Application of Near-Infrared Spectroscopy and Hyperspectral Imaging Combined with Machine Learning Algorithms for Quality Inspection of Grape: A Review
Abstract
:1. Introduction
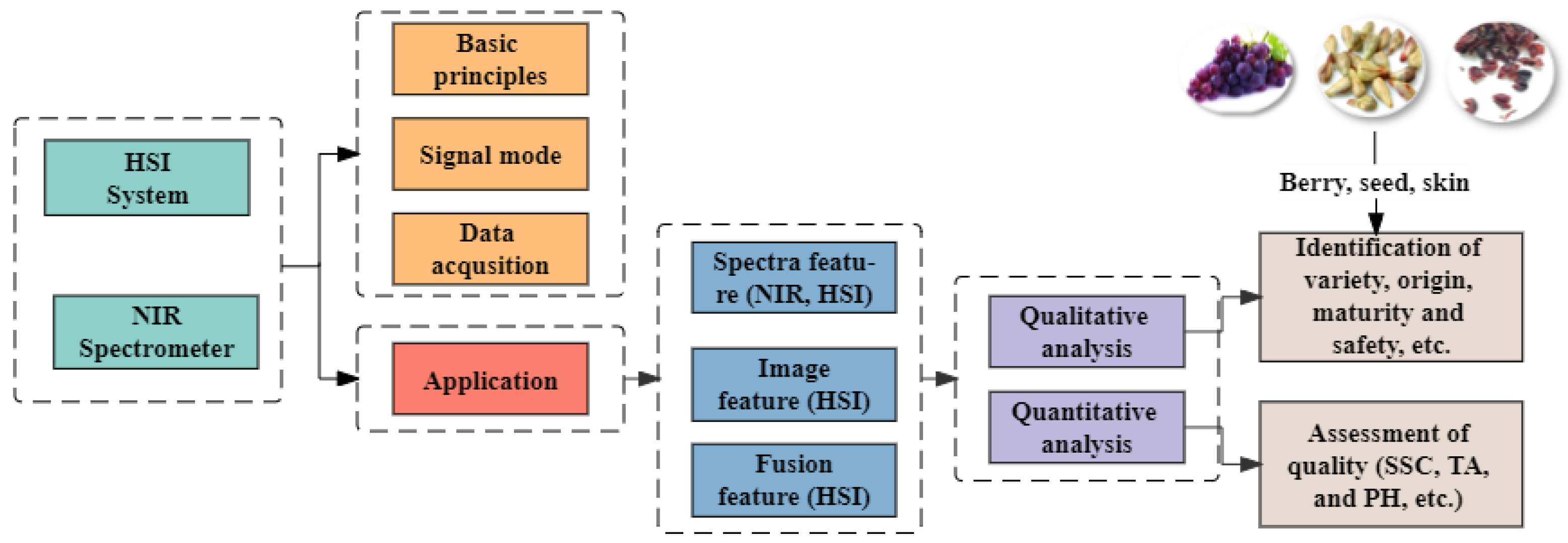
2. Introduction to NIRS and HSI Technologies
2.1. Basic Principles and Signal Mode
2.2. Data Acquisition
2.3. Data Analysis and Processing
- (1)
- Division of sample set. When the sample set is divided, the sample’s content distribution, gradient, and physical and chemical properties should be considered to improve the calibration model’s stability and expand the model’s practical application. The main dividing methods include Kolmogorov-Smirnov (KS) [27], sample set partitioning based on joint x-y distance (SPXY) [28] and random ratio.
- (2)
- Collection and extraction of data. The NIRS and HSI data are obtained, and the chemical analysis values are measured. NIRS acquires spectral data and directly processes it later. Regarding HSI data, it will be corrected with a black and white reference image to eliminate random noise signals caused by a light source or power supply [29]. The region of interest (ROI) is extracted using masking to remove the background.
- (3)
- Data preprocessing. The spectral signal obtained by the detector includes various non-target factors, such as high-frequency random noise, baseline drift, stray light, etc. Therefore, the obtained spectra should be reasonably pretreated before data analysis for the specific spectral measurement and sample. Normalization, Savitzky-Golay (SG) [30], Standard Normal Variate (SNV) [31], and Multiplicative Scatter Correction (MSC) [32] have been used widely to reduce noise. Normalization is to map the data to the range to unify the dimension and speed up the calculation. Besides, it could reduce the spectral difference caused by the varying height of the sample surface. SG can eliminate spectral noise, such as baseline offset, tilt, reverse, etc. SNV is commonly used to attenuate the slope variation of spectra. MSC is applied to remove the undesirable scatter effect. Besides, derivative processing, Fourier Transform (FT), Wavelet Transform (WT), etc., are applied in some cases.
- (4)
- Establishment of calibration models. For the qualitative analysis, the calibrations are conducted by the classification model using the sample label (variety, origin, year, etc.) as the dependent (Y) variable and grape spectra as the independent (X) variable [33]. Classification calibrations models are built, such as Partial least squares discriminant analysis (PLS-DA) [34], K-nearest Neighbor (KNN) [35], Support Vector Machine (SVM) [36], K-means [37], Artificial neural networks (ANN) [38], etc. For quantitative analysis, calibrations were developed by the regression model using the fruit physicochemical attribute as the dependent (Y) variable and grape spectra as the independent (X) variable [33]. Regression calibrations models are established, such as Partial Least-square Regression (PLSR) [39], Multiple Linear Regression (MLR) [40], SVM [36], ANN [38], Principle component regression (PCR) [41], etc. For NIRS, the input data is the principal component of the grape spectra. Regarding HSI, it is spectra, images, or a combination of spectra and image features.
- (5)
- Evaluation of the calibration model. The model conducted is evaluated for its reliability and generalization capability with external validation data sets or/and cross-validation techniques. There are some evaluation indices: Accuracy (acc), precision, recall, and F-score, etc., for qualitative analysis; the correlation coefficient (R), coefficient of determination (R2/RSQ), root mean squared error (RMSE), residual predictive deviation (RPD), etc., for quantitative analysis.
- (6)
- Prediction of unknown samples [42]. The unknown samples were scanned to obtain NIRS and HSI data, and their contents were calculated by models established and evaluated.
