The Bacterial Microbiota of Edible Insects Acheta domesticus and Gryllus assimilis Revealed by High Content Analysis
Abstract
:1. Introduction
2. Materials and Methods
2.1. Crickets
2.2. Sample Preparation
2.2.1. Sampling of Microorganisms from the Surface of Crickets
2.2.2. Sampling of Microorganisms from the Whole-Body of Crickets
2.3. DNA Extraction
2.4. Amplicon Sequencing
2.5. Processing and Analysis of the Sequencing Data
2.6. Processing of Crickets
2.7. Microbial Analysis of Crickets and Feeding Substrate
2.8. Viable Bacteria Identification
3. Results
3.1. Diversity and Richness of Acheta domesticus and Gryllus assimilis Bacterial Communities
3.2. Bacterial Community Profiling of Jamaican and House Crickets
3.3. Comparison of Jamaican Field Cricket and House Cricket Bacterial Communities
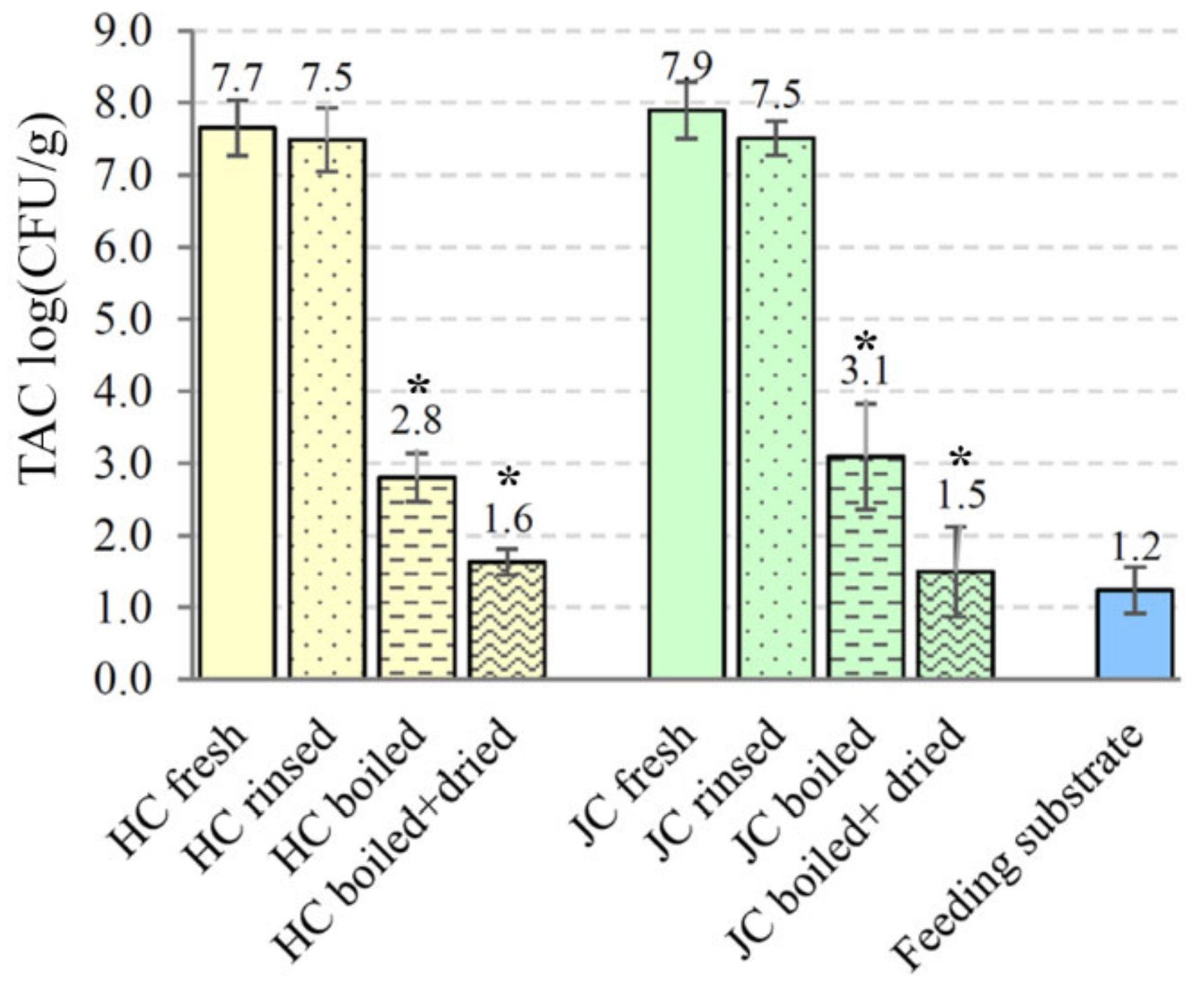
3.4. Microbial Analysis of Crickets and Feeding Substrate
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- United Nations, Department of Economic and Social Affairs, Population Division. World Population Prospects; Highlights (ST/ESA/SER.A/423); United Nations, Department of Economic and Social Affairs, Population Division: New York, NY, USA, 2019. [Google Scholar]
- McLaughlin, D.; Kinzelbach, W. Food security and sustainable resource management. Water Resour. Res. 2015, 51, 4966–4985. [Google Scholar] [CrossRef]
- Magara, H.J.O.; Niassy, S.; Ayieko, M.A.; Mukundamago, M.; Egonyu, J.P.; Tanga, C.M.; Kimathi, E.K.; Ongere, J.O.; Fiaboe, K.K.M.; Hugel, S.; et al. Edible Crickets (Orthoptera) around the World: Distribution, Nutritional Value, and Other Benefits—A Review. Front. Nutr. 2021, 7, 537915. [Google Scholar] [CrossRef] [PubMed]
- Gomiero, T.; Pimentel, D.; Paoletti, M.G. Is There a Need for a More Sustainable Agriculture? Crit. Rev. Plant Sci. 2011, 30, 6–23. [Google Scholar] [CrossRef]
- Garofalo, C.; Osimani, A.; Milanović, V.; Taccari, M.; Cardinali, F.; Aquilanti, L.; Riolo, P.; Ruschioni, S.; Isidoro, N.; Clementi, F. The microbiota of marketed processed edible insects as revealed by high-throughput sequencing. Food Microbiol. 2017, 62, 15–22. [Google Scholar] [CrossRef]
- Godfray, H.C.J.; Beddington, J.R.; Crute, I.R.; Haddad, L.; Lawrence, D.; Muir, J.F.; Pretty, J.; Robinson, S.; Thomas, S.M.; Toulmin, C. Food Security: The Challenge of Feeding 9 Billion People. Science 2010, 327, 812–818. [Google Scholar] [CrossRef] [Green Version]
- Van Huis, A.; Oonincx, D.G.A.B. The environmental sustainability of insects as food and feed. A review. Agron. Sustain. Dev. 2017, 37, 43. [Google Scholar] [CrossRef] [Green Version]
- Van Huis, A. Potential of insects as food and feed in assuring food security. Annu. Rev. Entomol. 2013, 58, 563–583. [Google Scholar] [CrossRef]
- Orkusz, A. Edible insects versus meat—Nutritional comparison: Knowledge of their composition is the key to good health. Nutrients 2021, 13, 1207. [Google Scholar] [CrossRef]
