Interaction Mechanism between OVA and Flavonoids with Different Hydroxyl Groups on B-Ring and Effect on Antioxidant Activity
Abstract
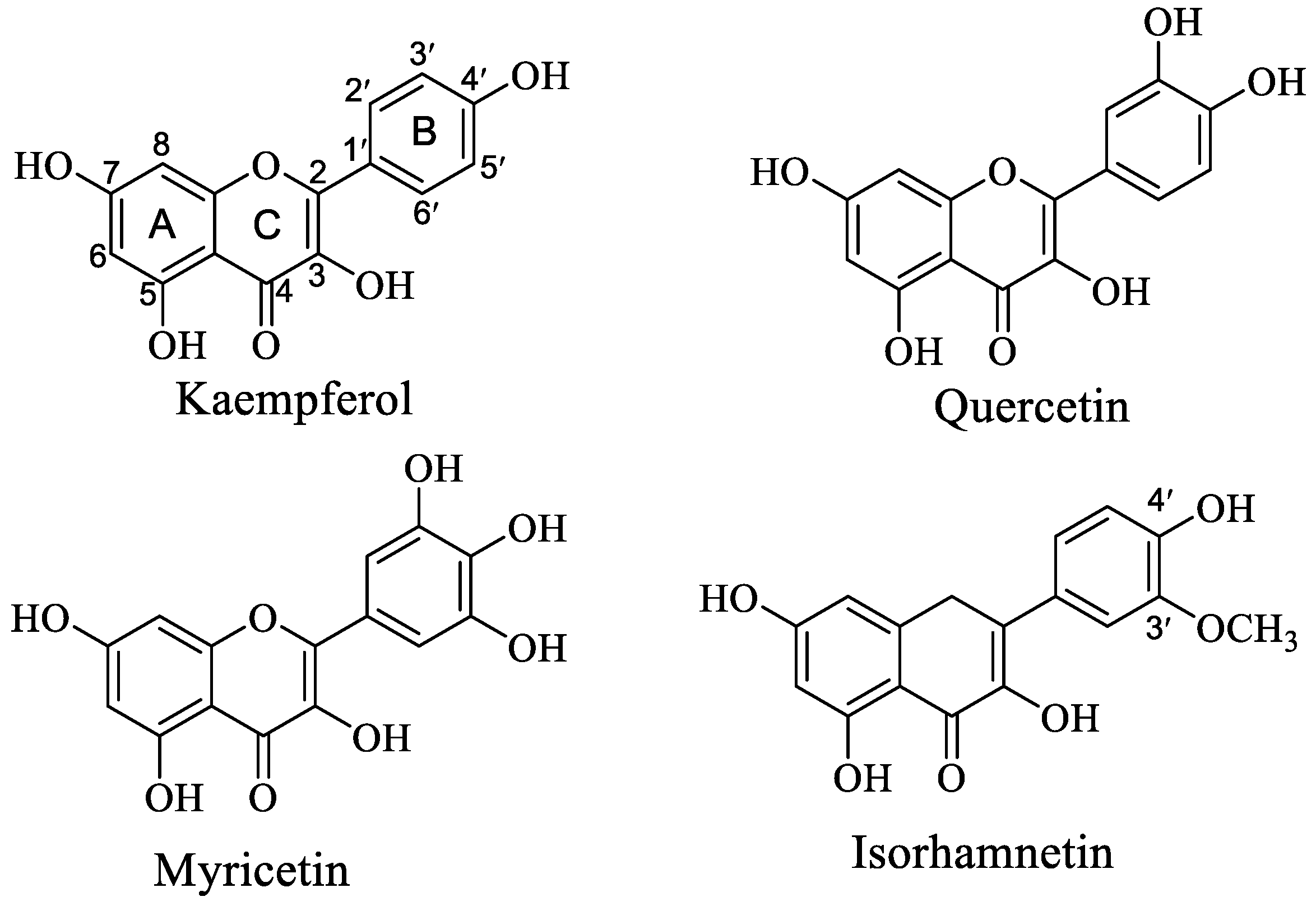
:1. Introduction
2. Materials and Methods
2.1. Materials
2.2. Sample Preparation
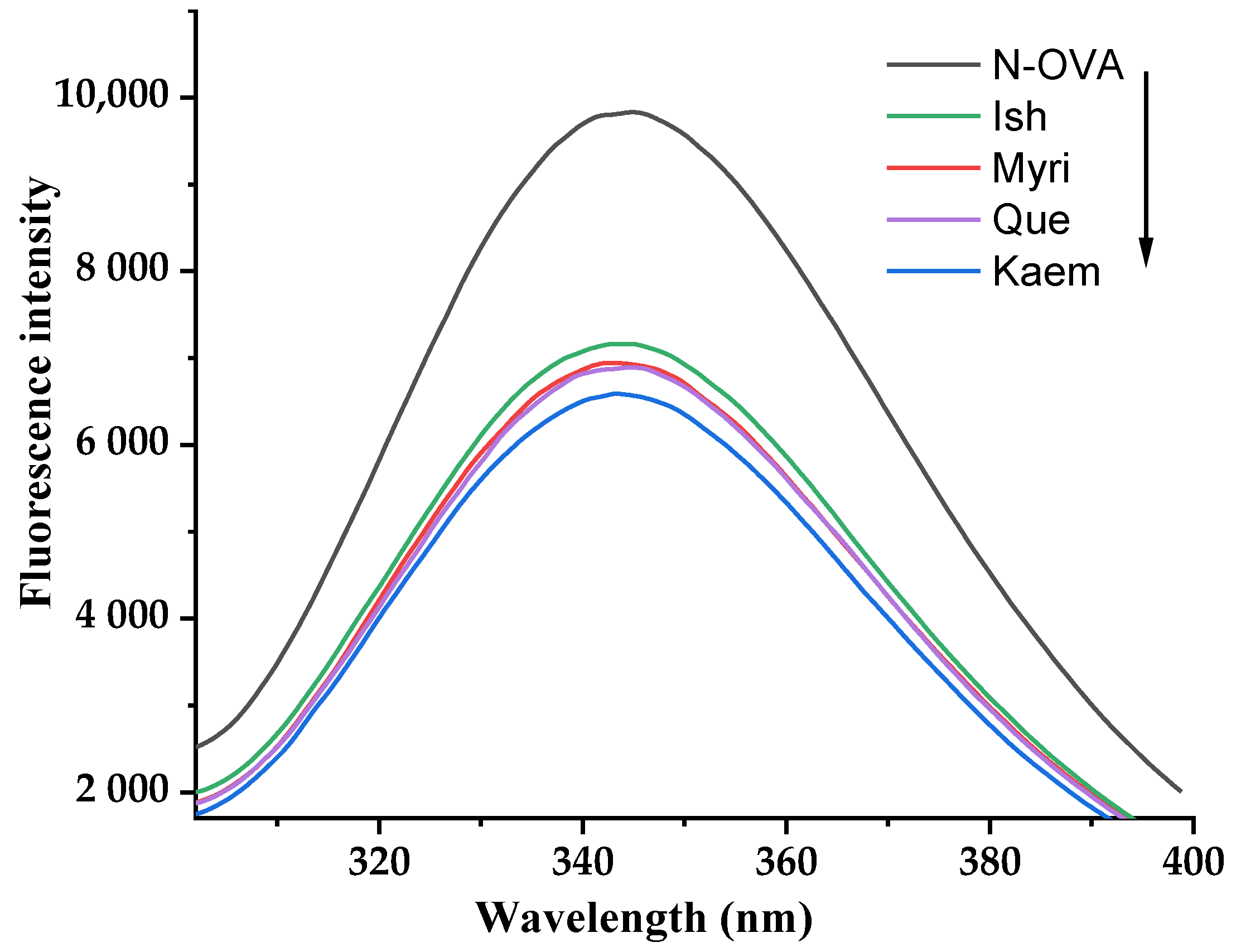
2.3. Intrinsic Fluorescence Analysis
2.4. FT−IR Spectroscopy
2.5. SDS−PAGE Analysis
2.6. Circular Dichroism Spectroscopy Analysis
2.7. Fluorescence Titration Experiment
2.8. Molecular Docking Analysis
2.9. Antioxidant Abilities Assays
2.10. Statistical Analyses
3. Results and Discussion
3.1. Intrinsic Fluorescence Spectra of OVA Affected by Que/Myri/Kaem/Ish
3.2. Infrared Spectroscopy and SDS−PAGE Analysis
3.3. CD Spectra Analysis
3.4. Interaction Mechanism of OVA with Que/Kaem/Ish/Myri
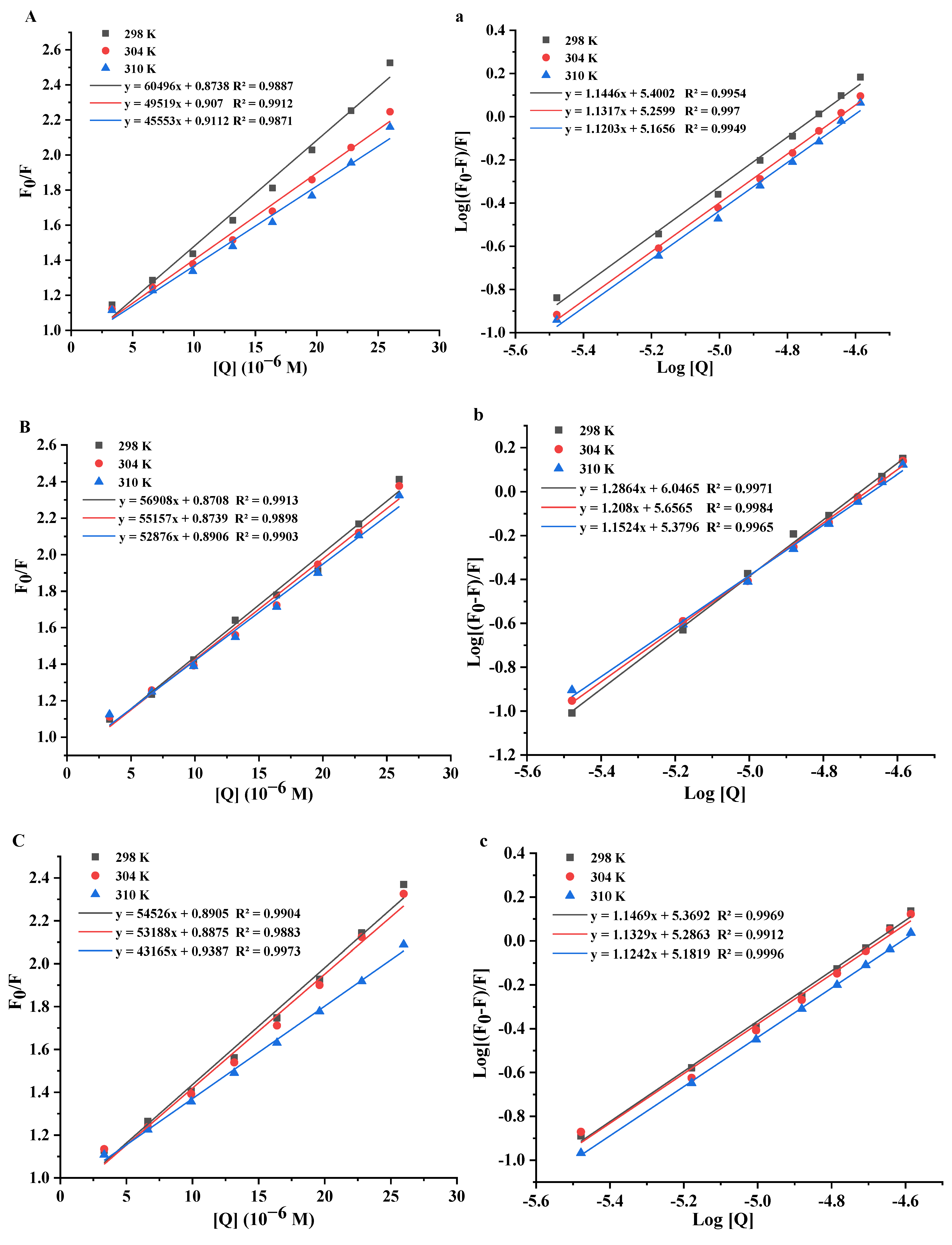
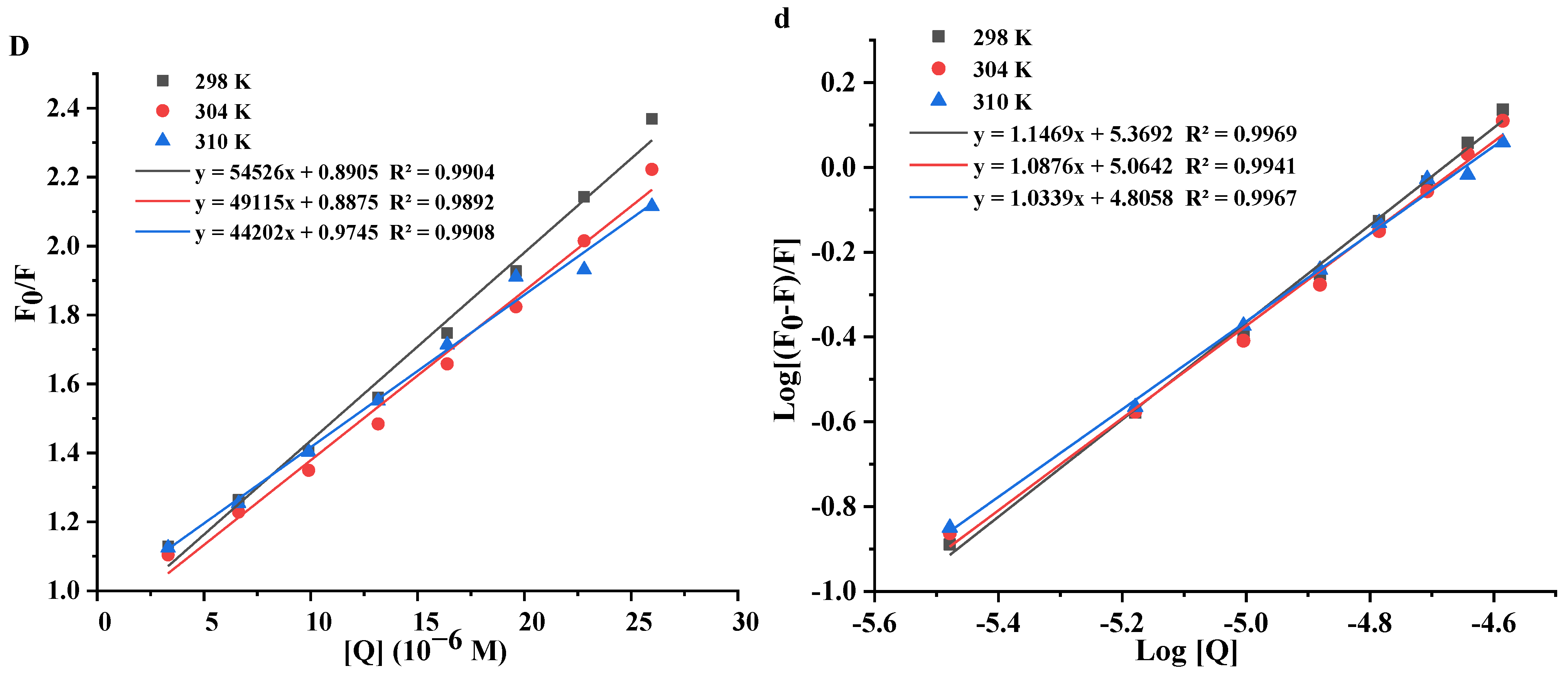
3.4.1. Fluorescence Quenching Mechanism
3.4.2. Binding Constants and Binding Sites
3.4.3. Thermodynamic Parameters
| T (K) | Ksv (104 L/moL) | Kq (1012 L/moL/s) | n | Ka (104 L/moL) | ΔH° (kJ/mol) | ΔS° (kJ/mol/K) | R | ΔG° (kJ/mol) | |
|---|---|---|---|---|---|---|---|---|---|
| Que | 298 K | 60.50 | 60.50 | 1.145 | 25.13 | −30.77 | |||
| 304 K | 4.95 | 4.95 | 1.132 | 18.19 | −34.61 | −0.01 | 0.9898 | −30.69 | |
| 310 K | 4.56 | 4.56 | 1.120 | 14.64 | −30.61 | ||||
| Kaem | 298 K | 5.69 | 5.69 | 1.286 | 111.30 | −34.40 | |||
| 304 K | 5.52 | 5.52 | 1.208 | 45.34 | −98.38 | −0.21 | 0.9933 | −33.11 | |
| 310 K | 5.29 | 5.29 | 1.152 | 23.75 | −31.82 | ||||
| Ish | 298 K | 5.45 | 5.45 | 1.147 | 23.40 | −24.50 | |||
| 304 K | 5.32 | 5.32 | 1.133 | 19.33 | −27.58 | −0.01 | 0.994 | −24.44 | |
| 310 K | 4.32 | 4.32 | 1.124 | 15.20 | −24.38 | ||||
| Myri | 298 K | 5.45 | 5.45 | 1.147 | 23.40 | −30.60 | |||