3. Applications
3.1. Spectral Feature Analysis
3.1.1. Qualitative Analysis
Variety Identification
| Variety | No | Mod | S/I | Attribute | Ext | Object | Model | Application | Best Result (Accuracy%) | Reference |
|---|---|---|---|---|---|---|---|---|---|---|
| ‘Kyoho’ | 86 | inter | S | seed or seedless | No | berry | PLS-DA | identify seed or seedless | acc = 93.10% | [49] |
| Graciano (two origins) | 84 | refl | S | phenolic in skin and seed | No | seed, skin, berry | DPLS | identify the origin | acc = 95%, 66%, 93% (DPLS, seed, berry, skin) | [50] |
| Manicure Finger, Ugni Blanc | 341 | inter | S | SSC, TP, CIELAB | No | cluster | PLS-DA | quality grade | 77.00–94.00% | [51] |
| Tempranillo, Syrah (two years) | 400 | drefl | S | TP, anth, flav | No | berry, skin | LDA, DPLS, Pearson | quality assessment | acc = 87.0, 91.3, 91.3 (LDA), others are poor result | [52] |
| Syrah, Cabernet Sauvignon | 1008 | refl | S | TSS, yellow flavonoids, anth | No | berry | PCA-LDA, PCA-QDA, LDA_Mahalanobis, PLS-DA | maturity evaluation | acc = 93.15% (PLS-DA), 92.86% (LDA), 92.26% (QDA), 92.26% (LDA_Mahalanobis) | [53] |
| Manicure Finger Ugni Blanc | 540 | drefl | S | L*a*b, SSC, TP | SPA, CARS | berry | PCA SVM-DA | maturity evaluation | acc = 90.00% (MF) acc = 100.00% (UB) (SSC-CARS-SVM-DA) | [54] |
| Sangiovese | 400 | absorb | S | SSC, TA, DI anth | No | berry | PCA | maturity evaluation | clear clusters (PC1 for 93.42%, PC2 for 4.72%) | [55] |
| Pedro Ximénez, Cabernet Sauvignon | 24 | refl | S | SSC, PH, TA, MA, reducing-sugar, tartaric acid | No | bunch | PLS-DA | maturity evaluation | acc = 79.00–100.00% | [56] |
| Variety | No | Mod | S/I | Attribute | Ext | Object | Model | Application | Best Result (Accuracy%) | Reference |
|---|---|---|---|---|---|---|---|---|---|---|
| Garnacha (two vineyards), Graciano, Mazuelo, Tempranillo | 50 | refl | SI | Chromatographic, color, NIR, fusion data | No | berry | Stepwise-LDA | identify grape variety | acc = 88%, 54%, 100%, 100% (internal validation) acc = 86%, 52%, 86%, 86% (external validation) | [57] |
| Six white and red wine grapes | 5640 | refl | S | No | PCA | berry | AdaBoost, SVM, RF | identify grape variety | acc = 81–93.00% | [43] |
| Hutai, Kyoho, Muscat, Summer black | 480(120 * 4 varieties) | refl | S | No | CARS, CARS-SPA, MCCV | berry | SVM | identity grape variety | acc = 99.3125% (CARS-SPA) | [44] |
| Tempranillo, Syrah, Zalem-a (two soils) | 56 | refl | S | No | PCA | seed | GDA | identity grape seed variety | acc = 100% (full wavelength), ≥96% (selected wavelength) | [47] |
| Hongtizi, Meirenzhi, Jufeng | 500 | refl | S | No | PCA | seed | SVM | identity grape seed variety | acc = 88.70% | [46] |
| Tempranillo | 1232 | refl | S | Flavanolic | PCA | seed | k-means | predict flavanolic | k-means clustering great | [48] |
| Variety | No | Mod | S/I | Attribute | Ext | Object | Model | Application | Best Result (Accuracy%) | Reference |
|---|---|---|---|---|---|---|---|---|---|---|
| Chardonnay, Grillo, Inzolia, Viognier, Nero’d’Avola, Syrah | 1235 healthy, 1324 diseased | refl | S | Healthy and diseased status | No | bunch | PLS-DA | phytosanitary status evaluation | acc = 89.80–94.00% | [58] |
| table grape | 686 | refl | S | no, single and double dose of pesticide | PCA, LASSO, Elastic Net regularization | cluster | ANN, SVM, RF, XGBoost | identity pesticide level | acc = 91.98% (SVM-LASSO) | [59] |
| cabernet sauvignon, Red grape, Munage | 1071 | refl | S | four mixed pesticide levels | No | cluster | RF, LR, SVM, ResNet | identity pesticide level | acc > 93.00% | [60] |
| Variety | No | Mod | Attribute | Ext | Object | Model | Application | Best Result(R2) | Reference |
|---|---|---|---|---|---|---|---|---|---|
| Grape mash (36 varieties) | 168 | refl | Fructose, PH Glucose, TA, Glycerol, MA, Gluconic acid, Ergosterol, Ethanol, acetic acid, Tartaric acid, Laccase activity | No | berry | PLSR | predict grape mashes composition | R2 = 0.873 (Relative density), 0.836 (Glycerol), 0.851 (Ergosterol), 0.345 (TA), PH (0.393) | [25] |
| Tannat (3 years) | 56 | refl | glycosylated aroma compounds | No | homogenized, juice | PLSR | predict glycosylated aroma compounds | RPD > 1.5 (5 and 4 norisoprenoids compounds, in homogenized and juice) | [61] |
| Cabernet Sauvignon, Syrah | 1008 | refl | TSS, anth, yellow flavonoids | No | berry | PCR, MLR, PLSR | quality evaluation | ≥0.90 (TSS and anthocyanins); ≥0.70 (flavonoids) | [53] |
| Autumn royal, Timpson, Sweet scarlett | 450 | refl | Dry matter (DM), TSS/SSC | No | berry | PLSR | quality evaluation | R2 = 0.83,0.81 (DM), 0.97, 0.95 (TSS) for two spectrometers | [62] |
| Jufeng | 115 | dtran | SSC | No | bunch | PLSR | quality evaluation | R = 0.83 | [63] |
| Tempranillo (laboratory, field) | 1643 | refl | TSS | No | berry | PLSR | quality evaluation | RMSEP = 1.42°Brix, SEP = 1.40°Brix (laboratory);1.68°Brix, 1.67 Brix (field) | [64] |
| Sangiovese | 9600 | drefl | Brix, Babo, TS, glucose, fructose, density, TA, tartaric acid, pH, MA, anth, TP, gluconic acid, assumable nitrogenm | No | berry | PLSR | quality evaluation | R2 = 0.93 (°Brix), 0.93 (°Babo), 0.94 (TS), 0.93 (glucose), 0.55 (TA), 0.92 (fructose), 0.91 (density), 0.66 (PH), 0.76 (anth) | [65] |
| Tempranillo | 144 | refl | TSS, anth, total polyphenols | PCA | bunch | PLSR | predict TSS, anth, total polyphenols | R2 = 0.95, 0.79, 0.43 | [66] |
| Grenache | 128 | refl | TSS, amino acid | No | cluster | PLSR | predict amino acids and TSS | R2~0.60 (asparagine, tyrosine proline in 570–1000; lysine, tyrosine, proline in 1100–2100), 0.90 (TSS) | [67] |
| Ruby Seedless grape | 700 | refl | SSC | No | berry | PLSR, LS-SVM | predict SSC | R2 = 0.889~0.918 (LS-SVM); 0.874~0.907 (P-LSR) | [68] |
| Syrah, Tempranillo | 400 | drefl | TP, anth, flava | No | berry, skin | MPLSR | quality evaluation | poor results | [52] |
| table grape cv Italia | 682 | drefl | SSC | No | berry | PLSR | sensory analysis | R2 = 0.85 (cross-validation); 0.82 (external validation) | [69] |
| Autumn Royal, Victoria | 350 | refl | TSS/SSC, TA | No | berry | PLSR | predict consumer preference driving factors | R2 = 0.5732 (TA), 0.8304 (TSS) | [70] |