- Oonincx, D.G.A.B.; van Itterbeeck, J.; Heetkamp, M.J.W.; van den Brand, H.; van Loon, J.J.A.; van Huis, A. An exploration on greenhouse gas and ammonia production by insect species suitable for animal or human consumption. PLoS ONE 2010, 5, e14445. [Google Scholar] [CrossRef] [Green Version]
- Poveda, J. Insect frass in the development of sustainable agriculture. A review. Agron. Sustain. Dev. 2021, 41, 5. [Google Scholar] [CrossRef]
- Fowles, T.M.; Nansen, C. Insect-Based Bioconversion: Value from Food Waste. In Food Waste Management; Närvänen, E., Mesiranta, N., Mattila, M., Heikkinen, A., Eds.; Palgrave Macmillan: Cham, Switzerland, 2020; pp. 321–346. [Google Scholar] [CrossRef] [Green Version]
- Candia, F.; Manzanares, A. Insect market outlook: Are insect producers ready to deliver? In The Basics of Edible Insect Rearing—Handbook for the Production Chain, 1st ed.; Veldkamp, T., Claeys, J., Haenen, O.L.M., van Loon, J.J.A., Spranghers, T., Eds.; Wageningen Academic Publishers: Wageningen, The Netherlands, 2022; p. 261. [Google Scholar]
- Gere, A.; Radványi, D.; Héberger, K. Which insect species can best be proposed for human consumption? Innov. Food Sci. Emerg. Technol. 2019, 52, 358–367. [Google Scholar] [CrossRef] [Green Version]
- Da Rosa Machado, K.; Thys, R.C.S. Cricket powder (Gryllus assimilis) as a new alternative protein source for gluten-free breads. Innov. Food Sci. Emerg. Technol. 2019, 56, 102180. [Google Scholar] [CrossRef]
- Khatun, H.; Claes, J.; Smets, R.; de Winne, A.; Akhtaruzzaman, M.; van Der Borght, M. Characterization of freeze-dried, oven-dried and blanched house crickets (Acheta domesticus) and Jamaican field crickets (Gryllus assimilis) by means of their physicochemical properties and volatile compounds. Eur. Food Res. Technol. 2021, 247, 1291–1305. [Google Scholar] [CrossRef]
- Mlček, J.; Adámková, A.; Adámek, M.; Borkovcová, M.; Bednářová, M.; Kouřimská, L. Selected nutritional values of field cricket (Gryllus assimilis) and its possible use as a human food. Indian J. Tradit. Knowl. 2018, 17, 518–524. [Google Scholar]
- Lähteenmäki-Uutela, A.; Marimuthu, S.B.; Meijer, N. Regulations on insects as food and feed: A global comparison. J. Insects Food Feed. 2021, 7, 849–856. [Google Scholar] [CrossRef]
- Van der Fels-Klerx, H.J.; Camenzuli, L.; Belluco, S.; Meijer, N.; Ricci, A. Food safety issues related to uses of insects for feeds and foods. Compr. Rev. Food Sci. Food Saf. 2018, 17, 1172–1183. [Google Scholar] [CrossRef] [Green Version]
- Belluco, S.; Mantovani, A.; Ricci, A. Edible Insects in a Food Safety Perspective. In Edible Insects in Sustainable Food Systems; Halloran, A., Flore, R., Vantomme, P., Roos, N., Eds.; Springer: Cham, Switzerland, 2018; pp. 109–126. [Google Scholar] [CrossRef]
