L.H. Biochemistry of cheese ripening. Int. J. Dairy Technol. 2004, 57, 127–144. [Google Scholar] [CrossRef]
- Binetti, A.; Carrasco, M.; Reinheimer, J.; Suárez, V. Yeasts from autochthonal cheese starters: Technological and functional properties. J. Appl. Microbiol. 2013, 115, 434–444. [Google Scholar] [CrossRef]
- Fröhlich-Wyder, M.A.-O.; Arias-Roth, E.; Jakob, E. Cheese yeasts. Yeast 2019, 3, 129–141. [Google Scholar] [CrossRef] [PubMed]
- Zheng, X.; Liu, F.; Li, K.; Shi, X.; Ni, Y.; Li, B.; Zhuge, B. Evaluating the microbial ecology and metabolite profile in Kazak artisanal cheeses from Xinjiang, China. Food Res. Int. 2018, 111, 130–136. [Google Scholar] [CrossRef] [PubMed]
- Dank, A.; van Mastrigt, O.; Yang, Z.; Dinesh, V.M.; Lillevang, S.K.; Weij, C.; Smid, E.J. The cross-over fermentation concept and its application in a novel food product: The dairy miso case study. LWT-Food Sci. Technol. 2021, 142, 111041. [Google Scholar] [CrossRef]
- McSweeney, P.L.H. Ripening. In Encyclopedia of Dairy Sciences, 2nd ed.; Fuquay, J.W., Ed.; Academic Press: San Diego, CA, USA, 2011; pp. 667–674. [Google Scholar]
- Zheng, X.; Shi, X.; Wang, B. A Review on the General Cheese Processing Technology, Flavor Biochemical Pathways and the Influence of Yeasts in Cheese. Front. Microbiol. 2021, 2186, 703284. [Google Scholar] [CrossRef] [PubMed]
- Yvon, M.; Rijnen, L. Cheese flavour formation by amino acid catabolism. Int. Dairy J. 2001, 11, 185–201. [Google Scholar] [CrossRef]
- Adesulu-Dahunsi, A.T.; Dahunsi, S.O.; Olayanju, A. Synergistic microbial interactions between lactic acid bacteria and yeasts during production of Nigerian indigenous fermented foods and beverages. Food Control. 2020, 110, 106963. [Google Scholar] [CrossRef] [Green Version]
- Liu, N.; Miao, S.; Qin, L. Screening and application of lactic acid bacteria and yeasts with l-lactic acid-producing and antioxidant capacity in traditional fermented rice acid. Food Sci. Nutr. 2020, 8, 6095–6111. [Google Scholar] [CrossRef] [PubMed]
- Luan, C.; Zhang, M.; Devahastin, S.; Liu, Y. Effect of two-step fermentation with lactic acid bacteria and Saccharomyces cerevisiae on key chemical properties, molecular structure and flavor characteristics of horseradish sauce. LWT-Food Sci. Technol. 2021, 147, 111637. [Google Scholar] [CrossRef]
- Boudaoud, S.; Aouf, C.; Devillers, H.; Sicard, D.; Segond, D. Sourdough yeast-bacteria interactions can change ferulic acid metabolism during fermentation. Food Microbiol. 2021, 98, 103790. [Google Scholar] [CrossRef]
- Zhang, Z.; Lan, Q.; Yu, Y.; Zhou, J.; Lu, H. Comparative metabolome and transcriptome analyses of the properties of Kluyveromyces marxianus and Saccharomyces yeasts in apple cider fermentation. Food Chem. Mol. Sci. 2022, 4, 100095. [Google Scholar] [CrossRef]
- Khattab, A.R.; Guirguis, H.A.; Tawfik, S.M.; Farag, M.A. Cheese ripening: A review on modern technologies towards flavor enhancement, process acceleration and improved quality assessment. Trends Food Sci. Technol. 2019, 88, 343–360. [Google Scholar] [CrossRef]