| Thompson seedless, Regal seedless, Prime seedless | 338 | drefl | TSS, TA, PH, TSS/TA, BrimA | No | bunch | PLSR | predict maturity and sensory parameters | R2 = 0.71, 0.33, 0.57, 0.28, 0.77 | [71] |
| Graciano red grape (two vineyards) | 150 | refl | taste, texture, visual, olfactory feature | No | seed skin | MPLSR | predict sensory parameters and harvest time | seed (4.5% for hardness, 8.7% for colour), skin (9.8% for tannic intensity, 13.7% for astringency | [72] |
| Corvina | 300 | refl | TSS, DI, weight loss | No | berry | PLSR, PCA | predict withering quality | R2 = 0.62, RPD =1.87 (TSS); 0.56, 1.79 (firmness) | [73] |
| Manicure Finger (MF), Ugni Blanc (UB) | 540 | drefl | L*a*b, SSC TP | SPA CARS | berry | PLSR, LS-SVM | quality evaluation | R2 = 0.531~0.929 (LS-SVM), 0.520~0.897 (PLS); 0.897, 0.929 ( SSC, UB) | [54] |
| Sangiovese | 400 | absorb | SSC, TA, DI, anth | No | berry | Pearson | quality evaluation | R2 = 0.92 (SSC), 0.87 (TA), 0.89 (DI), 0.68~0.97 (anth) | [55] |
| ‘Kyoho’ grape | 172 | inter | DI, SSC, PH, | No | berry | PLSR | quality evaluation | R2 = 0.7427, 0.7804 (DI); 0.6276, 0.7676 (PH); 0.6926, 0.8052 (SSC) | [49] |
| Manicure Finger, Ugni Blanc | 341 | inter | SSC, TP, LAB | No | berry | PLSR | quality evaluation | R2 = 0.735, 0.823 (SSC, TP) | [51] |
| Variety | No | Mod | S/I | Attribute | Ext | Object | Model | Application | Best Result (R2) | Reference |
|---|---|---|---|---|---|---|---|---|---|---|
| Zalema, Te-mpranillo | 95 | refl | S | flav | No | seed | PLSR | predict flavanols in grape seeds | R2 = 0.88 (1 variety); 0.85 (2 varieties) | [45] |
| Syrah, Tempranillo | 99 | refl | S | anth | No | berry | MPLSR | Screen anthocyanins | R2 = 0.86 | [33] |
| Cabernet Sauvignon | 46 | refl | S | anth | PCA | skin | PLSR | detect anthocyanin concentration | R2 = 0.65 | [74] |
| Cabernet Sauvignon | 120 | refl | S | anth | PLSR | berry | PLSR SVR | predict the anthocyanin content | R2 = 0.94 (SVR) | [75] |
| Touriga Franca, Tin-ta Barroca, Touriga Nacional | 552 | refl | S | anth, PH sugar | PCA | bunch | SVR | prediction of oenological parameters for different vintages and varieties | R2 = 0.89 (anth);0.81 (PH); 0.90 (sugar) | [76] |
| Syrah, Tempranillo | 200 | refl | S | TP, anth, flav | PCA | skin | MPLSR | screen of extractable polyphenols in red grape skins | R2 = 0.82 (TP), 0.79 (anth); 0.82 (flavanol), | [77] |
| Tempranillo | 144 | refl | S | SSC/TSS, anth | PCA | berry | SVM | Evaluate TSS and anthocyanin concentration | R2 = 0.92 (TSS); 0.83 (anth) | [78] |
| Sangiovese | 429 | refl | S | SSC | VIP | berry | PLSR, PLS-DA | Evaluate SSC and assess harvest time | R2 = 0.77 (PLSR) acc = 0.86–91% (PLS-DA) | [79] |
| Kyoho grapes | 240 | refl | S | SSC/TSS | CARS, IRIV, V-MDRC | berry | LSSVM, PLSR | detect TSS | R2 P = 0.93 (VMD-RC-LSSVM) | [80] |
| Sangiovese | 33 | drefl | S | SSC | No | berry | PLSR | predict SSC in the field | R2 = 0.75, RMSECV = 0.84 | [81] |
| Tempranillo | 144 | refl | S | TSS, TA, PH, anth, MA, total polyphenols, ftartaric acid | No | cluster | PLSR | predict internal parameters | R2 = 0.82 (TSS), 0.81 (TA), 0.61(PH), 0.62 (Tartaric acid), 0.84 (MA), 0.88 (anth), 0.55 (Total polyphenols) | [82] |
| Sugarone Superior, Thompson, Victoria, Sable, Lival, Alphonse Lavallée, Black Magic | 350 | refl | S | flav, anth, TSS, | VIP, regression coefficient (PLS) | berry | PLS (full bands), MLR (selected bands) | predict TSS, anth, total flavonoid | MLR: (flav, anth, TSS, selected, β-coefficient) R2 = 0.93, 0.97, 0.97; 0.93, 0.98, 0.86 VIP-PLS: R2 = 0.95, 0.99, 0.94 | [83] |
| Touriga Franca, Tint-a Barroca, Touriga Nacional | 2665 | refl | S | sugar | No | berry | RR, NN, PLSR, 1DCNN | predict sugar content | R2 = 0.94 (1DCNN) | [84] |
| Touriga Franca (2012 and 2013) | 324 | refl | S | sugar | No | bunch | PLSR, NN | predict sugar content in new vintages | R2 = 0.93,0.92 (PLSR, NN, 2012); 0.95, 0.92 (PLSR, NN, 2013) for external | [85] |
| Touriga Franca (2012 and 2013) | 324 | refl | S | sugar | No | bunch | NN | predict sugar content (satisfactory generalization) | R2 = 0.906, RSME = 1.165 (2012); 0.959, RSME = 1.026 (2013) | [86] |
| Touriga Franca | 240 | refl | S | sugar, PH, anth | No | berry | NN | predict maturity parameters | R2 = 0.73 (PH), 0.92 (sugar), 0.95 (anth) | [87] |
| Touriga franca (TF, 2012 + 2013); Touriga nacional (TN, 2013); Tinta barroca (TB, 2013) | 465 | refl | S | PH, anth | No | berry | NN | predict PH and anthocyanin for new vintages and varieties | R2 = 0.72 (2013, TF, PH), 0.90 (2013.TF, anthocyanin) | [88] |
| Zalema, Syrah, Tempranillo | 213 | refl | S | TP, TA, sugar, PH | No | skin | MPLSR | screen and control maturity parameters | RSQ = 0.89 (TP), 0.99 (sugar), 0.98 (TA), 0.94(PH) | [89] |
| Globe grapes | 360 | drefl | SI | SSC | CARS, S-PA, UVE, GA, CA-RS-SPA, UVE-SPA | berry | PLSR | predict SCC | R2 c = 0.9775, R2 P = 0.9762 | [90] |
| 4 white and 3 red/black varieties | 140 | refl | SI | PH, TA, SSC | No | berry | PLSR | predict physical-chemical content and sensory | R2 = 0.95, 0.82 (TA); 0.94, 0.93 (SSC); 0.80, 0.90 (PH) for white and red/black grape | [91] |
| Kyoho grape | 240 | refl | S | DI, PH | SAE, SPA, CARS | berry | LSSVM, PLS | predict DI and PH | R2 = 0.923 (SAE-LSSVM) | [92] |
Maturity Identification
Seeded and Seedless and Geographical Origin Identification
Safety Inspection
3.1.2. Quantitative Analysis
Quality Assessment
Parameters Sensory Prediction
3.1.3. Conclusions
3.2. Image Feature Analysis
3.2.1. Qualitative Analysis
3.2.2. Quantitative Analysis
3.2.3. Conclusions
3.3. Fusion Data Analysis
3.3.1. Qualitative Analysis
3.3.2. Quantitative Analysis
3.3.3. Conclusions
4. Challenges and Prospects
5. Conclusions
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| 1D-CNN | One-dimensional convolutional neural networks |
| ANN/NN | Artificial neural networks |
| Anth | Total anthocyanins |
| CARS | Competitive adaptive reweighted sample |
| DI | Durofel index (berry firmness) |
| DL | Deep learning |
| DM | Dry matter |