- Fernandez-Cassi, X.; Supeanu, A.; Vaga, M.; Jansson, A.; Boqvist, S.; Vagsholm, I. The house cricket (Acheta domesticus) as a novel food: A risk profile. J. Insects Food Feed. 2019, 5, 137–157. [Google Scholar] [CrossRef]
- EFSA (European Food Safety Authority). Risk profile related to production and consumption of insects as food and feed. EFSA J. 2015, 13, 4257. [Google Scholar] [CrossRef] [Green Version]
- Vandeweyer, D.; Crauwels, S.; Lievens, B.; van Campenhout, L. Microbial counts of mealworm larvae (Tenebrio molitor) and crickets (Acheta domesticus and Gryllodes sigillatus) from different rearing companies and different production batches. Int. J. Food Microbiol. 2017, 242, 13–18. [Google Scholar] [CrossRef]
- Fernandez-Cassi, X.; Söderqvist, K.; Bakeeva, A.; Vaga, M.; Dicksved, J.; Vagsholm, I.; Jansson, A.; Boqvist, S. Microbial communities and food safety aspects of crickets (Acheta domesticus) reared under controlled conditions. J. Insects Food Feed 2020, 6, 429–440. [Google Scholar] [CrossRef]
- Ng’ang’a, J.; Imathiu, S.; Fombong, F.; Vanden Broeck, J.; Kinyuru, J. Effect of dietary supplementation with powder derived from Moringa oleifera and Azadirachta indica leaves on growth and microbial load of edible crickets. J. Insects Food Feed 2021, 7, 419–431. [Google Scholar] [CrossRef]
- Inácio, A.C.; Vågsholm, I.; Jansson, A.; Vaga, M.; Boqvist, S.; Fraqueza, M.J. Impact of starvation on fat content and microbial load in edible crickets (Acheta domesticus). J. Insects Food Feed 2021, 7, 1143–1147. [Google Scholar] [CrossRef]
- Fröhling, A.; Bußler, S.; Durek, J.; Schlüter, O.K. Thermal Impact on the Culturable Microbial Diversity Along the Processing Chain of Flour From Crickets (Acheta domesticus). Front. Microbiol. 2020, 11, 1–13. [Google Scholar] [CrossRef] [PubMed]
- Caparros Megido, R.; Desmedt, S.; Blecker, C.; Béra, F.; Haubruge, É.; Alabi, T.; Francis, F. Microbiological load of edible insects found in Belgium. Insects 2017, 8, 12. [Google Scholar] [CrossRef]
- Larouche, J.; Deschamps, M.H.; Saucier, L.; Lebeuf, Y.; Doyen, A.; Vandenberg, G.W. Effects of Killing Methods on Lipid Oxidation, Colour and Microbial Load of Black Soldier Fly. Animals 2019, 9, 182. [Google Scholar] [CrossRef] [Green Version]
- Grabowski, N.T.; Klein, G. Microbiology of cooked and dried edible Mediterranean field crickets (Gryllus bimaculatus) and superworms (Zophobas atratus) submitted to four different heating treatments. Food Sci. Technol. 2017, 23, 17–23. [Google Scholar] [CrossRef]
- Bawa, M.; Songsermpong, S.; Kaewtapee, C.; Chanput, W. Effects of microwave and hot air oven drying on the nutritional, microbiological load, and color parameters of the house crickets (Acheta domesticus). J. Food Process. Preserv. 2020, 44, 1–12. [Google Scholar] [CrossRef]