| 304 K | 4.91 | 4.91 | 1.088 | 11.59 | −83.07 | −0.18 | 0.9987 | −29.54 | |
| 310 K | 4.42 | 4.42 | 1.034 | 6.39 | −28.48 |
3.4.4. Molecular Docking
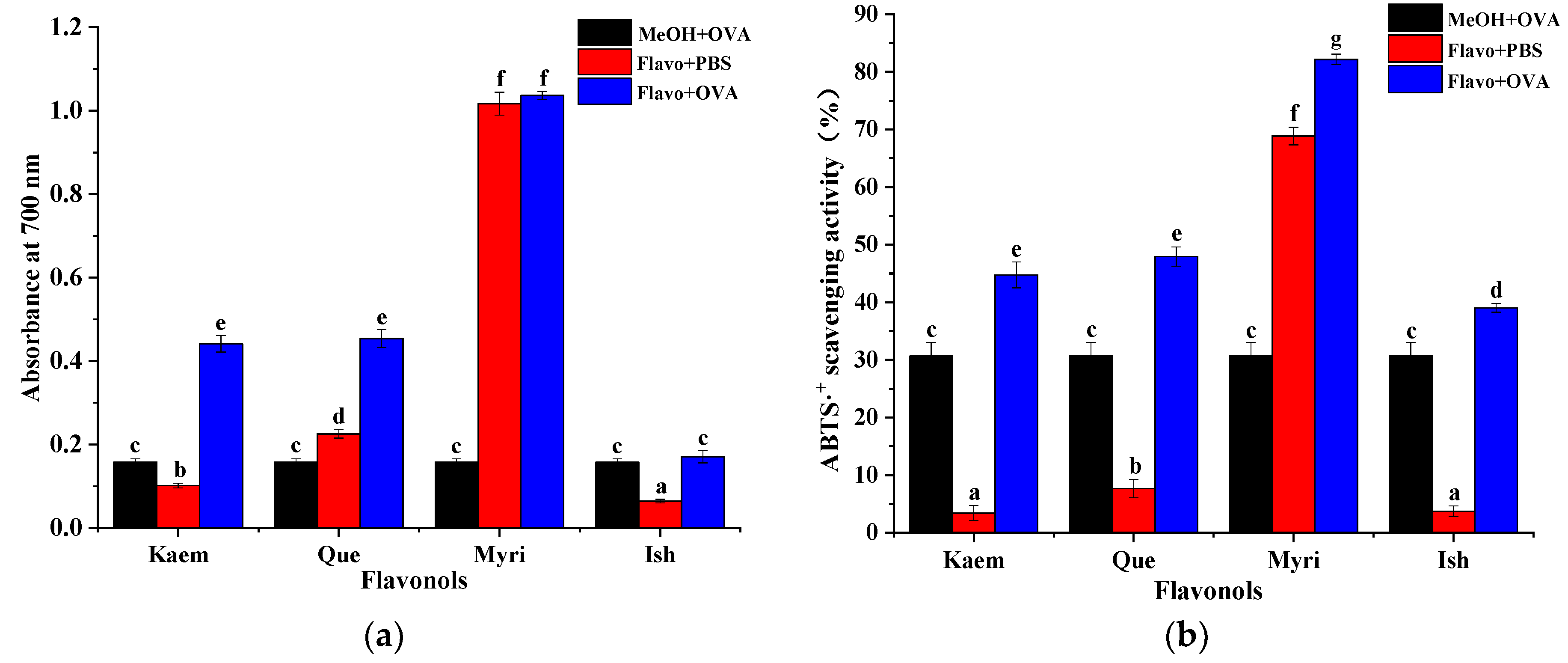
3.5. Antioxidant Activities of Que/Kaem/Ish/Myri−OVA Complexes
4. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Jiang, B.; Song, J.-L.; Jin, Y.-C. A flavonoid monomer tricin in Gramineous plants: Metabolism, bio/chemosynthesis, biological properties, and toxicology. Food Chem. 2020, 320, 126617. [Google Scholar] [CrossRef] [PubMed]
- Lin, S.-Y.; Zhang, G.-W.; Liao, Y.-J.; Pan, J.-H.; Gong, D.-M. Dietary flavonoids as xanthine oxidase inhibitors: Structure-affinity and structure-activity relationships. J. Agric. Food Chem. 2015, 63, 7784–7794. [Google Scholar] [CrossRef] [PubMed]
- Lerman, J.S.; Haslem, J.; Kim, L.; Ponderelli, J.; Ma, M.; Felson, R.; Sanderson, K.; Murphy, K.; Hopkins, K.; Wright, E.; et al. Collected research on phytonutrients: Flavonoids. J. Culin. Sci. Technol. 2015, 13, 214–241. [Google Scholar] [CrossRef]
- Survay, N.S.; Upadhyaya, C.P.; Kumar, B.; Young, K.E.; Yoon, D.Y.; Park, S.W. New genera of flavonols and flavonol derivatives as therapeutic molecules. J. Korean Soc. Appl. Biol. 2011, 54, 1–18. [Google Scholar] [CrossRef]
- Dabeek, W.M.; Marra, M.V. Dietary quercetin and kaempferol: Bioavailability and potential cardiovascular-related bioactivity in humans. Nutrients 2019, 11, 2288. [Google Scholar] [CrossRef] [Green Version]
- Zhang, Q.-Z.; Cheng, Z.-Z.; Wang, Y.-B.; Fu, L.-L. Dietary protein-phenolic interactions: Characterization, biochemical-physiological consequences, and potential food applications. Crit. Rev. Food Sci. 2020, 61, 3589–3615. [Google Scholar] [CrossRef]
