Applications of Enzyme Technology to Enhance Transition to Plant Proteins: A Review
Abstract
:1. Introduction
1.1. Why Plant Proteins?
1.2. What Is Protein Functionality?
1.3. Seed Storage Proteins Are Major Candidates but Come with Challenges
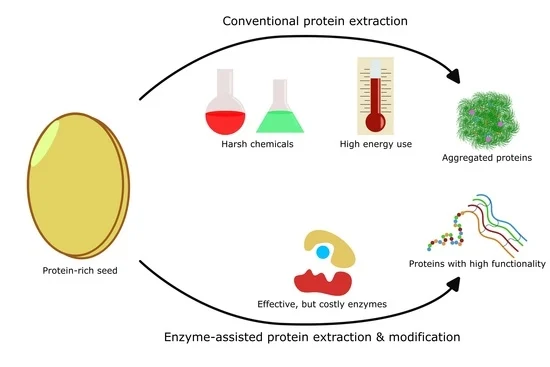
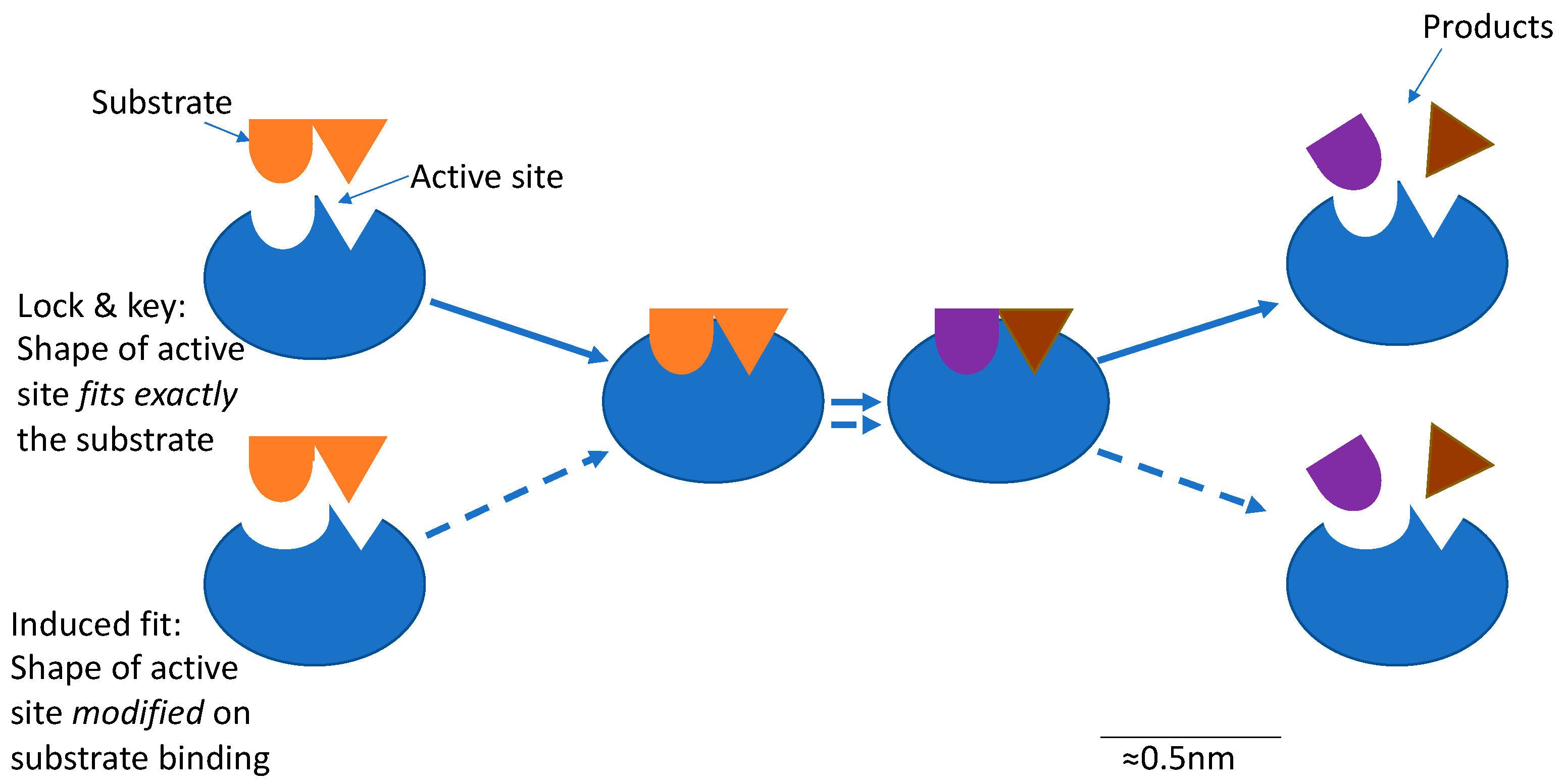
1.4. Enzymes Can Assist in Promoting Plant Protein Utilization
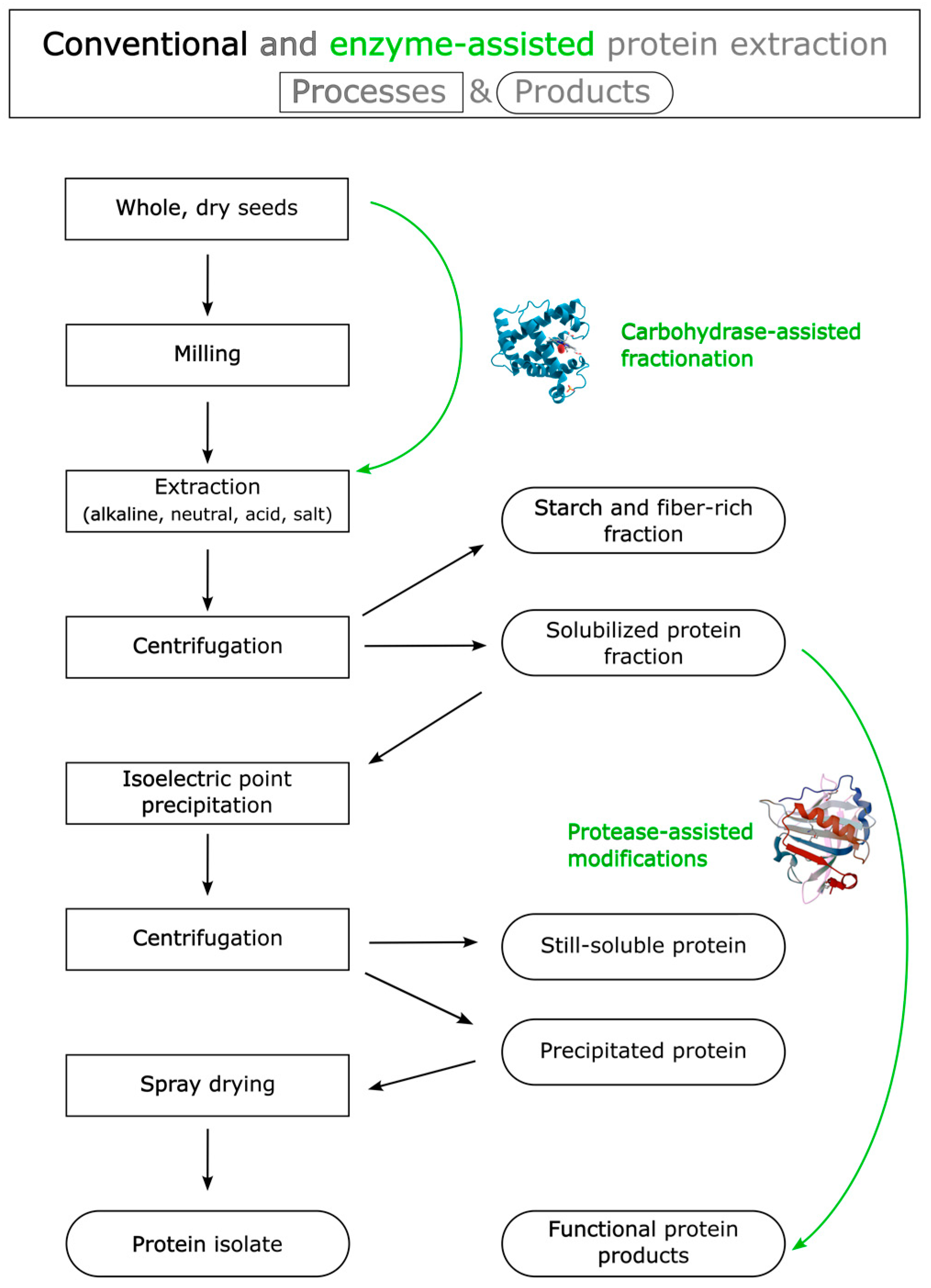
2. Potential of Enzyme-Assisted Plant Protein Extraction
3. Enzymatic Modifications to Improve Protein Functionality
3.1. Plant Protein Hydrolysis
3.2. Cross-Linking
3.3. Deamidation
4. Challenges and Opportunities
4.1. Plant Proteins as Substrates: Large, Aggregated, Variable Mixtures
4.2. Plant Proteins May Contain Protease Inhibitors
4.3. Understanding Substrate Presentation in Complex Systems
4.4. Enzyme Inactivation
4.5. From Enzymatic Reactions to Food Products
4.6. Optimizing Reaction Conditions
4.7. Challenges on Scaling up Enzymes
4.8. Synergies with Other Techniques