- Hernández-Álvarez, A.J.; Mondor, M.; Piña-Domínguez, I.A.; Sánchez-Velázquez, O.A.; Melgar Lalanne, G. Drying technologies for edible insects and their derived ingredients. Dry 2021, 39, 1991–2009. [Google Scholar] [CrossRef]
- Nyangena, D.N.; Mutungi, C.; Imathiu, S.; Kinyuru, J.; Affognon, H.; Ekesi, S.; Nakimbugwe, D.; Fiaboe, K.K.M. Effects of Traditional Processing Techniques on the Nutritional and Microbiological Quality of Four East Africa. Foods 2020, 9, 574. [Google Scholar] [CrossRef]
- Grabowski, N.T.; Klein, G. Microbiological analysis of raw edible insects. J. Insects Food Feed. 2017, 3, 7–14. [Google Scholar] [CrossRef]
- Fasolato, L.; Cardazzo, B.; Carraro, L.; Fontana, F.; Novelli, E.; Balzan, S. Edible processed insects from e-commerce: Food safety with a focus on the Bacillus cereus group. Food Microbiol. 2018, 76, 296–303. [Google Scholar] [CrossRef] [PubMed]
- Laroche, M.; Raoult, D.; Parola, P. Insects and the Transmission of Bacterial Agents. Microbiol. Spectr. 2018, 6, MTBP-0017-2016. [Google Scholar] [CrossRef] [PubMed]
- Hugenholtz, P.; Goebel, B.M.; Pace, N.R. Impact of culture-independent studies on the emerging phylogenetic view of bacterial diversity. J. Bacteriol. 1998, 180, 4765–4774. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Vandeweyer, D.; Crauwels, S.; Lievens, B.; van Campenhout, L. Metagenetic analysis of the bacterial communities of edible insects from diverse production cycles at industrial rearing companies. Int. J. Food Microbiol. 2017, 261, 11–18. [Google Scholar] [CrossRef] [PubMed]
- Messina, C.M.; Gaglio, R.; Morghese, M.; Tolone, M.; Arena, R.; Moschetti, G.; Santulli, A.; Francesca, N.; Settanni, L. Microbiological profile and bioactive properties of insect powders used in food and feed formulations. Foods 2019, 8, 400. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Galli, V.; Venturi, M.; Pini, N.; Granchi, L. Technological feature assessment of lactic acid bacteria isolated from cricket powder’s spontaneous fermentation as potential starters for cricket-wheat bread production. Foods 2020, 9, 1322. [Google Scholar] [CrossRef] [PubMed]
- Hanboonsong, A.; Durst, P. Guidance on Sustainable Cricket Farming—A Practical Manual; FAO: Bangkok, Thailand, 2020. [Google Scholar] [CrossRef]
- Morales-Ramos, J.A.; Guadalupe Rojas, M.; Dossey, A.T.; Berhow, M. Self-selection of food ingredients and agricultural by-products by the house cricket, Acheta domesticus (Orthoptera: Gryllidae): A holistic approach to develop optimized diets. PLoS ONE 2020, 15, e0227400. [Google Scholar] [CrossRef] [PubMed]
- Klindworth, A.; Pruesse, E.; Schweer, T.; Peplles, J.; Quast, C.; Horn, M.; Glöckner, F.O. Evaluation of general 16S ribosomal RNA gene PCR primers for classical and next-generation sequencing-based diversity studies. Nucleic Acids Res. 2013, 41, e1. [Google Scholar] [CrossRef]