- Teng, H.; Zheng, Y.-M.; Cao, H.; Huang, Q.; Xiao, J.-B.; Chen, L. Enhancement of bioavailability and bioactivity of diet-derived flavonoids by application of nanotechnology: A review. Crit. Rev. Food Sci. 2021. [Google Scholar] [CrossRef]
- Tsuchiya, H. Structure-dependent membrane interaction of flavonoids associated with their bioactivity. Food Chem. 2010, 120, 1089–1096. [Google Scholar] [CrossRef]
- Kovacs-Nolan, J.; Phillips, M.; Mine, Y. Advances in the value of eggs and egg components for human health. J. Agr. Food Chem. 2005, 53, 8421–8431. [Google Scholar] [CrossRef]
- Mine, Y. Recent advances in egg protein functionality in the food system. World Poult. Sci. J. 2002, 58, 31–39. [Google Scholar] [CrossRef]
- Gou, S.-Q.; Chen, Q.-B.; Liu, Y.; Zeng, L.; Song, H.-L.; Xu, Z.-G.; Kang, Y.-J.; Li, H.-M.; Xiao, B. Green fabrication of ovalbumin nanoparticles as natural polyphenol carriers for ulcerative Colitis Therapy. ACS Sustain. Chem. Eng. 2018, 6, 12658–12667. [Google Scholar] [CrossRef]
- Liu, Y.-J.; Ying, D.-Y.; Cai, Y.-X.; Le, X.-Y. Improved antioxidant activity and physicochemical properties of curcumin by adding ovalbumin and its structural characterization. Food Hydrocolloid 2017, 72, 304–311. [Google Scholar] [CrossRef]
- Zhang, L.; Zhou, W.-N.; Tu, Z.-C.; Yang, S.-H.; Xu, L.; Yuan, T. Influence of hydroxyl substitution on the suppression of flavonol in harmful glycation product formation and the inhibition mechanism revealed by spectroscopy and mass spectrometry. J. Agric. Food Chem. 2020, 68, 8263–8273. [Google Scholar] [CrossRef] [PubMed]
- Zhang, L.; Lu, Y.; Ye, Y.-H.; Yang, S.-H.; Tu, Z.-C.; Chen, J.; Wang, H.; Wang, H.-H.; Yuan, T. Insights into the mechanism of quercetin against BSA-fructose glycation by spectroscopy and high-resolution mass spectrometry: Effect on physicochemical properties. J. Agric. Food Chem. 2019, 67, 236–246. [Google Scholar] [CrossRef]
