Application of a Loop-Mediated Isothermal Amplification (LAMP) Assay for the Detection of Listeria monocytogenes in Cooked Ham
Abstract
:1. Introduction
2. Materials and Methods
2.1. Bacterial Strains and Culture Conditions
2.2. Bacterial DNA Extraction
2.3. Primers
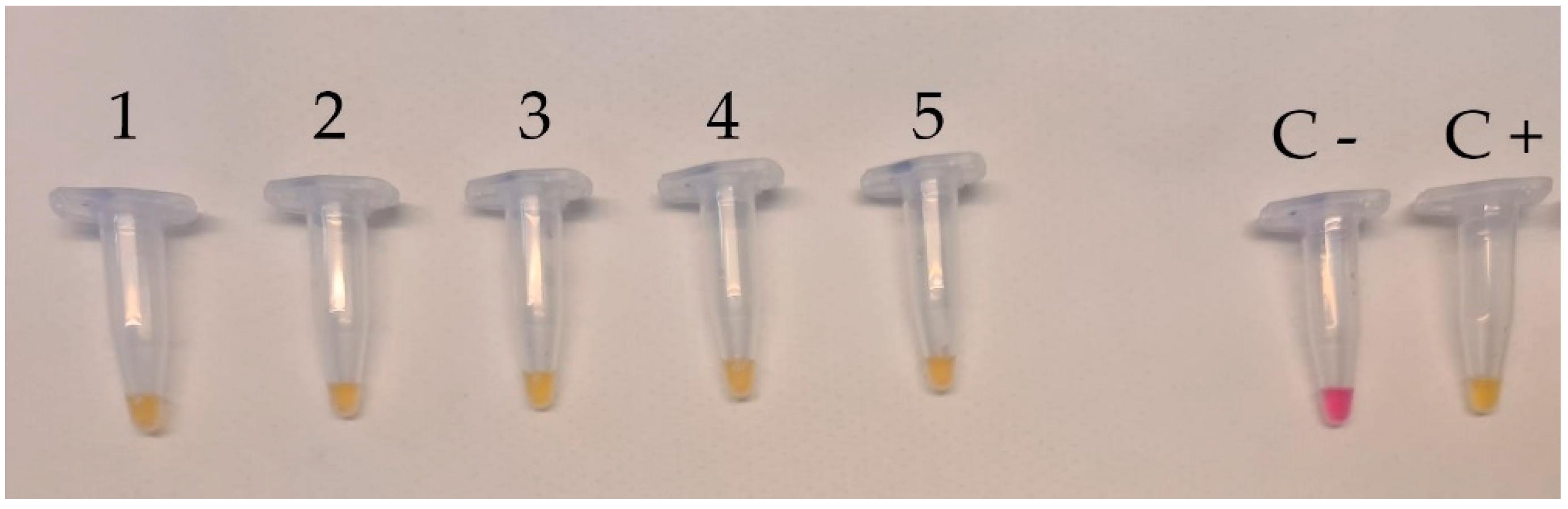
2.4. Colorimetric LAMP Assay
2.5. Colorimetric LAMP Assay: Specificity and Sensitivity Determination
2.6. Real-Time LAMP PCR
2.7. Real-Time PCR
2.8. Food Samples, Experimental Contamination, and Procedure
3. Results
3.1. Specificity and Sensitivity of Colorimetric LAMP
3.2. Specificity of Real-Time LAMP PCR
3.3. Detection of L. monocytogenes in Artificially Contaminated Samples
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- EFSA and ECDC. European Food Safety Authority and European Centre for Disease Prevention and Control. The European Union One Health 2020 Zoonoses Report. EFSA J. 2021, 19, 6971. [Google Scholar] [CrossRef]
- Lundén, J.; Autio, T.; Makula, A.; Hellström, S.; Korkeala, H. Adaptive and cross-adaptive responses of persistent and non-persistent Listeria monocytogenes strains to disinfectants. Int. J. Food Microbiol. 2003, 82, 265–272. [Google Scholar] [CrossRef] [PubMed]
- Tang, M.J.; Zhou, S.; Zhang, X.Y.; Pu, J.H.; Ge, Q.L.; Tang, X.J.; Gao, Y.S. Rapid and sensitive detection of Listeria monocytogenes by Loop-Mediated Isothermal Amplification. Curr. Microbiol. 2011, 63, 511–516. [Google Scholar] [CrossRef] [PubMed]
- Liu, H.; Lu, L.; Pan, Y.; Sun, X.; Hwang, C.; Zhao, Y.; Wu, V.C.H. Rapid detection and differentiation of Listeria monocytogenes and Listeria species in deli meats by a new multiplex PCR method. Food Control 2015, 52, 78–84. [Google Scholar] [CrossRef]
- Matle, I.; Mbatha, K.R.; Madoroba, E. A review of Listeria monocytogenes from meat and meat products: Epidemiology, virulence factors, antimicrobial resistance and diagnosis. Onderstepoort J. Vet. Res. 2020, 87, a1869. [Google Scholar] [CrossRef] [PubMed]
- Commission Regulation (EC) No 2073/2005 of 15 November 2005 on Microbiological Criteria for Foodstuffs. Available online: http://data.europa.eu/eli/reg/2005/2073/oj (accessed on 9 May 2022).
