Lipids in a Nutshell: Quick Determination of Lipid Content in Hazelnuts with NIR Spectroscopy
Abstract
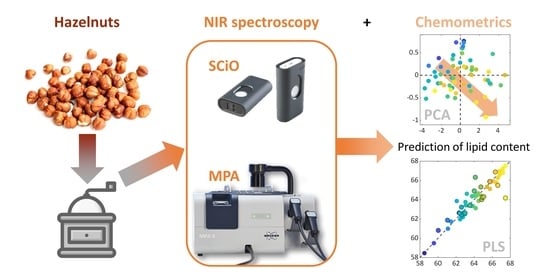
:1. Introduction
2. Materials and Methods
2.1. Hazelnut Samples
2.2. Lipid Extraction—Randall/Soxtec Extraction
2.3. NIR Instrumentation and Spectra Acquisitions
2.4. Multivariate Data Analysis
2.4.1. Exploratory Data Analysis
2.4.2. Regression Models
3. Results and Discussion
3.1. Exploratory Analysis Results
3.2. PLS Regression Results
4. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Oliveira, I.; Sousa, A.; Morais, J.S.; Ferreira, I.C.F.R.; Bento, A.; Estevinho, L.; Pereira, J.A. Chemical composition, and antioxidant and antimicrobial activities of three hazelnut (Corylus avellana L.) cultivars. Food Chem. Toxicol. 2008, 46, 1801–1807. [Google Scholar] [CrossRef] [PubMed]
- Tüfekci, F.; Karataş, Ş. Determination of geographical origin Turkish hazelnuts according to fatty acid composition. Food Sci. Nutr. 2018, 6, 557–562. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Köksal, A.I.; Artik, N.; Şimşek, A.; Güneş, N. Nutrient composition of hazelnut (Corylus avellana L.) varieties cultivated in Turkey. Food Chem. 2006, 99, 509–515. [Google Scholar] [CrossRef]
- Ros, E. Health benefits of nut consumption. Nutrients 2010, 2, 652–682. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jang, H.-G.; Heo, B.-G.; Park, Y.S.; Namiesnik, J.; Barasch, D.; Katrich, E.; Vearasilp, K.; Trakhtenberg, S.; Gorinstein, S. Chemical Composition, Antioxidant and Anticancer Effects of the Seeds and Leaves of Indigo (Polygonum tinctorium Ait.) Plant. Appl. Biochem. Biotechnol. 2012, 167, 1986–2004. [Google Scholar] [CrossRef]
- Alasalvar, C.; Shahidi, F.; Liyanapathirana, C.M.; Ohshima, T. Turkish Tombul Hazelnut (Corylus avellana L.). 1. Compositional Characteristics. J. Agric. Food Chem. 2003, 51, 3790–3796. [Google Scholar] [CrossRef]
- Pannico, A.; Cirillo, C.; Giaccone, M.; Scognamiglio, P.; Romano, R.; Caporaso, N.; Sacchi, R.; Basile, B. Fruit position within the canopy affects kernel lipid composition of hazelnuts. J. Sci. Food Agric. 2017, 97, 4790–4799. [Google Scholar] [CrossRef] [Green Version]
- Cristofori, V.; Ferramondo, S.; Bertazza, G.; Bignami, C. Nut and kernel traits and chemical composition of hazelnut (Corylus avellana L.) cultivars. J. Sci. Food Agric. 2008, 88, 1091–1098. [Google Scholar] [CrossRef]
- Król, K.; Gantner, M.; Piotrowska, A. Morphological Traits, Kernel Composition and Sensory Evaluation of Hazelnut (Corylus avellana L.) Cultivars Grown in Poland. Agronomy 2019, 9, 703. [Google Scholar] [CrossRef] [Green Version]
- Cristofori, V.; Bertazza, G.; Bignami, C. Changes in kernel chemical composition during nut development of three Italian hazelnut cultivars. Fruits 2015, 70, 311–322. [Google Scholar] [CrossRef]
- Ciemniewska-Zytkiewicz, H.; Pasini, F.; Verardo, V.; Bryś, J.; Koczoń, P.; Caboni, M.F. Changes of the lipid fraction during fruit development in hazelnuts (Corylus avellana L.) grown in Poland. Eur. J. Lipid Sci. Technol. 2015, 117, 710–717. [Google Scholar] [CrossRef]
- Romero-Aroca, A.; Rovira, M.; Cristofori, V.; Silvestri, C. Hazelnut Kernel Size and Industrial Aptitude. Agriculture 2021, 11, 1115. [Google Scholar] [CrossRef]