| DPLS | Discriminant partial least square |
| DWT | Discrete wavelet transform |
| EEMD | Ensemble empirical mode decomposition |
| Flav | Flavanols |
| GLCM | Grey-level co-occurrence matrix |
| GDA | General discriminant analysis |
| IRIV | Iteratively retains informative variables |
| LAB | color space values |
| LDA | Linear discriminant analysis |
| LV | Latent variable/factors |
| MA | Malic acid contents |
| MCCV | Monte Carlo cross-validation |
| ML | Machine learning |
| MPLSR | Modified partial least squares regression |
| MSC | Multivariate scattering correction |
| PCA | Principal component analysis |
| Pearson | Pearson’s similarity index (1/(1 − R2)) |
| PLS | Partial least squares analysis |
| PLS-DA | PLS discriminant analysis |
| PLSR | Partial least-square regression |
| QDA | Quadratic discriminant analysis |
| RF | Random forest |
| ResNet | Residual Network |
| RR | Ridge regression |
| RSQ/R2 | Coefficient of determination |
| SAE | Stacked auto-encoders |
| SG | Savitzky-Golay smoothing |
| SNV | Standard normal variate transform |
| SSC | Soluble solids content |
| SVR | Support vector regression |
| TA | Titratable acidity |
| TB | Tinta Barroca |
| TF | Touriga Franca |
| TN | Touriga Nacional |
| TP | Total phenolic |
| TS | Total sugars |
| TSS | Total soluble solids |
| VIP | Variable importance in projection |
| VMD-RC | Variational mode decomposition (VMD)-regression coefficients |
References
- Santos, J.A.; Fraga, H.; Malheiro, A.C.; Moutinho-Pereira, J.; Dinis, L.-T.; Correia, C.; Moriondo, M.; Leolini, L.; Dibari, C.; Costafreda-Aumedes, S.; et al. A Review of the Potential Climate Change Impacts and Adaptation Options for European Viticulture. Appl. Sci. 2020, 10, 3092. [Google Scholar] [CrossRef]
- Kalekhan, F.; Bala, N.; Rao, S.; Pais, M.L.J.; Adnan, M.; Sajan, S.; Baliga, M.S. Usefulness of grape seed polyphenols in the prevention of skin cancer: A mini review. In Functional Foods in Cancer Prevention and Therapy; Academic Press: Cambridge, MA, USA, 2020; pp. 159–167. [Google Scholar]
- Shiraishi, M.; Fujishima, H.; Chijiwa, H. Evaluation of table grape genetic resources for sugar, organic acid, and amino acid composition of berries. Euphytica 2009, 174, 1–13. [Google Scholar] [CrossRef]
- Sharma, M.; Usmani, Z.; Gupta, V.K.; Bhat, R. Valorization of fruits and vegetable wastes and by-products to produce natural pigments. Crit. Rev. Biotechnol. 2021, 41, 535–563. [Google Scholar] [CrossRef]
- Park, E.; Luo, Y.; Marine, S.C.; Everts, K.L.; Micallef, S.A.; Bolten, S.; Stommel, J. Consumer preference and physicochemical evaluation of organically grown melons. Postharvest. Biol. Technol. 2018, 141, 77–85. [Google Scholar] [CrossRef]
- Mohd Ali, M.; Hashim, N. Non-destructive methods for detection of food quality. In Future Foods; Elsevier: Amsterdam, The Netherlands, 2022; pp. 645–667. [Google Scholar]
- Reuhs, B.L. High-Performance Liquid Chromatography. In Food Analysis; Food Science Text Series; Springer: Berlin/Heidelberg, Germany, 2017; pp. 213–226. [Google Scholar]
- Feng, T.; Sun, M.; Song, S.; Zhuang, H.; Yao, L. Gas chromatography for food quality evaluation. In Evaluation Technologies for Food Quality; Woodhead Publishing: Sawston, UK, 2019; pp. 219–265. [Google Scholar]
- Aiello, D.; De Luca, D.; Gionfriddo, E.; Naccarato, A.; Napoli, A.; Romano, E.; Russo, A.; Sindona, G.; Tagarelli, A. Review: Multistage mass spectrometry in quality, safety and origin of foods. Eur. J. Mass Spectrom. 2011, 17, 1–31. [Google Scholar] [CrossRef]
- Putri, S.P.; Ikram, M.M.M.; Sato, A.; Dahlan, H.A.; Rahmawati, D.; Ohto, Y.; Fukusaki, E. Application of gas chromatography-mass spectrometry-based metabolomics in food science and technology. J. Biosci. Bioeng. 2022, 133, 425–435. [Google Scholar] [CrossRef]
- Reddy, P.; Guthridge, K.M.; Panozzo, J.; Ludlow, E.J.; Spangenberg, G.C.; Rochfort, S.J. Near-Infrared Hyperspectral Imaging Pipelines for Pasture Seed Quality Evaluation: An Overview. Sensors 2022, 22, 1981. [Google Scholar] [CrossRef]
- Caporaso, N.; Whitworth, M.B.; Fisk, I.D. Near-Infrared spectroscopy and hyperspectral imaging for non-destructive quality assessment of cereal grains. Appl. Spectrosc. Rev. 2018, 53, 667–687. [Google Scholar] [CrossRef] [Green Version]
- Kohler, L.H.; Kohler, H.; Kohler, S.; Langer, S.; Nuwayhid, R.; Gockel, I.; Spindler, N.; Osterhoff, G. Hyperspectral Imaging (HSI) as a new diagnostic tool in free flap monitoring for soft tissue reconstruction: A proof of concept study. BMC Surg. 2021, 21, 222. [Google Scholar] [CrossRef]
- Zhang, S.; Liu, S.; Shen, L.; Chen, S.; He, L.; Liu, A. Application of near-infrared spectroscopy for the nondestructive analysis of wheat flour: A review. Curr. Res. Food Sci. 2022, 5, 1305–1312. [Google Scholar] [CrossRef]
- Cen, H.; He, Y. Theory and application of near infrared reflectance spectroscopy in determination of food quality. Trends Food Sci. Technol. 2007, 18, 72–83. [Google Scholar] [CrossRef]
- Nicolaï, B.M.; Beullens, K.; Bobelyn, E.; Peirs, A.; Saeys, W.; Theron, K.I.; Lammertyn, J. Nondestructive measurement of fruit and vegetable quality by means of NIR spectroscopy: A review. Postharvest Biol. Technol. 2007, 46, 99–118. [Google Scholar] [CrossRef]
- Hussain, A.; Pu, H.; Sun, D.-W. Innovative nondestructive imaging techniques for ripening and maturity of fruits—A review of recent applications. Trends Food Sci. Technol. 2018, 72, 144–152. [Google Scholar] [CrossRef]
- Saha, D.; Manickavasagan, A. Machine learning techniques for analysis of hyperspectral images to determine quality of food products: A review. Curr. Res. Food Sci. 2021, 4, 28–44. [Google Scholar] [CrossRef] [PubMed]
- Porep, J.U.; Kammerer, D.R.; Carle, R. On-line application of near infrared (NIR) spectroscopy in food production. Trends Food Sci. Technol. 2015, 46, 211–230. [Google Scholar] [CrossRef]
- Norris, K.H. Reflectance spectroscopy. In Modern Methods of Food Analysis; Springer: Dordrecht, The Netherlands, 1984; pp. 167–186. [Google Scholar] [CrossRef]
- Xie, L.; Wang, A.; Xu, H.; Fu, X.; Ying, Y. Applications of Near-Infrared Systems for Quality Evaluation of Fruits: A Review. Trans. ASABE 2016, 59, 399–419. [Google Scholar] [CrossRef]