- Bolyen, E.; Rideout, J.R.; Dillon, M.R.; Bokulich, N.A.; Abnet, C.C.; Al-Ghalith, G.A.; Caporaso, J.G. Reproducible, interactive, scalable and extensible microbiome data science using QIIME 2. Nat. Biotechnol. 2019, 37, 852–857. [Google Scholar] [CrossRef]
- Martin, M. Cutadapt Removes Adapter Sequences from High-Throughput Sequencing Reads. EMBnet. J. 2011, 17, 10–12. [Google Scholar] [CrossRef]
- Callahan, B.J.; McMurdie, P.J.; Rosen, M.J.; Han, A.W.; Johnson, A.J.A.; Holmes, S.P. DADA2: High-Resolution Sample Inference from Illumina Amplicon Data. Nat. Methods 2016, 13, 581–583. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Caporaso, J.G.; Bittinger, K.; Bushman, F.D.; DeSantis, T.Z.; Andersen, G.L.; Knight, R. PyNAST: A flexible tool for aligning sequences to a template alignment. Bioinformatics 2010, 26, 266–267. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bokulich, N.A.; Kaehler, B.D.; Rideout, J.R.; Dillon, M.; Bolyen, E.; Knight, R.; Huttley, G.A.; Gregory Caporaso, J. Optimizing Taxonomic Classification of Marker-Gene Amplicon Sequences with QIIME 2′sQ2-Feature-Classifier Plugin. Microbiome 2018, 6, 90. [Google Scholar] [CrossRef] [PubMed]
- Price, M.N.; Dehal, P.S.; Arkin, A.P. FastTree 2-Approximately Maximum-Likelihood Trees for Large Alignments. PLoS ONE 2010, 5, e9490. [Google Scholar] [CrossRef]
- Lozupone, C.; Knight, R. UniFrac: A new phylogenetic method for comparing microbial communities. Appl. Environ. Microbiol. 2005, 71, 8228–8235. [Google Scholar] [CrossRef] [Green Version]
- Vazquez-Baeza, Y.; Pirrung, M.; Gonzalez, A.; Knight, R. Emperor: A tool for visualizing high-throughput microbial community data. Gigascience 2013, 2, 2047-217X-2-16. [Google Scholar] [CrossRef] [Green Version]
- Warnes, G.; Bolker, B.; Bonebakker, L.; Gentleman, R.; Huber, W.; Liaw, A.; Lumley, T.; Mächler, M.; Magnusson, A.; Möller, S. Gplots: Various R Programming Tools for Plotting Data. 2005. Available online: https://CRAN.R-project.org/package=gplots (accessed on 28 January 2022).
- Jones, R.T.; Sanchez, L.G.; Fierer, N.A. Cross-Taxon Analysis of Insect-Associated Bacterial Diversity. PLoS ONE 2013, 8, e61218. [Google Scholar] [CrossRef]
- Yun, J.H.; Roh, S.W.; Whon, T.W.; Jung, M.J.; Kim, M.S.; Park, D.S.; Yoon, C.; Nam, Y.D.; Kim, Y.J.; Choi, J.H.; et al. Insect gut bacterial diversity determined by environmental habitat, diet, developmental stage, and phylogeny of host. Appl. Environ. Microbiol. 2014, 80, 5254–5264. [Google Scholar] [CrossRef] [Green Version]
- Vandeweyer, D.; Wynants, E.; Crauwels, S.; Verreth, C.; Viaene, N.; Claes, J.; Lievens, B.; van Campenhout, L. Microbial Dynamics during Industrial Rearing, Processing, and Storage of Tropical House Crickets (Gryllodes sigillatus) for Human Consumption. Appl. Environ. Microbiol. 2018, 84, e00255-18. [Google Scholar] [CrossRef] [Green Version]