- Boccard, J.; Rutledge, D.N. A consensus orthogonal partial least squares discriminant analysis (OPLS-DA) strategy for multiblock Omics data fusion. Anal. Chim. Acta 2013, 769, 30–39. [Google Scholar] [CrossRef] [PubMed]
- Dos Santos, F.A.; Sousa, I.P.; Furtado, N.A.J.C.; Da Costa, F.B. Combined OPLS-DA and decision tree as a strategy to identify antimicrobial biomarkers of volatile oils analyzed by gas chromatography–mass spectrometry. Rev. Bras. Farmacogn. 2018, 28, 647–653. [Google Scholar] [CrossRef]
- Mozzi, F.; Ortiz, M.E.; Bleckwedel, J.; De Vuyst, L.; Pescuma, M. Metabolomics as a tool for the comprehensive understanding of fermented and functional foods with lactic acid bacteria. Food Res. Int. 2013, 54, 1152–1161. [Google Scholar] [CrossRef]
- Zheng, X.; Ge, Z.; Lin, K.; Zhang, D.; Chen, Y.; Xiao, J.; Wang, B.; Shi, X. Dynamic changes in bacterial microbiota succession and flavour development during milk fermentation of Kazak artisanal cheese. Int. Dairy J. 2021, 113, 104878. [Google Scholar] [CrossRef]
- Afshari, R.; Pillidge, C.J.; Dias, D.A.; Osborn, A.M.; Gill, H. Biomarkers associated with cheese quality uncovered by integrative multi-omic analysis. Food Control. 2021, 123, 107752. [Google Scholar] [CrossRef]
- Suzuki-Iwashima, A.; Matsuura, H.; Iwasawa, A.; Shiota, M. Metabolomics analyses of the combined effects of lactic acid bacteria and Penicillium camemberti on the generation of volatile compounds in model mold-surface-ripened cheeses. J. Biosci. Bioeng. 2020, 129, 333–347. [Google Scholar] [CrossRef]
- Xiao, J.; Chen, Y.; Li, J.; Shi, X.; Deng, L.; Wang, B. Evaluation of the Effect of Auxiliary Starter Yeasts With Enzyme Activities on Kazak Cheese Quality and Flavor. Front. Microbiol. 2020, 11, 614208. [Google Scholar] [CrossRef]
- Buňka, F.; Kříž, O.; Veličková, A.; Buňková, L.; Kráčmar, S. Effect of acid hydrolysis time on amino acid determination in casein and processed cheeses with different fat content. J. Food Compos. Anal. 2009, 22, 224–232. [Google Scholar] [CrossRef]
- Song, H.; Liu, J. GC-O-MS technique and its applications in food flavor analysis. Food Res. Int. 2018, 114, 187–198. [Google Scholar] [CrossRef]
- Liu, Q.; Li, B.; Li, Y.; Wei, Y.; Huang, B. Altered faecal microbiome and metabolome in IgG4-related sclerosing cholangitis and primary sclerosing cholangitis. Gut Microbiota 2021, 1–11. [Google Scholar] [CrossRef] [PubMed]
- Cai, Y.; Weng, K.; Guo, Y.; Peng, J.; Zhu, Z.-J. An integrated targeted metabolomic platform for high-throughput metabolite profiling and automated data processing. Metabolomics 2015, 11, 1575–1586. [Google Scholar] [CrossRef]
- Fleet, G.H. Yeasts in dairy products. J. Appl. Bacteriol. 1990, 68, 199–211. [Google Scholar] [CrossRef] [PubMed]
- De Freitas, I.; Pinon, N.; Berdagué, J.L.; Tournayre, P.; Lortal, S.; Thierry, A. Kluyveromyces lactis but Not Pichia fermentans Used as Adjunct Culture Modifies the Olfactory Profiles of Cantalet Cheese. J. Dairy Sci. 2008, 91, 531–543. [Google Scholar] [CrossRef]