- Lu, Y.; Saeys, W.; Kim, M.; Peng, Y.; Lu, R. Hyperspectral imaging technology for quality and safety evaluation of horticultural products: A review and celebration of the past 20-year progress. Postharvest Biol. Technol. 2020, 170, 111318. [Google Scholar] [CrossRef]
- Qu, J.H.; Liu, D.; Cheng, J.H.; Sun, D.W.; Ma, J.; Pu, H.; Zeng, X.A. Applications of near-infrared spectroscopy in food safety evaluation and control: A review of recent research advances. Crit. Rev. Food Sci. Nutr. 2015, 55, 1939–1954. [Google Scholar] [CrossRef]
- Ferrer-Gallego, R.; Hernandez-Hierro, J.M.; Rivas-Gonzalo, J.C.; Escribano-Bailon, M.T. Feasibility study on the use of near infrared spectroscopy to determine flavanols in grape seeds. Talanta 2010, 82, 1778–1783. [Google Scholar] [CrossRef]
- Porep, J.U.; Erdmann, M.E.; Körzendörfer, A.; Kammerer, D.R.; Carle, R. Rapid determination of ergosterol in grape mashes for grape rot indication and further quality assessment by means of an industrial near infrared/visible (NIR/VIS) spectrometer—A feasibility study. Food Control 2014, 43, 142–149. [Google Scholar] [CrossRef]
- Mehta, N.; Shaik, S.; Devireddy, R.; Gartia, M.R. Single-Cell Analysis Using Hyperspectral Imaging Modalities. J. Biomech. Eng. 2018, 140, 0208021–02080216. [Google Scholar] [CrossRef] [PubMed]
- Hoffman, J.I.E. Categorical and Cross-Classified Data: McNemar’s and Bowker’s Tests, Kolmogorov-Smirnov Tests, Concordance. In Basic Biostatistics for Medical and Biomedical Practitioners; Academic Press: Cambridge, MA, USA, 2019; pp. 233–247. [Google Scholar]
- Galvao, R.K.; Araujo, M.C.; Jose, G.E.; Pontes, M.J.; Silva, E.C.; Saldanha, T.C. A method for calibration and validation subset partitioning. Talanta 2005, 67, 736–740. [Google Scholar] [CrossRef] [PubMed]
- Jia, B.; Wang, W.; Ni, X.; Lawrence, K.C.; Zhuang, H.; Yoon, S.-C.; Gao, Z. Essential processing methods of hyperspectral images of agricultural and food products. Chemom. Intell. Lab. Syst. 2020, 198, 103936. [Google Scholar] [CrossRef]
- Luo, J.; Ying, K.; Bai, J. Savitzky–Golay smoothing and differentiation filter for even number data. Signal Process. 2005, 85, 1429–1434. [Google Scholar] [CrossRef]
- Grisanti, E.; Totska, M.; Huber, S.; Krick Calderon, C.; Hohmann, M.; Lingenfelser, D.; Otto, M. Dynamic Localized SNV, Peak SNV, and Partial Peak SNV: Novel Standardization Methods for Preprocessing of Spectroscopic Data Used in Predictive Modeling. J. Spectrosc. 2018, 2018, 1–14. [Google Scholar] [CrossRef]
- Isaksson, T.; Næs, T. The Effect of Multiplicative Scatter Correction (MSC) and Linearity Improvement in NIR Spectroscopy. Appl. Spectrosc. 1988, 42, 1273–1284. [Google Scholar] [CrossRef]
- Hernandez-Hierro, J.M.; Nogales-Bueno, J.; Rodriguez-Pulido, F.J.; Heredia, F.J. Feasibility study on the use of near-infrared hyperspectral imaging for the screening of anthocyanins in intact grapes during ripening. J. Agric. Food Chem. 2013, 61, 9804–9809. [Google Scholar] [CrossRef]
- Lee, L.C.; Liong, C.Y.; Jemain, A.A. Partial least squares-discriminant analysis (PLS-DA) for classification of high-dimensional (HD) data: A review of contemporary practice strategies and knowledge gaps. Analyst 2018, 143, 3526–3539. [Google Scholar] [CrossRef]
- Adeniyi, D.A.; Wei, Z.; Yongquan, Y. Automated web usage data mining and recommendation system using K-Nearest Neighbor (KNN) classification method. Appl. Comput. Inform. 2016, 12, 90–108. [Google Scholar] [CrossRef]
- Cervantes, J.; Garcia-Lamont, F.; Rodríguez-Mazahua, L.; Lopez, A. A comprehensive survey on support vector machine classification: Applications, challenges and trends. Neurocomputing 2020, 408, 189–215. [Google Scholar] [CrossRef]
- Gong, W.; Zhao, R.; Grünewald, S. Structured sparse K-means clustering via Laplacian smoothing. Pattern Recognit. Lett. 2018, 112, 63–69. [Google Scholar] [CrossRef]
- Abiodun, O.I.; Jantan, A.; Omolara, A.E.; Dada, K.V.; Mohamed, N.A.; Arshad, H. State-of-the-art in artificial neural network applications: A survey. Heliyon 2018, 4, e00938. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cheng, J.-H.; Sun, D.-W. Partial Least Squares Regression (PLSR) Applied to NIR and HSI Spectral Data Modeling to Predict Chemical Properties of Fish Muscle. Food Eng. Rev. 2016, 9, 36–49. [Google Scholar] [CrossRef]
- Krzywinski, M.; Altman, N. Multiple linear regression. Nat. Methods 2015, 12, 1103–1104. [Google Scholar] [CrossRef] [PubMed]
- Artigue, H.; Smith, G.; Lu, Z. The principal problem with principal components regression. Cogent Math. Stat. 2019, 6, 1622190–1622201. [Google Scholar] [CrossRef]
- Khare, S.; Aggarwal, S. Predicting Unknown Classes on Hyperspectral Image Data Using Deep Learning Techniques. In Proceedings of the 2021 IEEE International India Geoscience and Remote Sensing Symposium (InGARSS), Ahmedabad, India, 6–10 December 2021; pp. 475–478. [Google Scholar]
- Liu, X.; Zhang, E. Identification of Wine Grape Varieties Based on Near-infrared Hyperspectral Imaging. Appl. Eng. Agric. 2019, 35, 959–967. [Google Scholar] [CrossRef]
- Xu, M.; Sun, J.; Zhou, X.; Tang, N.; Shen, J.; Wu, X. Research on nondestructive identification of grape varieties based on EEMD-DWT and hyperspectral image. J. Food Sci. 2021, 86, 2011–2023. [Google Scholar] [CrossRef]
- Rodriguez-Pulido, F.J.; Hernandez-Hierro, J.M.; Nogales-Bueno, J.; Gordillo, B.; Gonzalez-Miret, M.L.; Heredia, F.J. A novel method for evaluating flavanols in grape seeds by near infrared hyperspectral imaging. Talanta 2014, 122, 145–150. [Google Scholar] [CrossRef]
- Zhao, Y.; Zhang, C.; Zhu, S.; Gao, P.; Feng, L.; He, Y. Non-Destructive and Rapid Variety Discrimination and Visualization of Single Grape Seed Using Near-Infrared Hyperspectral Imaging Technique and Multivariate Analysis. Molecules 2018, 23, 1352. [Google Scholar] [CrossRef]
- Rodríguez-Pulido, F.J.; Barbin, D.F.; Sun, D.-W.; Gordillo, B.; González-Miret, M.L.; Heredia, F.J. Grape seed characterization by NIR hyperspectral imaging. Postharvest Biol. Technol. 2013, 76, 74–82. [Google Scholar] [CrossRef]