- Patrick, S. Bacteroides. In Molecular Medical Microbiology, 2nd ed.; Tang, Y.W., Sussman, M., Liu, D., Poxton, I., Schwartzman, J., Eds.; Academic Press: Cambridge, MA, USA, 2015; Volume 2, pp. 917–944. [Google Scholar] [CrossRef]
- Deng, X.; Tian, H.; Yang, R.; Han, Y.; Wei, K.; Zheng, C.; Liu, Z.; Chen, T. Oral Probiotics Alleviate Intestinal Dysbacteriosis for People Receiving Bowel Preparation. Front. Med. 2020, 7, 73. [Google Scholar] [CrossRef]
- Giraffa, G. Enterococcus. In Encyclopedia of Food Microbiology; Robinson, R.K., Ed.; Elsevier Ltd.: Amsterdam, The Netherlands, 1999; pp. 617–624. [Google Scholar] [CrossRef]
- Doughari, H.J.; Ndakidemi, P.A.; Human, I.S.; Benade, S. The ecology, biology and pathogenesis of Acinetobacter spp.: An overview. Microbes Environ. 2011, 26, 101–112. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, T.; Li, Q.; Cheng, L.; Buch, H.; Zhang, F. Akkermansia muciniphila is a promising probiotic. Microb. Biotechnol. 2019, 12, 1109–1125. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jung, M.Y.; Lee, C.; Seo, M.J.; Roh, S.W.; Lee, S.H. Characterization of a potential probiotic bacterium Lactococcus raffinolactis WiKim0068 isolated from fermented vegetable using genomic and in vitro analyses. BMC Microbiol. 2020, 20, 136. [Google Scholar] [CrossRef] [PubMed]
- Cani, P.D.; de Vos, W.M. Next-generation beneficial microbes: The case of Akkermansia muciniphila. Front. Microbiol. 2017, 8, 1765. [Google Scholar] [CrossRef]
- Kort, R.; Schlösser, J.; Vazquez, A.R.; Atukunda, P.; Muhoozi, G.K.M.; Wacoo, A.P.; Sybesma, W.F.H.; Westerberg, A.C.; Iversen, P.O.; Schoen, E.D. Model Selection Reveals the Butyrate-Producing Gut Bacterium Coprococcus eutactus as Predictor for Language Development in 3-Year-Old Rural Ugandan Children. Front. Microbiol. 2021, 12, 1406. [Google Scholar] [CrossRef]
- Qiu, Y.L.; Kuang, X.Z.; Shi, X.S.; Yuan, X.Z.; Guo, R.B. Paludibacter jiangxiensis sp. nov., a strictly anaerobic, propionate-producing bacterium isolated from rice paddy field. Arch. Microbiol. 2014, 196, 149–155. [Google Scholar] [CrossRef]
- Song, A.A.; In, L.L.A.; Lim, S.H.E.; Rahim, R.A. A review on Lactococcuslactis: From food to factory. Microb. Cell Fact. 2017, 16, 55. [Google Scholar] [CrossRef] [Green Version]
- Bauer, S.; Tholen, A.; Overmann, J.; Brune, A. Characterization of abundance and diversity of lactic acid bacteria in the hindgut of wood- and soil-feeding termites by molecular and culture- dependent techniques. Arch. Microbiol. 2000, 173, 126–137. [Google Scholar] [CrossRef]
- Yang, S.Y.; Zheng, Y.; Huang, Z.; Wang, X.M.; Yang, H. Lactococcus nasutitermitis sp. nov. isolated from a termite gut. Int. J. Syst. Evol. 2016, 66, 518–522. [Google Scholar] [CrossRef] [Green Version]