- ISO 11290-1:2017; Microbiology of the Food Chain—Horizontal Method for the Detection and Enumeration of Listeria monocytogenes and of Listeria spp.—Part 1: Detection Method. International Organization for Standardization: Geneva, Switzerland, 2017.
- Rodriguez-Lazaro, D.; Gonzalez-García, P.; Gattuso, A.; Gianfranceschi, M.V.; Hernandez, M. Reducing time in the analysis of Listeria monocytogenes in meat, dairy and vegetable products. Int. J. Food Microbiol. 2014, 184, 98–105. [Google Scholar] [CrossRef] [PubMed]
- Notomi, T.; Okayama, H.; Masubuchi, H.; Yonekawa, T.; Watanabe, K.; Amino, N.; Hase, T. Loop-mediated isothermal amplification of DNA. Nucleic Acids Res. 2000, 28, e63. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, L.; Li, Y.; Chu, J.; Xu, Z.; Zhong, Q. Development and application of a simple loop-mediated isothermal amplification method on rapid detection of Listeria monocytogenes strains. Mol. Biol. Rep. 2012, 39, 445–449. [Google Scholar] [CrossRef] [PubMed]
- Aleyasin, N.; Karimaei, S.; Talebi, M.; Karimaei, M.; Irajian, G.R. Rapid Detection of Listeria monocytogenes Strains Isolated from Clinical and Non-Clinical Samples by Loop-Mediated Isothermal Amplification Method (LAMP). Infect. Epidemiol. Microbiol. 2018, 4, 87–92. Available online: https://civilica.com/doc/992036/ (accessed on 9 May 2022).
- Gianfranceschi, M.V.; Rodriguez-Lazaro, D.; Hernandez, M.; González-García, P.; Comin, D.; Gattuso, A.; Delibato, E.; Sonnessa, M.; Pasquali, F.; Prencipe, V.; et al. European validation of a real-time PCR-based method for detection of Listeria monocytogenes in soft cheese. Int. J. Food Microbiol. 2014, 184, 128–133. [Google Scholar] [CrossRef] [PubMed]
- Wu, R.; Liu, X.; Guo, B.; Chen, F.; Wang, X. Development of Double Loop-Mediated Isothermal Amplification to Detect Listeria monocytogenes in Food. Curr. Microbiol. 2014, 69, 839–845. [Google Scholar] [CrossRef] [PubMed]
- Rodriguez-Lazaro, D.; Hernandez, M.; Pla, M. Simultaneous quantitative detection of Listeria spp. and Listeria monocytogenes using a duplex real-time PCR-based assay. FEMS Microbiol. Lett. 2004, 233, 257–267. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wachiralurpan, S.; Sriyapai, T.; Areekit, S.; Sriyapai, P.; Thongphueak, D.; Santiwatanakule, S.; Chansiri, K. A one-step rapid screening test of Listeria monocytogenes in food samples using a real-time loop-mediated isothermal amplification turbidity assay. Anal. Methods 2017, 9, 6403–6410. [Google Scholar] [CrossRef] [Green Version]
- Wang, D.; Zhang, G.; Lu, C.; Deng, R.; Zhi, A.; Guo, J.; Zhao, D.; Xu, Z. Rapid Detection of Listeria monocytogenes in Raw Milk with Loop-Mediated Isothermal Amplification and Chemosensor. Food Sci. 2011, 76, M611–M615. [Google Scholar] [CrossRef] [PubMed]
- Shan, X.; Zhang, Y.; Zhang, Z.; Chen, M.; Su, Y.; Yuan, Y.; Alam, M.J.; Yan, H.; Shi, L. Rapid detection of food-borne Listeria monocytogenes by Real-Time quantitative Loop-Mediated Isothermal Amplification. Food Sci. Biotechnol. 2012, 21, 101–106. [Google Scholar] [CrossRef]
- Chaouch, M. Loop-mediated isothermal amplification (LAMP): An effective molecular point-of-care technique for the rapid diagnosis of coronavirus SARS-CoV-2. Rev. Med. Virol. 2020, 31, e2215. [Google Scholar] [CrossRef] [PubMed]



| Bacterial Strains | Source |
|---|---|
| L. monocytogenes | …………… |
| L. monocytogenes | * ATCC 13932 |
| L. monocytogenes | ATCC 35152 |
| L. monocytogenes | ATCC 7644 |
| L. monocytogenes | ATCC 19111 |
| L. monocytogenes | ATCC 19115 |
| Listeria spp | …………… |