- Quijada-Morin, N.; Garcia-Estevez, I.; Nogales-Bueno, J.; Rodriguez-Pulido, F.J.; Heredia, F.J.; Rivas-Gonzalo, J.C.; Escribano-Bailon, M.T.; Hernandez-Hierro, J.M. Trying to set up the flavanolic phases during grape seed ripening: A spectral and chemical approach. Talanta 2016, 160, 556–561. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kanchanomai, C.; Ohashi, S.; Naphrom, D.; Nemoto, W.; Maniwara, P.; Nakano, K. Non-destructive analysis of Japanese table grape qualities using near-infrared spectroscopy. Hortic. Environ. Biotechnol. 2020, 61, 725–733. [Google Scholar] [CrossRef]
- Ferrer-Gallego, R.; Hernandez-Hierro, J.M.; Rivas-Gonzalo, J.C.; Escribano-Bailon, M.T. A comparative study to distinguish the vineyard of origin by NIRS using entire grapes, skins and seeds. J. Sci. Food Agric. 2013, 93, 967–972. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Xiao, H.; Feng, L.; Song, D.; Tu, K.; Peng, J.; Pan, L. Grading and Sorting of Grape Berries Using Visible-Near Infrared Spectroscopy on the Basis of Multiple Inner Quality Parameters. Sensors 2019, 19, 2600. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Baca-Bocanegra, B.; Hernandez-Hierro, J.M.; Nogales-Bueno, J.; Heredia, F.J. Feasibility study on the use of a portable micro near infrared spectroscopy device for the “in vineyard” screening of extractable polyphenols in red grape skins. Talanta 2019, 192, 353–359. [Google Scholar] [CrossRef]
- Dos Santos Costa, D.; Oliveros Mesa, N.F.; Santos Freire, M.; Pereira Ramos, R.; Teruel Mederos, B.J. Development of predictive models for quality and maturation stage attributes of wine grapes using vis-nir reflectance spectroscopy. Postharvest Biol. Technol. 2019, 150, 166–178. [Google Scholar] [CrossRef]
- Xiao, H.; Li, A.; Li, M.; Sun, Y.; Tu, K.; Wang, S.; Pan, L. Quality assessment and discrimination of intact white and red grapes from Vitis vinifera L. at five ripening stages by visible and near-infrared spectroscopy. Sci. Hortic. 2018, 233, 99–107. [Google Scholar] [CrossRef]
- Ribera-Fonseca, A.; Noferini, M.; Jorquera-Fontena, E.; Rombolà, A.D. Assessment of technological maturity parameters and anthocyanins in berries of cv. Sangiovese (Vitis vinifera L.) by a portable vis/NIR device. Sci. Hortic. 2016, 209, 229–235. [Google Scholar] [CrossRef]
- González-Caballero, V.; Sánchez, M.-T.; Fernández-Novales, J.; López, M.-I.; Pérez-Marín, D. On-Vine Monitoring of Grape Ripening Using Near-Infrared Spectroscopy. Food Anal. Methods 2012, 5, 1377–1385. [Google Scholar] [CrossRef]
- Nogales-Bueno, J.; Rodriguez-Pulido, F.J.; Heredia, F.J.; Hernandez-Hierro, J.M. Comparative study on the use of anthocyanin profile, color image analysis and near-infrared hyperspectral imaging as tools to discriminate between four autochthonous red grape cultivars from La Rioja (Spain). Talanta 2015, 131, 412–416. [Google Scholar] [CrossRef]
- Beghi, R.; Giovenzana, V.; Brancadoro, L.; Guidetti, R. Rapid evaluation of grape phytosanitary status directly at the check point station entering the winery by using visible/near infrared spectroscopy. J. Food Eng. 2017, 204, 46–54. [Google Scholar] [CrossRef]
- Kim, M.S.; Chao, K.; Chin, B.A.; Cho, B.-K.; Mohite, J.; Karale, Y.; Pappula, S.; Shabeer, T.P.A.; Sawant, S.D. Detection of pesticide (Cyantraniliprole) residue on grapes using hyperspectral sensing. In Proceedings of the Sensing for Agriculture and Food Quality and Safety IX, Anaheim, CA, USA, 13 April 2017. [Google Scholar]
- Ye, W.; Yan, T.; Zhang, C.; Duan, L.; Chen, W.; Song, H.; Zhang, Y.; Xu, W.; Gao, P. Detection of Pesticide Residue Level in Grape Using Hyperspectral Imaging with Machine Learning. Foods 2022, 11, 1609. [Google Scholar] [CrossRef] [PubMed]
- Boido, E.; Fariña, L.; Carrau, F.; Dellacassa, E.; Cozzolino, D. Characterization of Glycosylated Aroma Compounds in Tannat Grapes and Feasibility of the Near Infrared Spectroscopy Application for Their Prediction. Food Anal. Methods 2012, 6, 100–111. [Google Scholar] [CrossRef]
- Donis-González, I.R.; Valero, C.; Momin, M.A.; Kaur, A.; Slaughter, C.D. Performance Evaluation of Two Commercially Available Portable Spectrometers to Non-Invasively Determine Table Grape and Peach Quality Attributes. Agronomy 2020, 10, 148. [Google Scholar] [CrossRef] [Green Version]
- Yu, J.; Wang, H.; Sun, X.; Huang, W. Parameter optimization in soluble solid content prediction of entire bunches of grape based on near infrared spectroscopic technique. J. Food Meas. Charact. 2017, 11, 1676–1680. [Google Scholar] [CrossRef]
- Urraca, R.; Sanz-Garcia, A.; Tardaguila, J.; Diago, M.P. Estimation of total soluble solids in grape berries using a hand-held NIR spectrometer under field conditions. J. Sci. Food Agric. 2016, 96, 3007–3016. [Google Scholar] [CrossRef] [Green Version]
- Barnaba, F.E.; Bellincontro, A.; Mencarelli, F. Portable NIR-AOTF spectroscopy combined with winery FTIR spectroscopy for an easy, rapid, in-field monitoring of Sangiovese grape quality. J. Sci. Food Agric. 2014, 94, 1071–1077. [Google Scholar] [CrossRef]
- Fernandez-Novales, J.; Tardaguila, J.; Gutierrez, S.; Paz Diago, M. On-The-Go VIS + SW—NIR Spectroscopy as a Reliable Monitoring Tool for Grape Composition within the Vineyard. Molecules 2019, 24, 2795. [Google Scholar] [CrossRef] [Green Version]
- Fernandez-Novales, J.; Garde-Cerdan, T.; Tardaguila, J.; Gutierrez-Gamboa, G.; Perez-Alvarez, E.P.; Diago, M.P. Assessment of amino acids and total soluble solids in intact grape berries using contactless Vis and NIR spectroscopy during ripening. Talanta 2019, 199, 244–253. [Google Scholar] [CrossRef]
- Xiao, H.; Sun, K.; Sun, Y.; Wei, K.; Tu, K.; Pan, L. Comparison of Benchtop Fourier-Transform (FT) and Portable Grating Scanning Spectrometers for Determination of Total Soluble Solid Contents in Single Grape Berry (Vitis vinifera L.) and Calibration Transfer. Sensors 2017, 17, 2693. [Google Scholar] [CrossRef] [Green Version]
- Parpinello, G.P.; Nunziatini, G.; Rombolà, A.D.; Gottardi, F.; Versari, A. Relationship between sensory and NIR spectroscopy in consumer preference of table grape (cv Italia). Postharvest Biol. Technol. 2013, 83, 47–53. [Google Scholar] [CrossRef]