- Bachmann, R.; Klockmann, S.; Haerdter, J.; Fischer, M.; Hackl, T. 1H NMR Spectroscopy for Determination of the Geographical Origin of Hazelnuts. J. Agric. Food Chem. 2018, 66, 11873–11879. [Google Scholar] [CrossRef]
- Klockmann, S.; Reiner, E.; Bachmann, R.; Hackl, T.; Fischer, M. Food Fingerprinting: Metabolomic Approaches for Geographical Origin Discrimination of Hazelnuts (Corylus avellana) by UPLC-QTOF-MS. J. Agric. Food Chem. 2016, 64, 9253–9262. [Google Scholar] [CrossRef]
- Shakiba, N.; Gerdes, A.; Holz, N.; Wenck, S.; Bachmann, R.; Schneider, T.; Seifert, S.; Fischer, M.; Hackl, T. Determination of the geographical origin of hazelnuts (Corylus avellana L.) by Near-Infrared spectroscopy (NIR) and a Low-Level Fusion with nuclear magnetic resonance (NMR). Microchem. J. 2022, 174, 107066. [Google Scholar] [CrossRef]
- Kumaravelu, C.; Gopal, A. A review on the applications of Near-Infrared spectrometer and Chemometrics for the agro-food processing industries. In Proceedings of the 2015 IEEE Technological Innovation in ICT for Agriculture and Rural Development (TIAR), Chennai, India, 10–12 July 2015; pp. 8–12. [Google Scholar] [CrossRef]
- Kandala, C.V.; Sundaram, J.; Puppala, N. Analysis of moisture content, total oil and fatty acid composition by NIR reflectance spectroscopy: A review. Lect. Notes Electr. Eng. 2012, 146, 59–80. [Google Scholar] [CrossRef]
- Nobari Moghaddam, H.; Tamiji, Z.; Akbari Lakeh, M.; Khoshayand, M.R.; Haji Mahmoodi, M. Multivariate analysis of food fraud: A review of NIR based instruments in tandem with chemometrics. J. Food Compos. Anal. 2022, 107, 104343. [Google Scholar] [CrossRef]
- Moscetti, R.; Radicetti, E.; Monarca, D.; Cecchini, M.; Massantini, R. Near infrared spectroscopy is suitable for the classification of hazelnuts according to Protected Designation of Origin. J. Sci. Food Agric. 2015, 95, 2619–2625. [Google Scholar] [CrossRef]
- Biancolillo, A.; De Luca, S.; Bassi, S.; Roudier, L.; Bucci, R.; Magrì, A.D.; Marini, F. Authentication of an Italian PDO hazelnut (“Nocciola Romana”) by NIR spectroscopy. Environ. Sci. Pollut. Res. 2018, 25, 28780–28786. [Google Scholar] [CrossRef]
- Srigley, C.T.; Mossoba, M.M. Current Analytical Techniques for Food Lipids. Food Saf. Innov. Anal. Tools Saf. Assess. 2016, 33–64. [Google Scholar] [CrossRef] [Green Version]
- AOAC. AOAC, Official Method 948.22. Fat (crude) in Nuts and Nut Products. Gravimetric Methods. In Official Methods of Analysis of AOAC International, 19th ed.; AOAC International: Gaithersburg, MD, USA, 2012. [Google Scholar]
- Barnes, R.J.; Dhanoa, M.S.; Lister, S.J. Standard Normal Variate Transformation and De-Trending of Near-Infrared Diffuse Reflectance Spectra. Appl. Spectrosc. 2016, 43, 772–777. [Google Scholar] [CrossRef]
- Bro, R.; Smilde, A.K. Principal component analysis. Anal. Methods 2014, 6, 2812–2831. [Google Scholar] [CrossRef] [Green Version]
- Abdi, H.; Williams, L.J. Principal component analysis. Wiley Interdiscip. Rev. Comput. Stat. 2010, 2, 433–459. [Google Scholar] [CrossRef]
- Savage, G.P.; McNeil, D.L.; Dutta, P.C. Lipid composition and oxidative stability of oils in hazelnuts (Corylus avellana L.) grown in New Zealand. J. Am. Oil Chem. Soc. 1997, 74, 755–759. [Google Scholar] [CrossRef]