- Shen, F.; Niu, F.-G.; Li, J.-H.; Su, Y.-J.; Liu, Y.-T.; Yang, Y.-J. Interactions between tea polyphenol and two kinds of typical egg white proteins—ovalbumin and lysozyme: Effect on the gastrointestinal digestion of both proteins in vitro. Food Res. Int. 2014, 59, 100–107. [Google Scholar] [CrossRef]
- Stojadinovic, M.; Radosavljevic, J.; Ognjenovic, J.; Vesic, J.; Prodic, I.; Stanic-Vucinic, D. Binding affinity between dietary polyphenols and β-lactoglobulin negatively correlates with the protein susceptibility to digestion and total antioxidant activity of complexes formed. Food Chem. 2013, 136, 1263–1271. [Google Scholar] [CrossRef]
- Liu, Y.-J.; Cai, Y.-X.; Ying, D.-Y.; Fu, Y.-L.; Xiong, Y.-H.; Le, X.-Y. Ovalbumin as a carrier to significantly enhance the aqueous solubility and photostability of curcumin: Interaction and binding mechanism study. Int. J. Biol. Macromol. 2018, 116, 893–900. [Google Scholar] [CrossRef]
- Li, T.; Hu, P.; Dai, T.-T.; Li, P.-Y.; Ye, X.-Q.; Chen, J.; Liu, C.-M. Comparing the binding interaction between β-lactoglobulin and flavonoids with different structure by multi-spectroscopy analysis and molecular docking. Spectrochim. Acta A 2018, 201, 197–206. [Google Scholar] [CrossRef]
- Zhang, L.; Tu, Z.-C.; Xie, X.; Lu, Y.; Wang, Z.-X.; Wang, H.; Sha, X.-M. Antihyperglycemic, antioxidant activities of two Acer palmatum cultivars, and identification of phenolics profile by UPLC-QTOF-MS/MS: New natural sources of functional constituents. Ind. Crop. Prod. 2016, 89, 522–532. [Google Scholar] [CrossRef]
- Zhang, L.; Tu, Z.-C.; Wang, H.; Kou, Y.; Wen, Q.-H.; Fu, Z.-F.; Chang, H.-X. Response surface optimization and physicochemical properties of polysaccharides from Nelumbo nucifera leaves. Int. J. Biol. Macromol. 2015, 74, 103–110. [Google Scholar] [CrossRef]