- Basile, T.; Marsico, A.D.; Cardone, M.F.; Antonacci, D.; Perniola, R. FT-NIR Analysis of Intact Table Grape Berries to Understand Consumer Preference Driving Factors. Foods 2020, 9, 98. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Daniels, A.J.; Poblete-Echeverria, C.; Opara, U.L.; Nieuwoudt, H.H. Measuring Internal Maturity Parameters Contactless on Intact Table Grape Bunches Using NIR Spectroscopy. Front. Plant Sci. 2019, 10, 1517. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ferrer-Gallego, R.; Hernández-Hierro, J.M.; Rivas-Gonzalo, J.C.; Escribano-Bailón, M.T. Evaluation of sensory parameters of grapes using near infrared spectroscopy. J. Food Eng. 2013, 118, 333–339. [Google Scholar] [CrossRef]
- Beghi, R.; Giovenzana, V.; Marai, S.; Guidetti, R. Rapid monitoring of grape withering using visible near-infrared spectroscopy. J. Sci. Food Agric. 2015, 95, 3144–3149. [Google Scholar] [CrossRef]
- Fernandes, A.M.; Oliveira, P.; Moura, J.P.; Oliveira, A.A.; Falco, V.; Correia, M.J.; Melo-Pinto, P. Determination of anthocyanin concentration in whole grape skins using hyperspectral imaging and adaptive boosting neural networks. J. Food Eng. 2011, 105, 216–226. [Google Scholar] [CrossRef]
- Chen, S.; Zhang, F.; Ning, J.; Liu, X.; Zhang, Z.; Yang, S. Predicting the anthocyanin content of wine grapes by NIR hyperspectral imaging. Food Chem. 2015, 172, 788–793. [Google Scholar] [CrossRef]
- Silva, R.; Gomes, V.; Mendes-Faia, A.; Melo-Pinto, P. Using Support Vector Regression and Hyperspectral Imaging for the Prediction of Oenological Parameters on Different Vintages and Varieties of Wine Grape Berries. Remote Sens. 2018, 10, 312. [Google Scholar] [CrossRef] [Green Version]
- Nogales-Bueno, J.; Baca-Bocanegra, B.; Rodriguez-Pulido, F.J.; Heredia, F.J.; Hernandez-Hierro, J.M. Use of near infrared hyperspectral tools for the screening of extractable polyphenols in red grape skins. Food Chem. 2015, 172, 559–564. [Google Scholar] [CrossRef]
- Gutiérrez, S.; Tardaguila, J.; Fernández-Novales, J.; Diago, M.P. On-the-go hyperspectral imaging for the in-field estimation of grape berry soluble solids and anthocyanin concentration. Aust. J. Grape Wine Res. 2019, 25, 127–133. [Google Scholar] [CrossRef] [Green Version]
- Benelli, A.; Cevoli, C.; Ragni, L.; Fabbri, A. In-field and non-destructive monitoring of grapes maturity by hyperspectral imaging. Biosyst. Eng. 2021, 207, 59–67. [Google Scholar] [CrossRef]
- Xu, M.; Sun, J.; Yao, K.; Wu, X.; Shen, J.; Cao, Y.; Zhou, X. Nondestructive detection of total soluble solids in grapes using VMD-RC and hyperspectral imaging. J. Food Sci. 2022, 87, 326–338. [Google Scholar] [CrossRef] [PubMed]
- Benelli, A.; Cevoli, C.; Fabbri, A. In-field Vis/NIR hyperspectral imaging to measure soluble solids content of wine grape berries during ripening. In Proceedings of the 2020 IEEE International Workshop on Metrology for Agriculture and Forestry (MetroAgriFor), Trento, Italy, 4–6 November 2020. [Google Scholar] [CrossRef]
- Fernández-Novales, J.; Barrio, I.; Diago, M.P. Non-Invasive Monitoring of Berry Ripening Using On-the-Go Hyperspectral Imaging in the Vineyard. Agronomy 2021, 11, 2534. [Google Scholar] [CrossRef]
- Gabrielli, M.; Lançon-Verdier, V.; Picouet, P.; Maury, C. Hyperspectral Imaging to Characterize Table Grapes. Chemosensors 2021, 9, 71. [Google Scholar] [CrossRef]
- Gomes, V.; Reis, M.S.; Rovira-Más, F.; Mendes-Ferreira, A.; Melo-Pinto, P. Prediction of Sugar Content in Port Wine Vintage Grapes Using Machine Learning and Hyperspectral Imaging. Processes 2021, 9, 1241. [Google Scholar] [CrossRef]
- Gomes, V.M.; Fernandes, A.M.; Faia, A.; Melo-Pinto, P. Comparison of different approaches for the prediction of sugar content in new vintages of whole Port wine grape berries using hyperspectral imaging. Comput. Electron. Agric. 2017, 140, 244–254. [Google Scholar] [CrossRef]
- Gomes, V.M.; Fernandes, A.M.; Faia, A.; Melo-Pinto, P. Determination of sugar content in whole Port Wine grape berries combining hyperspectral imaging with neural networks methodologies. In Proceedings of the 2014 IEEE Symposium on Computational Intelligence for Engineering Solutions (CIES), Orlando, FL, USA, 9–12 December 2014. [Google Scholar] [CrossRef]
- Fernandes, A.M.; Franco, C.; Mendes-Ferreira, A.; Mendes-Faia, A.; Costa, P.L.d.; Melo-Pinto, P. Brix, pH and anthocyanin content determination in whole Port wine grape berries by hyperspectral imaging and neural networks. Comput. Electron. Agric. 2015, 115, 88–96. [Google Scholar] [CrossRef]
- Gomes, V.; Fernandes, A.; Martins-Lopes, P.; Pereira, L.; Mendes Faia, A.; Melo-Pinto, P. Characterization of neural network generalization in the determination of pH and anthocyanin content of wine grape in new vintages and varieties. Food Chem. 2017, 218, 40–46. [Google Scholar] [CrossRef]
- Nogales-Bueno, J.; Hernandez-Hierro, J.M.; Rodriguez-Pulido, F.J.; Heredia, F.J. Determination of technological maturity of grapes and total phenolic compounds of grape skins in red and white cultivars during ripening by near infrared hyperspectral image: A preliminary approach. Food Chem. 2014, 152, 586–591. [Google Scholar] [CrossRef]
- Gao, S.; Xu, J.-h. Hyperspectral image information fusion-based detection of soluble solids content in red globe grapes. Comput. Electron. Agric. 2022, 196, 106822. [Google Scholar] [CrossRef]
- Baiano, A.; Terracone, C.; Peri, G.; Romaniello, R. Application of hyperspectral imaging for prediction of physico-chemical and sensory characteristics of table grapes. Comput. Electron. Agric. 2012, 87, 142–151. [Google Scholar] [CrossRef]
- Xu, M.; Sun, J.; Yao, K.; Cai, Q.; Shen, J.; Tian, Y.; Zhou, X. Developing deep learning based regression approaches for prediction of firmness and pH in Kyoho grape using Vis/NIR hyperspectral imaging. Infrared Phys. Technol. 2022, 120, 104003. [Google Scholar] [CrossRef]
- Le Moigne, M.; Maury, C.; Bertrand, D.; Jourjon, F. Sensory and instrumental characterisation of Cabernet Franc grapes according to ripening stages and growing location. Food Qual. Prefer. 2008, 19, 220–231. [Google Scholar] [CrossRef]