- Liu, S.-Q.; Tsao, M. Enhancement of survival of probiotic and non-probiotic lactic acid bacteria by yeasts in fermented milk under non-refrigerated conditions. Int. J. Food Microbiol. 2009, 135, 34–38. [Google Scholar] [CrossRef]
- Liu, N.; Qin, L.; Lu, X.; Zhao, Y.; Miao, S. Physicochemical components and flavor properties of acid rice soup (rice-acid) fermented with Lactobacillus paracasei and/or Kluyveromyces marxianus. Food Biosci. 2021, 43, 101278. [Google Scholar] [CrossRef]
- Fernández-Salguero, J.; Sanjuán, E. Influence of vegetable and animal rennet on proteolysis during ripening in ewes’ milk cheese. Food Chem. 1999, 64, 177–183. [Google Scholar] [CrossRef]
- Su, Y.; Seguinot, P.; Sanchez, I.; Ortiz-Julien, A.; Heras, J.M.; Querol, A.; Camarasa, C.; Guillamón, J.M. Nitrogen sources preferences of non-Saccharomyces yeasts to sustain growth and fermentation under winemaking conditions. Food Microbiol. 2020, 85, 103287. [Google Scholar] [CrossRef]
- Gobbetti, M.; Corsetti, A.; Rossi, J. The sourdough microflora. Interactions between lactic acid bacteria and yeasts: Metabolism of carbohydrates. Appl. Microbiol. Biotechnol. 1994, 41, 456–460. [Google Scholar]
- Öründü, S.; Tarakçı, Z. Effects of different starter culture applications pre- and post-scalding on the biochemical and sensory properties of pasta filata type cheeses. LWT-Food Sci. Technol. 2021, 136, 110288. [Google Scholar] [CrossRef]
- Diezhandino, I.; Fernández, D.; González, L.; McSweeney, P.L.H.; Fresno, J.M. Microbiological, physico-chemical and proteolytic changes in a Spanish blue cheese during ripening (Valdeón cheese). Food Chem. 2015, 168, 134–141. [Google Scholar] [CrossRef] [PubMed]
- Ozturkoglu-Budak, S.; Gursoy, A.; Aykas, D.P.; Koçak, C.; Dönmez, S.; de Vries, R.P.; Bron, P.A. Volatile compound profiling of Turkish Divle Cave cheese during production and ripening. J. Dairy Sci. 2016, 99, 5120–5131. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, L.; Huang, C.; Johansen, P.G.; Petersen, M.A.; Poojary, M.M.; Lund, M.N.; Jespersen, L.; Arneborg, N. The utilisation of amino acids by Debaryomyces hansenii and Yamadazyma triangularis associated with cheese. Int. Dairy J. 2021, 121, 105135. [Google Scholar] [CrossRef]
- Ardö, Y. Flavour formation by amino acid catabolism. Biotechnol. Adv. 2006, 24, 238–242. [Google Scholar] [CrossRef] [PubMed]
- Zhao, C.J.; Schieber, A.; Gänzle, M.G. Formation of taste-active amino acids, amino acid derivatives and peptides in food fermentations–A review. Food Res. Int. 2016, 89, 39–47. [Google Scholar] [CrossRef]
- Ganesan, B.; Weimer, B.C. 4-Amino acid metabolism in relationship to cheese flavor development. In Improving the Flavour of Cheese; Weimer, B.C., Ed.; Woodhead Publishing: Cambridge, UK, 2007; pp. 70–101. [Google Scholar]
- Hazelwood, L.; Daran, J.-M.; Van Maris, A.; Pronk, J.; Dickinson, J. The Ehrlich Pathway for Fusel Alcohol Production: A Century of Research on Saccharomyces cerevisiae Metabolism. Appl. Environ. Microbiol. 2008, 74, 2259–2266. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Swiegers, J.H.; Pretorius, I.S. Yeast Modulation of Wine Flavor. Adv. Appl. Microbiol. 2005, 57, 131–175. [Google Scholar]