- Cavallini, N.; Pennisi, F.; Giraudo, A.; Pezzolato, M.; Esposito, G.; Gavoci, G.; Magnani, L.; Pianezzola, A.; Geobaldo, F.; Savorani, F.; et al. Chemometric Differentiation of Sole and Plaice Fish Fillets Using Three Near-Infrared Instruments. Foods 2022, 11, 1643. [Google Scholar] [CrossRef]
- Snee, R.D. Validation of Regression Models: Methods and Examples. Technometrics 1977, 19, 415–428. [Google Scholar] [CrossRef]
- Fearn, T. Assessing Calibrations: SEP, RPD, RER and R2. NIR News 2002, 13, 12–13. [Google Scholar] [CrossRef]
- Williams, P. The RPD Statistic: A Tutorial Note. NIR News 2014, 25, 22–26. [Google Scholar] [CrossRef]
- Davrieux, F.; Allal, F.-O.; Piombo, G.; Kelly, B.; Okulo, J.B.; Thiam, M.; Diallo, O.B.; Bouvet, J.-M. Near Infrared Spectroscopy for High-Throughput Characterization of Shea Tree (Vitellaria paradoxa) Nut Fat Profiles. J. Agric. Food Chem. 2010, 58, 7811. [Google Scholar] [CrossRef]
- Mulvey, B.W. Determination of Fat Content in Foods Using a Near-Infrared Spectroscopy Sensor. In Proceedings of the 2020 IEEE SENSORS, Rotterdam, The Netherlands, 25–28 October 2020. [Google Scholar] [CrossRef]
- Wiedemair, V.; Langore, D.; Garsleitner, R.; Dillinger, K.; Huck, C. Investigations into the Performance of a Novel Pocket-Sized Near-Infrared Spectrometer for Cheese Analysis. Molecules 2019, 24, 428. [Google Scholar] [CrossRef]



| SCiO | MPA Sphere | MPA Probe | |
|---|---|---|---|
| LVs | 7 | 4 | 3 |
| R2CAL | 0.717 | 0.925 | 0.846 |
| R2CV | 0.461 | 0.903 | 0.793 |
| R2PRED | 0.550 | 0.713 | 0.897 |
| RMSECAL | 0.966 | 0.566 | 0.870 |
| RMSECV | 1.332 | 0.645 | 1.008 |
| RMSEPRED | 1.217 | 1.109 | 0.712 |
| RPDCAL | 1.904 | 3.709 | 2.583 |
| RPDCV | 1.380 | 3.254 | 2.229 |
| RPDPRED | 1.909 | 1.773 | 2.167 |
| SCiO | MPA (Sphere and Probe) | |
|---|---|---|
| C–H [30,31,32] | 920 nm, stretching third overtones | 1214 nm, second overtone CH2 1390 nm, stretching CH2 1730–1760 nm, first overtone 2300–2340 nm, stretching and deformation combinations CH2 |
| C=C [30,31,32] | / | 2100 nm, stretching |
| O–H [33] | 960–1050 nm, stretching second overtones | 1900 nm, combination band |
| Unassigned | 780 nm, 830–850 nm | 1600 nm |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Cazzaniga, E.; Cavallini, N.; Giraudo, A.; Gavoci, G.; Geobaldo, F.; Pariani, M.; Ghirardello, D.; Zeppa, G.; Savorani, F. Lipids in a Nutshell: Quick Determination of Lipid Content in Hazelnuts with NIR Spectroscopy. Foods 2023, 12, 34. https://doi.org/10.3390/foods12010034
Cazzaniga E, Cavallini N, Giraudo A, Gavoci G, Geobaldo F, Pariani M, Ghirardello D, Zeppa G, Savorani F. Lipids in a Nutshell: Quick Determination of Lipid Content in Hazelnuts with NIR Spectroscopy. Foods. 2023; 12(1):34. https://doi.org/10.3390/foods12010034
Chicago/Turabian StyleCazzaniga, Elena, Nicola Cavallini, Alessandro Giraudo, Gentian Gavoci, Francesco Geobaldo, Mattia Pariani, Daniela Ghirardello, Giuseppe Zeppa, and Francesco Savorani. 2023. "Lipids in a Nutshell: Quick Determination of Lipid Content in Hazelnuts with NIR Spectroscopy" Foods 12, no. 1: 34. https://doi.org/10.3390/foods12010034