- Huang, X.-Q.; Tu, Z.-C.; Xiao, H.; Wang, H.; Zhang, L.; Hu, Y.-M.; Zhang, Q.-T.; Niu, P.-P. Characteristics and antioxidant activities of ovalbumin glycated with different saccharides under heat moisture treatment. Food Res. Int. 2012, 48, 866–872. [Google Scholar] [CrossRef]
- Wang, R.-Q.; Yin, Y.-J.; Li, H.; Wang, Y.; Pu, J.-J.; Wang, R.; Dou, H.-J.; Song, C.-J.; Wang, R.-Y. Comparative study of the interactions between ovalbumin and three alkaloids by spectrofluorimetry. Mol. Biol. Rep. 2013, 40, 3409–3418. [Google Scholar] [CrossRef] [PubMed]
- Xiao, M.-S.; Han, L.-N.; Zhou, L.; Zhou, Y.-H.; Huang, X.-Q.; Ge, X.-F.; Wei, S.-H.; Zhou, J.-H.; Wu, H.-M.; Shen, J. Comparison and investigation of bovine hemoglobin binding to dihydroartemisinin and 9-hydroxy-dihydroartemisinin: Spectroscopic characterization. Spectrochim. Acta A 2014, 125, 120–125. [Google Scholar] [CrossRef] [PubMed]
- Tang, S.-T.; Yu, J.-L.; Lu, L.-Z.; Fu, X.; Cai, Z.-X. Interfacial and enhanced emulsifying behavior of phosphorylated ovalbumin. Int. J. Biol. Macromol. 2019, 131, 293–300. [Google Scholar] [CrossRef]
- Xiong, W.-F.; Wang, Y.-T.; Zhang, C.-L.; Wan, J.-W.; Shah, B.R.; Pei, Y.-Q.; Zhou, B.; Li, J.; Li, B. High intensity ultrasound modified ovalbumin: Structure, interface and gelation properties. Ultrason. Sonochem. 2016, 31, 302–309. [Google Scholar] [CrossRef]
- Huang, Q.; Ma, M.-H.; Cai, Z.-X.; Luo, Z.; Huang, X.; Sun, S.-G. Effect of s-configuration transformation on the microstructure of ovalbumin. Spectrosc. Spect. Anal. 2011, 31, 3319–3322. [Google Scholar] [CrossRef]
- Sheng, L.; Tang, G.-Y.; Wang, Q.; Zou, J.; Ma, M.-H.; Huang, X. Molecular characteristics and foaming properties of ovalbumin-pullulan conjugates through the Maillard reaction. Food Hydrocolloid 2020, 100, 105384. [Google Scholar] [CrossRef]