- Dickinson, J.R.; Lanterman, M.M.; Danner, D.J.; Pearson, B.M.; Sanz, P.; Harrison, S.J.; Hewlins, M.J.E. A 13C Nuclear Magnetic Resonance Investigation of the Metabolism of Leucine to Isoamyl Alcohol in Saccharomyces cerevisiae. J. Biol. Chem. 1997, 272, 26871–26878. [Google Scholar] [CrossRef] [Green Version]
- Seguinot, P.; Bloem, A.; Brial, P.; Meudec, E.; Ortiz-Julien, A.; Camarasa, C. Analysing the impact of the nature of the nitrogen source on the formation of volatile compounds to unravel the aroma metabolism of two non-Saccharomyces strains. Int. J. Food Microbiol. 2020, 316, 108441. [Google Scholar] [CrossRef]
- Zheng, X. Analysis of the Microbial Flora Structure and Characteristic Flavor of Xinjiang Kazakh Cheese. Ph.D Thesis, Jiangnan University, Wuxi, China, 2018. [Google Scholar]
- Duval, P.; Chatelard-Chauvin, C.; Gayard, C.; Rifa, E.; Bouchard, P.; Hulin, S.; Delile, A.; Pollet, B.; Montel, M.C.; Picque, D. Changes in biochemical and sensory parameters in industrial blue-veined cheeses in different packaging. Int. Dairy J. 2018, 77, 89–99. [Google Scholar] [CrossRef]
- Agarwal, D.; Mui, L.; Aldridge, E.; Mottram, R.; McKinney, J.; Fisk, I. The impact of nitrogen gas flushing on the stability of seasonings: Volatile compounds and sensory perception of cheese & onion seasoned potato crisps. Food Funct. 2018, 9, 4730–4741. [Google Scholar] [PubMed] [Green Version]
- Nzekoue, F.K.; Caprioli, G.; Fiorini, D.; Torregiani, E.; Vittori, S.; Sagratini, G. HS-SPME-GC-MS technique for FFA and hexanal analysis in different cheese packaging in the course of long term storage. Food Res. Int. 2019, 121, 730–737. [Google Scholar] [CrossRef] [PubMed]
- Delgado, F.J.; González-Crespo, J.; Cava, R.; García-Parra, J.; Ramírez, R. Characterisation by SPME–GC–MS of the volatile profile of a Spanish soft cheese P.D.O. Torta del Casar during ripening. Food Chem. 2010, 118, 182–189. [Google Scholar]
- Hanko, E.K.R.; Denby, C.M.; Nogué, V.S.I.; Lin, W.; Ramirez, K.J.; Singer, C.A.; Beckham, G.T.; Keasling, J.D. Engineering β-oxidation in Yarrowia lipolytica for methyl ketone production. Metab. Eng. 2018, 48, 52–62. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Boltar, I.; Majhenič, A.Č.; Jarni, K.; Jug, T.; Kralj, M.B. Erratum to: Volatile compounds in Nanos cheese: Their formation during ripening and seasonal variation. J. Food Sci. Technol. 2015, 52, 624. [Google Scholar] [CrossRef] [Green Version]
- Gan, H.H.; Yan, B.; Linforth, R.S.T.; Fisk, I.D. Development and validation of an APCI-MS/GC–MS approach for the classification and prediction of Cheddar cheese maturity. Food Chem. 2016, 190, 442–447. [Google Scholar] [CrossRef]
- Ianni, A.; Bennato, F.; Martino, C.; Grotta, L.; Martino, G. Volatile Flavor Compounds in Cheese as Affected by Ruminant Diet. Molecules 2020, 25, 461. [Google Scholar] [CrossRef] [Green Version]
- De Jesus Filho, M.; Klein, B.; Wagner, R.; Godoy, H.T. Key aroma compounds of Canastra cheese: HS-SPME optimization assisted by olfactometry and chemometrics. Food Res. Int. 2021, 150, 110788. [Google Scholar] [CrossRef]