- Sapountzis, P.; Gruntjes, T.; Otani, S.; Estevez, J.; da Costa, R.R.; Plunkett III, G.; Perna, N.T.; Poulsen, M. The enterobacterium Trabulsiella odontotermitis presents novel adaptations related to its association with fungus-growing termites. Appl. Environ. Microbiol. 2015, 81, 6577–6588. [Google Scholar] [CrossRef] [Green Version]
- Hongoh, Y.; Sharma, V.K.; Prakash, T.; Noda, S.; Toh, H.; Taylor, T.D.; Kudo, T.; Sakaki, Y.; Toyoda, A.; Hattori, M.; et al. Genome of an endosymbiont coupling N2 fixation to celluolysis within protist cells in termite gut. Science 2008, 322, 1108–1109. [Google Scholar] [CrossRef] [PubMed]
- Tikhe, C.V.; Husseneder, C. Metavirome sequencing of the termite gut reveals the presence of an unexplored bacteriophage community. Front. Microbiol. 2018, 8, 2548. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Oberpaul, M.; Zumkeller, C.M.; Culver, T.; Spohn, M.; Mihajlovic, S.; Leis, B.; Glaeser, S.P.; Plarre, R.; McMahon, D.P.; Hammann, P.; et al. High-Throughput Cultivation for the Selective Isolation of AcidobacteriaFrom Termite Nests. Front. Microbiol. 2020, 11, 597628. [Google Scholar] [CrossRef] [PubMed]
- Soukup, P.; Větrovský, T.; Stiblik, P.; Votýpková, K.; Chakraborty, A.; Sillam-Dussès, D.; Kolařík, M.; Odriozola, I.; Lo, N.; Baldrian, P.; et al. Termites Are Associated with External Species-Specific Bacterial Communities. Appl. Environ. Microbiol. 2021, 87, e02042-20. [Google Scholar] [CrossRef]
- Schauer, C.; Thompson, C.L.; Brune, A. The bacterial community in the gut of the cockroach Shelfordella lateralis reflects the close evolutionary relatedness of cockroaches and termites. Appl. Environ. Microbiol. 2012, 78, 2758–2767. [Google Scholar] [CrossRef] [Green Version]
- Mladenović, K.G.; Grujović, M.Ž.; Kiš, M.; Furmeg, S.; Tkalec, V.J.; Stefanović, O.D.; Kocić-Tanackov, S.D. Enterobacteriaceae in food safety with an emphasis on raw milk and meat. Appl. Microbiol. Biotechnol. 2021, 105, 8615–8627. [Google Scholar] [CrossRef]
- Klunder, H.C.; Wolkers-Rooijackers, J.; Korpela, J.M.; Nout, M.J.R. Microbiological aspects of processing and storage of edible insects. Food Control 2012, 26, 628–631. [Google Scholar] [CrossRef]
- Van Moll, L.; De Smet, J.; Cos, P.; Van Campenhout, L. Microbial symbionts of insects as a source of new antimicrobials: A review. Crit. Rev. Microbiol. 2021, 47, 562–579. [Google Scholar] [CrossRef]
- Evans, J.D.; Armstrong, T.N. Antagonistic interaction between honey bee bacterial symbionts and implications for disease. BMC Ecol. 2006, 6, 4. [Google Scholar] [CrossRef] [Green Version]
- Elshaghabee, F.M.F.; Rokana, N.; Gulhane, R.D.; Sharma, C.; Panwar, H. Bacillus As Potential Probiotics: Status, Concerns, and Future Perspectives. Front. Microbiol. 2017, 8, 1490. [Google Scholar] [CrossRef] [Green Version]