- Li, Q.; Li, G.; Zhang, J.; Yan, H.; Liu, W.; Min, S. A new strategy of applying modeling indicator determined method to high-level fusion for quantitative analysis. Spectrochim. Acta A Mol. Biomol. Spectrosc. 2019, 219, 274–280. [Google Scholar] [CrossRef] [PubMed]
- Sun, F.; Diao, Z.; Li, Y. Research on Data Fusion Method Based on Multisource Data Awareness of Internet of Things. J. Sens. 2022, 2022, 1–10. [Google Scholar] [CrossRef]
- Orlandi, G.; Calvini, R.; Foca, G.; Pigani, L.; Vasile Simone, G.; Ulrici, A. Data fusion of electronic eye and electronic tongue signals to monitor grape ripening. Talanta 2019, 195, 181–189. [Google Scholar] [CrossRef]
- Ghamisi, P.; Rasti, B.; Yokoya, N.; Wang, Q.M.; Hofle, B.; Bruzzone, L.; Bovolo, F.; Chi, M.M.; Anders, K.; Gloaguen, R.; et al. Multisource and Multitemporal Data Fusion in Remote Sensing. IEEE Geosci. Remote Sens. Mag. 2019, 7, 6–39. [Google Scholar] [CrossRef]
- Robert, C.; Jessep, W.; Sutton, J.J.; Hicks, T.M.; Loeffen, M.; Farouk, M.; Ward, J.F.; Bain, W.E.; Craigie, C.R.; Fraser-Miller, S.J.; et al. Evaluating low- mid- and high-level fusion strategies for combining Raman and infrared spectroscopy for quality assessment of red meat. Food Chem. 2021, 361, 130154. [Google Scholar] [CrossRef]
- Xu, Z.; Cheng, W.; Fan, S.; Liu, J.; Wang, H.; Li, X.; Liu, B.; Wu, Y.; Zhang, P.; Wang, Q. Data fusion of near-infrared diffuse reflectance spectra and transmittance spectra for the accurate determination of rice flour constituents. Anal. Chim. Acta 2022, 1193, 339384. [Google Scholar] [CrossRef]
- Xiao, Q.; Bai, X.; Gao, P.; He, Y. Application of Convolutional Neural Network-Based Feature Extraction and Data Fusion for Geographical Origin Identification of Radix Astragali by Visible/Short-Wave Near-Infrared and Near Infrared Hyperspectral Imaging. Sensors 2020, 20, 4940. [Google Scholar] [CrossRef]
- Wang, Q.Q.; Huang, H.Y.; Wang, Y.Z. Geographical Authentication of Macrohyporia cocos by a Data Fusion Method Combining Ultra-Fast Liquid Chromatography and Fourier Transform Infrared Spectroscopy. Molecules 2019, 24, 1320. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yang, R.; Lu, X.; Huang, J.; Zhou, J.; Jiao, J.; Liu, Y.; Liu, F.; Su, B.; Gu, P. A Multi-Source Data Fusion Decision-Making Method for Disease and Pest Detection of Grape Foliage Based on ShuffleNet V2. Remote Sens. 2021, 13, 5102. [Google Scholar] [CrossRef]
- Su, Z.; Zhang, C.; Yan, T.; Zhu, J.; Zeng, Y.; Lu, X.; Gao, P.; Feng, L.; He, L.; Fan, L. Application of Hyperspectral Imaging for Maturity and Soluble Solids Content Determination of Strawberry With Deep Learning Approaches. Front. Plant Sci. 2021, 12, 736334. [Google Scholar] [CrossRef] [PubMed]
- Wang, D.; Feng, Z.; Ji, S.; Cui, D. Simultaneous prediction of peach firmness and weight using vibration spectra combined with one-dimensional convolutional neural network. Comput. Electron. Agric. 2022, 201, 107341. [Google Scholar] [CrossRef]
- Mansuri, S.M.; Chakraborty, S.K.; Mahanti, N.K.; Pandiselvam, R. Effect of germ orientation during Vis-NIR hyperspectral imaging for the detection of fungal contamination in maize kernel using PLS-DA, ANN and 1D-CNN modelling. Food Control 2022, 139, 109077. [Google Scholar] [CrossRef]
- Yan, T.; Xu, W.; Lin, J.; Duan, L.; Gao, P.; Zhang, C.; Lv, X. Combining Multi-Dimensional Convolutional Neural Network (CNN) With Visualization Method for Detection of Aphis gossypii Glover Infection in Cotton Leaves Using Hyperspectral Imaging. Front. Plant Sci. 2021, 12, 604510. [Google Scholar] [CrossRef]
- Li, H.; Zhang, L.; Sun, H.; Rao, Z.; Ji, H. Discrimination of unsound wheat kernels based on deep convolutional generative adversarial network and near-infrared hyperspectral imaging technology. Spectrochim. Acta A Mol. Biomol. Spectrosc. 2022, 268, 120722. [Google Scholar] [CrossRef]
- Sarker, I.H. Deep Learning: A Comprehensive Overview on Techniques, Taxonomy, Applications and Research Directions. SN Comput. Sci. 2021, 2, 420. [Google Scholar] [CrossRef]
- He, X.; Chen, Y.; Ghamisi, P. Heterogeneous Transfer Learning for Hyperspectral Image Classification Based on Convolutional Neural Network. IEEE Trans. Geosci. Remote Sens. 2020, 58, 3246–3263. [Google Scholar] [CrossRef]
- Chai, J.; Chang, J.; Zhao, Y.; Liu, H. An Auto-ML Framework Based on GBDT for Lifelong Learning. arXiv 2019, arXiv:1908.11033. [Google Scholar] [CrossRef]


| Tech 1 | Difference | Connection | ||
|---|---|---|---|---|
| Instrument | Data | Application | Data Process | |
| NIRS | Lower cost; portables | Spectra | Evaluate chemical parameters; on-lining inspection, | Rely on ML 2, chemo-metric model |
| HSI | Higher cost; ponderous | Spectra and image | Evaluate chemical and physical parameters; visualize map, | Poor robustness and adaptability; difficulty in valid information mining |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ye, W.; Xu, W.; Yan, T.; Yan, J.; Gao, P.; Zhang, C. Application of Near-Infrared Spectroscopy and Hyperspectral Imaging Combined with Machine Learning Algorithms for Quality Inspection of Grape: A Review. Foods 2023, 12, 132. https://doi.org/10.3390/foods12010132
Ye W, Xu W, Yan T, Yan J, Gao P, Zhang C. Application of Near-Infrared Spectroscopy and Hyperspectral Imaging Combined with Machine Learning Algorithms for Quality Inspection of Grape: A Review. Foods. 2023; 12(1):132. https://doi.org/10.3390/foods12010132
Chicago/Turabian StyleYe, Weixin, Wei Xu, Tianying Yan, Jingkun Yan, Pan Gao, and Chu Zhang. 2023. "Application of Near-Infrared Spectroscopy and Hyperspectral Imaging Combined with Machine Learning Algorithms for Quality Inspection of Grape: A Review" Foods 12, no. 1: 132. https://doi.org/10.3390/foods12010132
APA StyleYe, W., Xu, W., Yan, T., Yan, J., Gao, P., & Zhang, C. (2023). Application of Near-Infrared Spectroscopy and Hyperspectral Imaging Combined with Machine Learning Algorithms for Quality Inspection of Grape: A Review. Foods, 12(1), 132. https://doi.org/10.3390/foods12010132