- Bi, H.-N.; Tang, L.; Gao, X.; Jia, J.-J.; Lv, H.-H. Spectroscopic analysis on the binding interaction between tetracycline hydrochloride and bovine proteins β-casein, α-lactalbumin. J. Lumin. 2016, 178, 72–83. [Google Scholar] [CrossRef]
- Chen, T.-T.; Zhu, S.-J.; Cao, H.; Shang, Y.-F.; Wang, M.-A.; Jiang, G.-Q.; Shi, Y.-J.; Lu, T.-H. Studies on the interaction of salvianolic acid B with human hemoglobin by multi-spectroscopic techniques. Spectrochim. Acta A 2011, 78, 1295–1301. [Google Scholar] [CrossRef]
- Ding, F.; Peng, W.; Peng, Y.-K. Biophysical exploration of protein-flavonol recognition: Effects of molecular properties and conformational flexibility. Phys. Chem. Chem. Phys. 2016, 18, 11959–11971. [Google Scholar] [CrossRef]
- Lu, Y.; Wang, Y.-L.; Gao, S.-H.; Wang, G.-K.; Yan, C.-L.; Chen, D.-J. Interaction of quercetin with ovalbumin: Spectroscopic and molecular modeling studies. J. Lumin. 2009, 129, 1048–1054. [Google Scholar] [CrossRef]
- Wu, S.-M.; Zhang, Y.-Y.; Ren, F.-Z.; Qin, Y.-H.; Liu, J.-X.; Liu, J.-W.; Wang, Q.-Y.; Zhang, H. Structure-affinity relationship of the interaction between phenolic acids and their derivatives and β-lactoglobulin and effect on antioxidant activity. Food Chem. 2018, 245, 613–619. [Google Scholar] [CrossRef] [PubMed]
- Wang, Q.; Liu, X.-Y.; Su, M.; Shi, Z.-H.; Sun, H.-W. Study on the interaction characteristics of cefamandole with bovine serum albumin by spectroscopic technique. Spectrochim. Acta A 2015, 136, 321–326. [Google Scholar] [CrossRef] [PubMed]
- Yu, X.; Cai, X.; Li, S.; Luo, L.-Y.; Wang, J.; Wang, M.; Zeng, L. Studies on the interactions of theaflavin-3,3′-digallate with bovine serum albumin: Multi- spectroscopic analysis and molecular docking. Food Chem. 2021, 366, 130422. [Google Scholar] [CrossRef]