- Zheng, X.; Liu, F.; Ren, Q.; Li, B.; Li, K.; Zhu, G. Based on SPME-GC-MS method to compare the cheese flavor differences in different residential areas of Kazakhs in Xinjiang. Food Sci. 2018, 39, 83–89. [Google Scholar]
- McSweeney, P.; Sousa-Gallagher, M.J. Biochemical pathways for the production of flavour compounds in cheeses during ripening: A review. Dairy Sci. Technol. 2000, 80, 293–324. [Google Scholar] [CrossRef]
- Hong, Q.; Liu, X.M.; Hang, F.; Zhao, J.X.; Zhang, H.; Chen, W. Screening of adjunct cultures and their application in ester formation in Camembert-type cheese. Food Microbiol. 2018, 70, 33–41. [Google Scholar] [CrossRef] [PubMed]
- Procopio, S.; Krause, D.; Hofmann, T.; Becker, T. Significant amino acids in aroma compound profiling during yeast fermentation analyzed by PLS regression. LWT-Food Sci. Technol. 2013, 51, 423–432. [Google Scholar] [CrossRef]
- Molimard, P.; Spinnler, H.E. Review: Compounds Involved in the Flavor of Surface Mold-Ripened Cheeses: Origins and Properties. J. Dairy Sci. 1996, 79, 169–184. [Google Scholar] [CrossRef]
- Padilla, B.; Belloch, C.; López-Díez, J.J.; Flores, M.; Manzanares, P. Potential impact of dairy yeasts on the typical flavour of traditional ewes’ and goats’ cheeses. Int. Dairy J. 2014, 35, 122–129. [Google Scholar] [CrossRef] [Green Version]
- Plutowska, B.; Wardencki, W. Aromagrams–Aromatic profiles in the appreciation of food quality. Food Chem. 2007, 101, 845–872. [Google Scholar] [CrossRef]
- Verstrepen, K.J.; Derdelinckx, G.; Dufour, J.-P.; Winderickx, J.; Thevelein, J.M.; Pretorius, I.S.; Delvaux, F.R. Flavor-active esters: Adding fruitiness to beer. J. Biosci. Bioeng. 2003, 96, 110–118. [Google Scholar] [CrossRef]
- Leng, P.; Hu, H.-W.; Cui, A.-H.; Tang, H.-J.; Liu, Y.-G. HS-GC-IMS with PCA to analyze volatile flavor compounds of honey peach packaged with different preservation methods during storage. LWT-Food Sci. Technol. 2021, 149, 111963. [Google Scholar] [CrossRef]
- Savijoki, K.; Ingmer, H.; Varmanen, P. Proteolytic systems of lactic acid bacteria. Appl. Microbiol. Biotechnol. 2006, 71, 394–406. [Google Scholar] [CrossRef]
- Balthazar, C.F.; Guimarães, J.T.; Rocha, R.S.; Neto, R.P.C.; Esmerino, E.A.; Silva, H.L.A.; Filho, E.G.A.; Pimentel, T.C.; Rodrigues, S.; Brito, E.S.; et al. Metabolic profiling of probiotic low-sodium prato cheese with flavour enhancers: Usefulness of NMR spectroscopy and chemometric tools. Int. Dairy J. 2021, 119, 104992. [Google Scholar] [CrossRef]
- Wang, W.; Zhou, X.; Liu, Y. Characterization and evaluation of umami taste: A review. TrAC Trends Anal. Chem. 2020, 127, 115876. [Google Scholar] [CrossRef]
- Konings, W.N. The cell membrane and the struggle for life of lactic acid bacteria. Antonie Van Leeuwenhoek 2002, 82, 3–27. [Google Scholar] [CrossRef] [PubMed]
- Li, Q.; Tao, Q.; Teixeira, J.S.; Su, M.S.-W.; Gänzle, M.G. Contribution of glutaminases to glutamine metabolism and acid resistance in Lactobacillus reuteri and other vertebrate host adapted lactobacilli. Food Microbiol. 2020, 86, 103343. [Google Scholar] [CrossRef] [PubMed]