- Rhayat, L.; Maresca, M.; Nicoletti, C.; Perrier, J.; Brinch, K.S.; Christian, S.; Devillard, E.; Eckhardt, E. Effect of Bacillus subtilis Strains on Intestinal Barrier Function and Inflammatory Response. Front. Immunol. 2019, 10, 564. [Google Scholar] [CrossRef] [PubMed]
- Ignasiak, K.; Maxwell, A. Antibiotic-resistant bacteria in the guts of insects feeding on plants: Prospects for discovering plant-derived antibiotics. BMC Microbiol. 2017, 17, 223. [Google Scholar] [CrossRef] [PubMed] [Green Version]
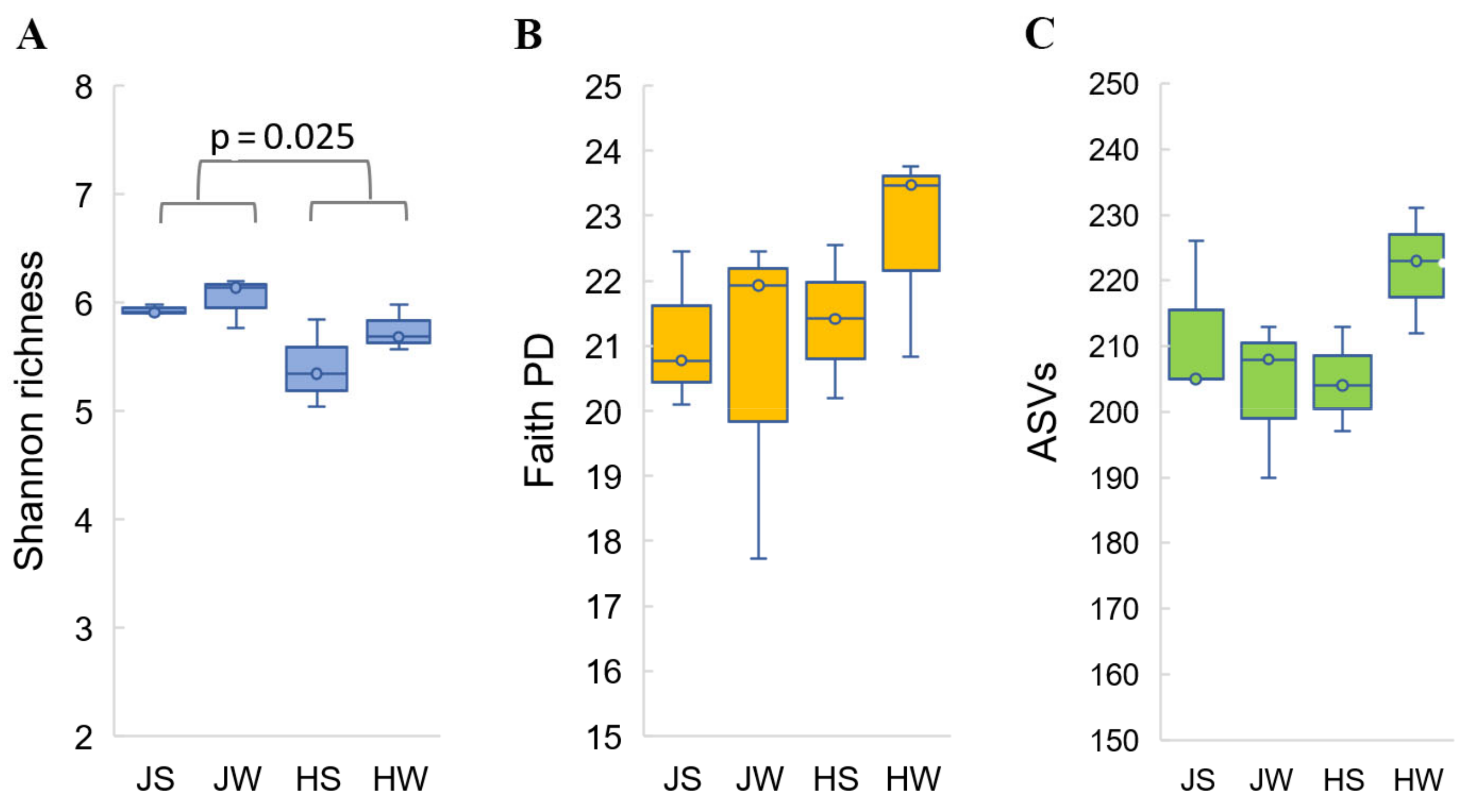
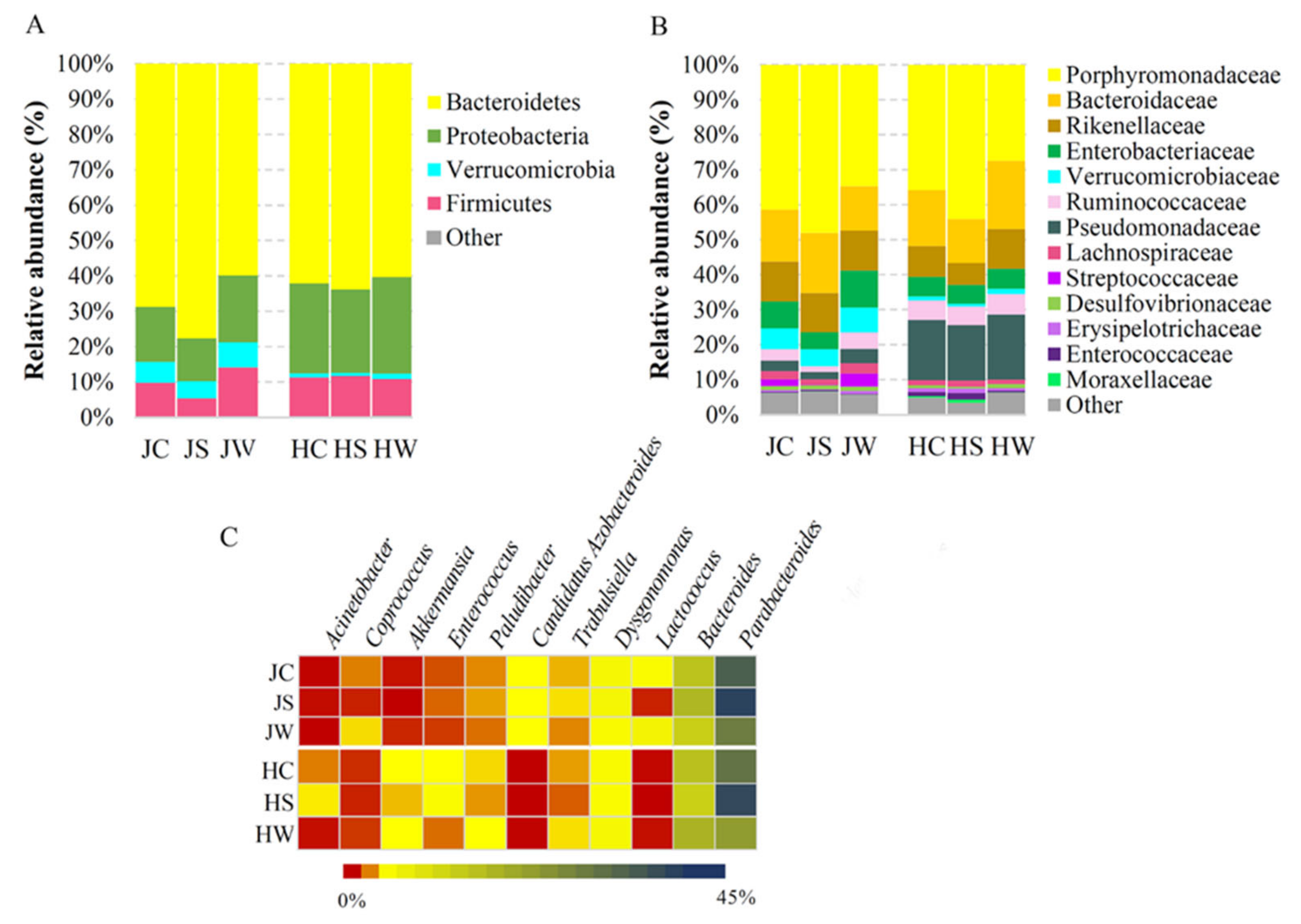
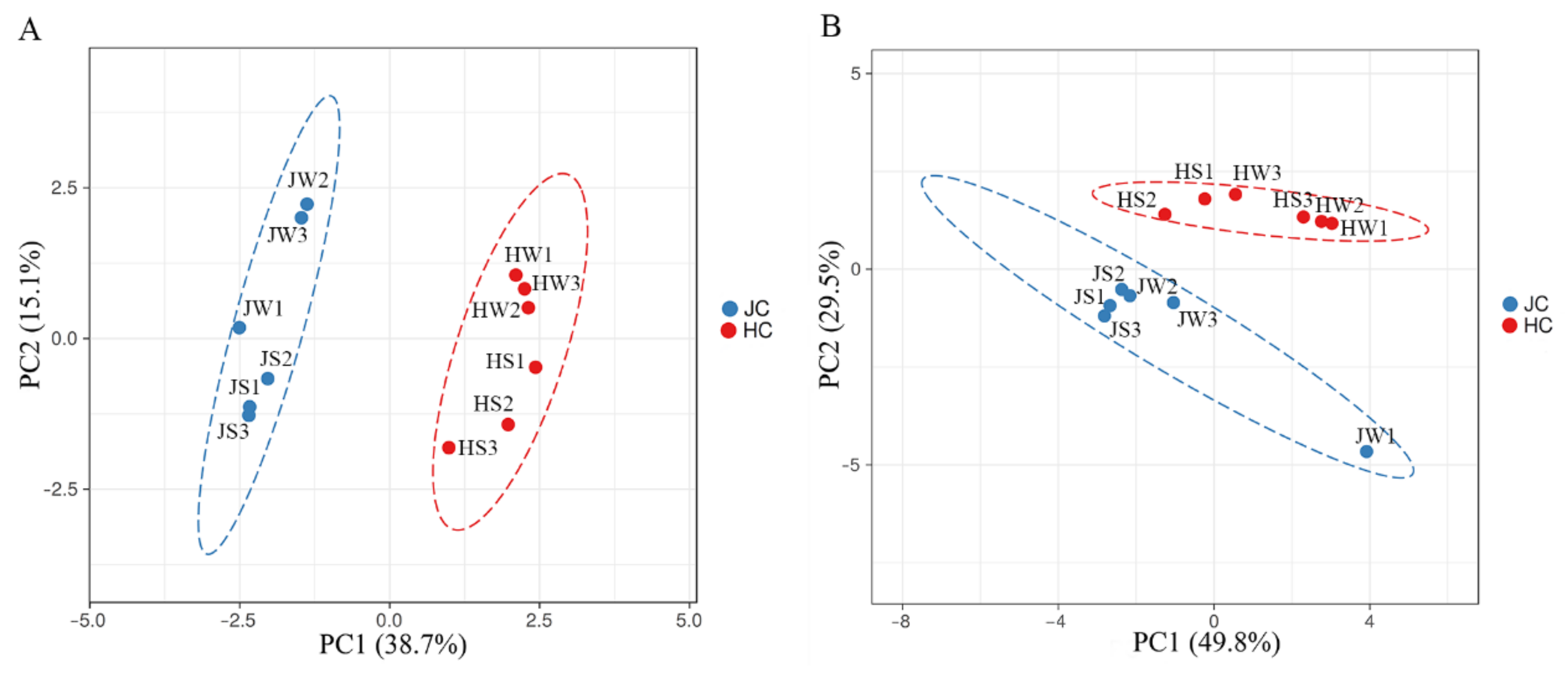
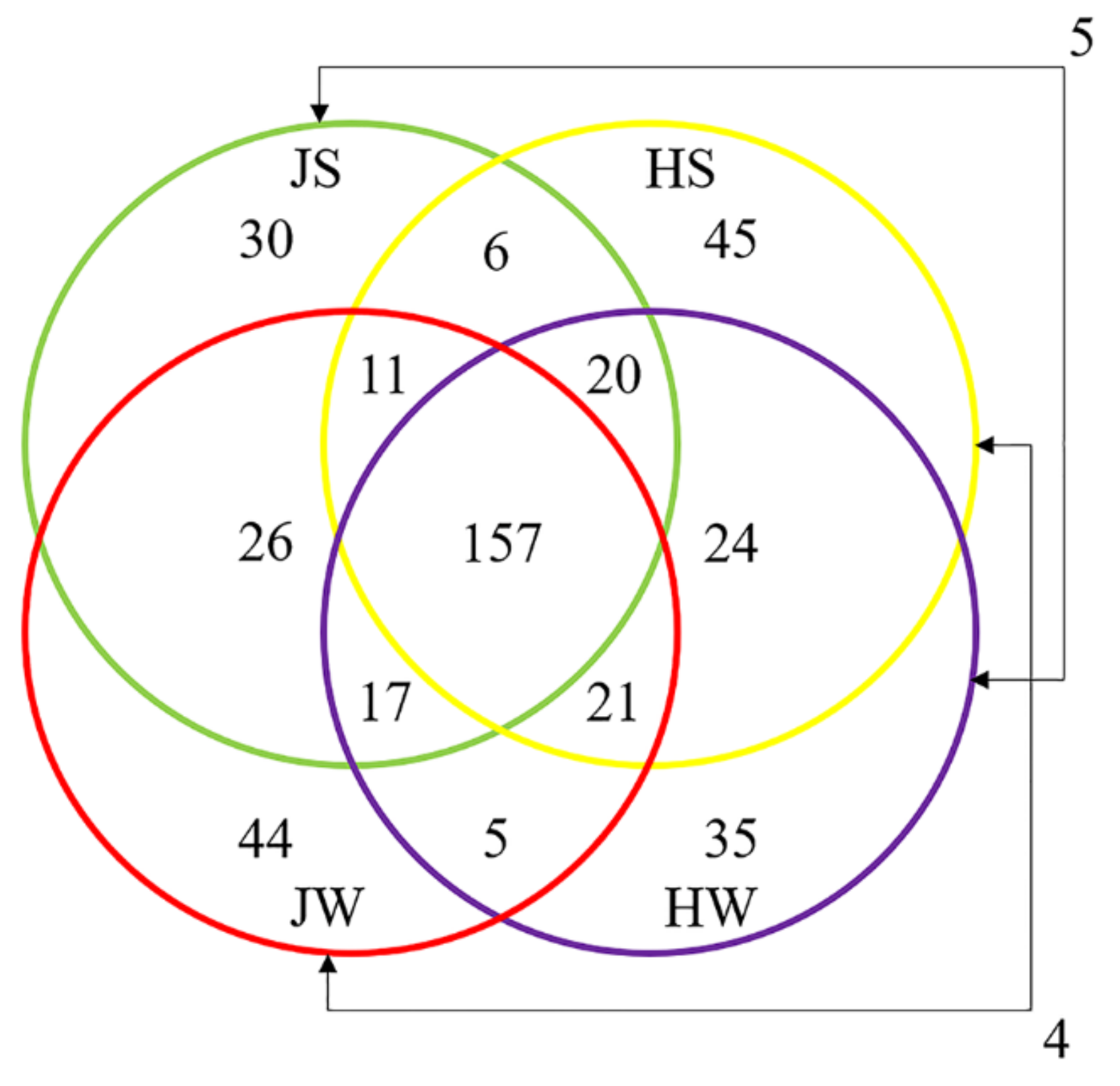

| Samples | Reads Obtained | High Quality Reads | ASVs | Shannon’s Entropy | Faith’s PD |
|---|---|---|---|---|---|
| JS1 | 206,102 | 51,061 | 205 | 5.90 | 20.10 |
| JS2 | 194,318 | 51,283 | 226 | 5.98 | 22.45 |
| JS3 | 224,948 | 55,243 | 205 | 5.91 | 20.78 |
| JW1 | 199,648 | 45,777 | 190 | 5.77 | 17.73 |
| JW2 | 194,848 | 43,851 | 213 | 6.19 | 21.93 |
| JW3 | 181,722 | 41,186 | 208 | 6.14 | 22.46 |
| HS1 | 190,144 | 47,757 | 197 | 5.04 | 20.20 |
| HS2 | 203,110 | 51,460 | 204 | 5.84 | 21.42 |
| HS3 | 185,310 | 40,950 | 213 | 5.34 | 22.54 |
| HW1 | 179,088 | 54,781 | 223 | 5.68 | 23.47 |
| HW2 | 183,930 | 54,392 | 212 | 5.57 | 20.83 |
| HW3 | 211,512 | 64,068 | 231 | 5.98 | 23.77 |
| Total | 2,354,680 | 601,809 | 2527 |
| Unweighted Pseudo-F | UniFrac p-Value | Weighted Pseudo-F | UniFrac p-Value | |
|---|---|---|---|---|
| JS vs. JW | 1.870 | 0.095 | 2.531 | 0.11 |
| JS vs. HS | 5.209 | 0.116 | 23.038 | 0.102 |
| JS vs. HW | 3.327 | 0.102 | 10.394 | 0.104 |
| JW vs. HS | 3.349 | 0.086 | 2.370 | 0.184 |
| JW vs. HW | 3.116 | 0.105 | 1.946 | 0.214 |
| HS vs. HW | 2.018 | 0.101 | 3.972 | 0.208 |
| Jamaican vs. house crickets | 4.443 | 0.004 | 4.229 | 0.018 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Aleknavičius, D.; Lukša, J.; Strazdaitė-Žielienė, Ž.; Servienė, E. The Bacterial Microbiota of Edible Insects Acheta domesticus and Gryllus assimilis Revealed by High Content Analysis. Foods 2022, 11, 1073. https://doi.org/10.3390/foods11081073
Aleknavičius D, Lukša J, Strazdaitė-Žielienė Ž, Servienė E. The Bacterial Microbiota of Edible Insects Acheta domesticus and Gryllus assimilis Revealed by High Content Analysis. Foods. 2022; 11(8):1073. https://doi.org/10.3390/foods11081073
Chicago/Turabian StyleAleknavičius, Dominykas, Juliana Lukša, Živilė Strazdaitė-Žielienė, and Elena Servienė. 2022. "The Bacterial Microbiota of Edible Insects Acheta domesticus and Gryllus assimilis Revealed by High Content Analysis" Foods 11, no. 8: 1073. https://doi.org/10.3390/foods11081073
APA StyleAleknavičius, D., Lukša, J., Strazdaitė-Žielienė, Ž., & Servienė, E. (2022). The Bacterial Microbiota of Edible Insects Acheta domesticus and Gryllus assimilis Revealed by High Content Analysis. Foods, 11(8), 1073. https://doi.org/10.3390/foods11081073