- Wen, Q.H.; Wang, L.H.; Zeng, X.A.; Niu, D.B.; Wang, M.S. Hydroxyl-related differences for three dietary flavonoids as inhibitors of human purine nucleoside phosphorylase. Int. J. Biol. Macromol. 2018, 118, 588–598. [Google Scholar] [CrossRef]
- Liu, J.-G.; Li, Y.-J.; Chen, S.; Lin, Y.-P.; Lai, H.-Q.; Chen, B.-L.; Chen, T.-F. Biomedical application of reactive oxygen species-responsive nanocarriers in cancer, inflammation, and neurodegenerative diseases. Front. Chem. 2020, 8, 838. [Google Scholar] [CrossRef]
- Pisoschi, A.M.; Pop, A.; Iordache, F.; Stanca, L.; Predoi, G.; Serban, A.I. Oxidative stress mitigation by antioxidants—An overview on their chemistry and influences on health status. Eur. J. Med. Chem. 2021, 209, 112891. [Google Scholar] [CrossRef]
- Vaganek, A.; Rimarcik, J.; Lukes, V.; Klein, E. On the energetics of homolytic and heterolytic O-H bond cleavage in flavonoids. Comput. Theor. Chem. 2012, 991, 192–200. [Google Scholar] [CrossRef]
- Seeram, N.P.; Nair, M.G. Inhibition of lipid peroxidation and structure-activity-related studies of the dietary constituents anthocyanins, anthocyanidins, and catechins. J. Agric. Food Chem. 2002, 50, 5308–5312. [Google Scholar] [CrossRef]
- Amorati, R.; Valgimigli, L. Modulation of the antioxidant activity of phenols by non-covalent interactions. Org. Biomol. Chem. 2012, 10, 4147–4158. [Google Scholar] [CrossRef]
- Qie, X.J.; Chen, W.P.; Zeng, M.M.; Wang, Z.J.; Chen, J.; Goff, H.D.; He, Z.Y. Interaction between beta-lactoglobulin and chlorogenic acid and its effect on antioxidant activity and thermal stability. Food Hydrocolloid 2021, 121, 107059. [Google Scholar] [CrossRef]