- Su, M.S.; Schlicht, S.; Gnzle, M.G. Contribution of glutamate decarboxylase in Lactobacillus reuteri to acid resistance and persistence in sourdough fermentation. Microb. Cell Fact. 2011, 10, 1–12. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Teixeira, J.S.; Seeras, A.; Sanchez-Maldonado, A.F.; Zhang, C.; Su, M.S.-W.; Gänzle, M.G. Glutamine, glutamate, and arginine-based acid resistance in Lactobacillus reuteri. Food Microbiol. 2014, 42, 172–180. [Google Scholar] [CrossRef] [PubMed]
- Gummalla, S.; Broadbent, J.R. Tyrosine and Phenylalanine Catabolism by Lactobacillus Cheese Flavor Adjuncts1. J. Dairy Sci. 2001, 84, 1011–1019. [Google Scholar] [CrossRef]
- Padilla, B.; Gil, J.V.; Manzanares, P. Past and Future of Non-Saccharomyces Yeasts: From Spoilage Microorganisms to Biotechnological Tools for Improving Wine Aroma Complexity. Front. Microbiol. 2016, 7, 411. [Google Scholar] [CrossRef] [Green Version]
- Rollero, S.; Bloem, A.; Brand, J.; Ortiz-Julien, A.; Camarasa, C.; Divol, B. Nitrogen metabolism in three non-conventional wine yeast species: A tool to modulate wine aroma profiles. Food Microbiol. 2021, 94, 103650. [Google Scholar] [CrossRef]





| Moisture (%) | pH | Protein (%) | Salt (%) | |
|---|---|---|---|---|
| S1 | 51.24 ± 0.57 Aa | 4.02 ± 0.02 Aa | 18.43 ± 0.18 Aa | 0.76 ± 0.02 Aa |
| S10 | 51.76 ± 0.33 Aa | 3.92 ± 0.01 Ab | 18.47 ± 0.17 Aa | 0.79 ± 0.03 Aa |
| S20 | 51.5 ± 0.51 Aa | 3.93 ± 0.02 Ab | 18.70 ± 0.26 Aa | 0.81 ± 0.02 Aa |
| S30 | 50.75 ± 0.48 Aa | 3.93 ± 0.01 Ab | 18.17 ± 0.28 Aa | 0.82 ± 0.02 Aa |
| S40 | 50.25 ± 0.38 Aa | 3.92 ± 0.01 Ab | 18.79 ± 0.16 Aa | 0.83 ± 0.02 Aa |
| M1 | 51.26 ± 0.33 Aa | 3.94 ± 0.01 Ba | 17.74 ± 0.15 Aa | 0.75 ± 0.03 Aa |
| M10 | 51.06 ± 0.63 Aa | 3.90 ± 0.02 Aa | 18.13 ± 0.16 Aa | 0.77 ± 0.03 Aa |
| M20 | 50.5 ± 0.52 Aa | 3.94 ± 0.02 Aa | 18.04 ± 0.19 Aa | 0.80 ± 0.02 Aa |
| M30 | 50.74 ± 0.36 Aa | 3.92 ± 0.01 Aa | 18.1 ± 0.15 Aa | 0.79 ± 0.02 Aa |
| M40 | 50.37 ± 0.44 Aa | 3.94 ± 0.02 Aa | 18.38 ± 0.14 Aa | 0.81 ± 0.03 Aa |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Li, Y.; Wang, J.; Wang, T.; Lv, Z.; Liu, L.; Wang, Y.; Li, X.; Fan, Z.; Li, B. Differences between Kazak Cheeses Fermented by Single and Mixed Strains Using Untargeted Metabolomics. Foods 2022, 11, 966. https://doi.org/10.3390/foods11070966
Li Y, Wang J, Wang T, Lv Z, Liu L, Wang Y, Li X, Fan Z, Li B. Differences between Kazak Cheeses Fermented by Single and Mixed Strains Using Untargeted Metabolomics. Foods. 2022; 11(7):966. https://doi.org/10.3390/foods11070966
Chicago/Turabian StyleLi, Yandie, Jianghan Wang, Tong Wang, Zhuoxia Lv, Linting Liu, Yuping Wang, Xu Li, Zhexin Fan, and Baokun Li. 2022. "Differences between Kazak Cheeses Fermented by Single and Mixed Strains Using Untargeted Metabolomics" Foods 11, no. 7: 966. https://doi.org/10.3390/foods11070966
APA StyleLi, Y., Wang, J., Wang, T., Lv, Z., Liu, L., Wang, Y., Li, X., Fan, Z., & Li, B. (2022). Differences between Kazak Cheeses Fermented by Single and Mixed Strains Using Untargeted Metabolomics. Foods, 11(7), 966. https://doi.org/10.3390/foods11070966