| Listeria innocua | ATCC 33090 |
| Listeria ivanovii | ATCC 19119 |
| Listeria seeligeri | ** CLISS 11 |
| Others strains | ………………. |
| Escherichia coli | ATCC 25922 |
| Salmonella Enteritidis | ATCC 13076 |
| Salmonella Typhimurium | ATCC 14028 |
| Staphylococcus aureus | ATCC 25923 |
| Enterococcus faecalis | ATCC 29212 |
| Enterobacter aerogenes | ATCC 13048 |
| Pseudomonasaeruginosa | ATCC 9027 |
| Bacillus cereus | ATCC 6633 |
| Citrobacter freundii | ATCC 8090 |
| Proteus vulgaris | ATCC 13315 |
| Primer | Primer Sequence 5′-3′ | References |
|---|---|---|
| LAMP/real-time LAMP PCR assay | ||
| FIP_14 A | TCGCTCCAGTTTTTATGTTGAACACCTTGGGATGAARTAAATTATGATCC | [13] |
| BIP_12 | AGCAAGCTAGCTCATTTCACATAGCGTAAACATTAATATTTCTCGC | [13] |
| F3_12 | GGAGGMTACGTTGCTCAA | [13] |
| B3_12 | AAGCTAAACCAGTGCATTC | [13] |
| LF_12 | ACTTCCATTKCTTTA | [13] |
| LB_12 | CGTCCATCTATTTGCCAGGTAAC | [13] |
| Real-time PCR | ||
| hlyF | CATGGCACCACCAGCATCT | [14] |
| hlyR | ATCCGCGTGTTTCTTTTCGA | [14] |
| hlyP | 6-FAM- CGCCTGCAAGTCCTAAGACGCCA-BHQ1 | [14] |
| Strains | Cycle Threshold | Melting Temperature |
|---|---|---|
| L. monocytogenes ATCC 13932 | 19.09 | 83 °C |
| L. monocytogenes ATCC 35152 | 18.90 | 83 °C |
| L. monocytogenes ATCC 7644 | 18.29 | 83 °C |
| L. monocytogenes ATCC 19111 | 17.96 | 83 °C |
| L. monocytogenes ATCC 19115 | 17.88 | 83 °C |
| Positive control | 18.25 | 83 °C |
| Negative control | No Ct | 79.7 °C |
| Strains | Cycle Threshold (Ct) | Melting Temperature |
|---|---|---|
| Listeria innocua ATCC 33090 | No Ct | 79.7 °C |
| Listeria ivanovii ATCC 19119 | No Ct | 79.7 °C |
| Listeria seeligeri CLISS 11 | No Ct | 79.7 °C |
| Positive control | 19.13 | 83 °C |
| Negative control | No Ct | 79.7 °C |
| Strains | Cycle Threshold (Ct) | Melting Temperature |
|---|---|---|
| Escherichia coli ATCC 25922 | No Ct | 79.2 °C |
| Salmonella Enteritidis ATCC 13076 | No Ct | 79.2 °C |
| Salmonella Typhimurium ATCC 14928 | No Ct | 79.2 °C |
| Staphylococcus aureus ATCC 25923 | No Ct | 79.7 °C |
| Enterococcus faecalis ATCC 29212 | No Ct | 79.7 °C |
| Enterobacter aerogenes ATCC 13048 | No Ct | 79.2 °C |
| Pseudomonas aeruginosa ATCC 9027 | No Ct | 79.7 °C |
| Bacillus cereus ATCC 6633 | No Ct | 79.7 °C |
| Citrobacter freundii ATCC 8090 | No Ct | 79.7 °C |
| Proteus vulgaris ATCC 13315 | No Ct | 79.7 °C |
| Positive control. | 18.94 | 83 °C |
| Negative control | No Ct | 79.7 °C |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Fiore, A.; Treglia, I.; Ciccaglioni, G.; Ortoffi, M.F.; Gattuso, A. Application of a Loop-Mediated Isothermal Amplification (LAMP) Assay for the Detection of Listeria monocytogenes in Cooked Ham. Foods 2023, 12, 193. https://doi.org/10.3390/foods12010193
Fiore A, Treglia I, Ciccaglioni G, Ortoffi MF, Gattuso A. Application of a Loop-Mediated Isothermal Amplification (LAMP) Assay for the Detection of Listeria monocytogenes in Cooked Ham. Foods. 2023; 12(1):193. https://doi.org/10.3390/foods12010193
Chicago/Turabian StyleFiore, Alfonsina, Ida Treglia, Gianni Ciccaglioni, Marco Francesco Ortoffi, and Antonietta Gattuso. 2023. "Application of a Loop-Mediated Isothermal Amplification (LAMP) Assay for the Detection of Listeria monocytogenes in Cooked Ham" Foods 12, no. 1: 193. https://doi.org/10.3390/foods12010193
APA StyleFiore, A., Treglia, I., Ciccaglioni, G., Ortoffi, M. F., & Gattuso, A. (2023). Application of a Loop-Mediated Isothermal Amplification (LAMP) Assay for the Detection of Listeria monocytogenes in Cooked Ham. Foods, 12(1), 193. https://doi.org/10.3390/foods12010193