- Geng, R.; Ma, L.; Liu, L.L.; Xie, Y.X. Influence of bovine serum albumin-flavonoid interaction on the antioxidant activity of dietary flavonoids: New evidence from electrochemical quantification. Molecules 2019, 24, 70. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liu, J.; Tu, Z.; Shan, Y.-H.; Wang, H.; Liu, G.-X.; Sha, X.-M.; Zhang, L.; Yang, P. Improved antioxidant activity and glycation of alpha-lactalbumin after ultrasonic pretreatment revealed by high-resolution mass spectrometry. J. Agric. Food Chem. 2017, 65, 10317–10324. [Google Scholar] [CrossRef] [PubMed]


| Sample | Secondary Structure | |||
|---|---|---|---|---|
| α-Helix (%) | β-Sheet (%) | Turns (%) | Unordered (%) | |
| OVA | 27.40 ± 1.60 b | 18.25 ± 2.22 a | 23.33 ± 0.37 a | 31.02 ± 0.29 a |
| OVA+Que | 25.84 ± 0.90 ab | 20.47 ± 1.02 ab | 23.38 ± 0.22 a | 30.35 ± 0.10 a |
| OVA+Kaem | 24.76 ± 0.31 a | 21.29 ± 0.19 a | 22.84 ± 0.45 a | 31.09 ± 0.60 a |
| OVA+Ish | 26.12 ± 0.79 ab | 19.66 ± 1.36 ab | 23.61 ± 0.65 a | 30.62 ± 0.73 a |
| OVA+Myri | 25.92 ± 0.83 ab | 20.63 ± 1.36 ab | 22.84 ± 0.23 a | 30.61 ± 0.69 a |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhou, W.; Peng, C.; Wang, D.; Li, J.; Tu, Z.; Zhang, L. Interaction Mechanism between OVA and Flavonoids with Different Hydroxyl Groups on B-Ring and Effect on Antioxidant Activity. Foods 2022, 11, 1302. https://doi.org/10.3390/foods11091302
Zhou W, Peng C, Wang D, Li J, Tu Z, Zhang L. Interaction Mechanism between OVA and Flavonoids with Different Hydroxyl Groups on B-Ring and Effect on Antioxidant Activity. Foods. 2022; 11(9):1302. https://doi.org/10.3390/foods11091302
Chicago/Turabian StyleZhou, Wenna, Chunyan Peng, Danshu Wang, Jinlin Li, Zongcai Tu, and Lu Zhang. 2022. "Interaction Mechanism between OVA and Flavonoids with Different Hydroxyl Groups on B-Ring and Effect on Antioxidant Activity" Foods 11, no. 9: 1302. https://doi.org/10.3390/foods11091302