4.9. Choosing Solvents
4.10. Addressing the Challenge of Costly Enzymes
5. Conclusions
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- McClements, D.J.; Grossmann, L. A Brief Review of the Science behind the Design of Healthy and Sustainable Plant-Based Foods. NPJ Sci. Food 2021, 5, 17. [Google Scholar] [CrossRef] [PubMed]
- Hoehnel, A.; Zannini, E.; Arendt, E.K. Targeted Formulation of Plant-Based Protein-Foods: Supporting the Food System’s Transformation in the Context of Human Health, Environmental Sustainability and Consumer Trends. Trends Food Sci. Technol. 2022, 128, 238–252. [Google Scholar] [CrossRef]
- FAO. Key Facts and Findings; FAO: Rome, Italy, 2019; p. 6. [Google Scholar] [CrossRef]
- FAO. The Future of Food and Agriculture—Alternative Pathways to 2050; FAO: Rome, Italy, 2018. [Google Scholar]
- United Nations. World Population Projected to Reach 9.8 Billion in 2050, and 11.2 Billion in 2100|United Nations. 2017. Available online: https://www.un.org/development/desa/en/news/population/world-population-prospects-2017.html (accessed on 23 April 2023).
- Henchion, M.; Hayes, M.; Mullen, A.M.; Fenelon, M.; Tiwari, B. Future Protein Supply and Demand: Strategies and Factors Influencing a Sustainable Equilibrium. Foods 2017, 2017, 53. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Van Dijk, M.; Morley, T.; Rau, M.L.; Saghai, Y. A Meta-Analysis of Projected Global Food Demand and Population at Risk of Hunger for the Period 2010–2050. Nat. Food 2021, 2, 494–501. [Google Scholar] [CrossRef] [PubMed]
- Andreoli, V.; Bagliani, M.; Corsi, A.; Frontuto, V. Drivers of Protein Consumption: A Cross-Country Analysis. Sustainability 2021, 13, 7399. [Google Scholar] [CrossRef]
- Ohanenye, I.C.; Tsopmo, A.; Ejike, C.E.C.C.; Udenigwe, C.C. Germination as a Bioprocess for Enhancing the Quality and Nutritional Prospects of Legume Proteins. Trends Food Sci. Technol. 2020, 101, 213–222. [Google Scholar] [CrossRef]
- Bloomberg. Plant-Based Foods Market to Hit $162 Billion in Next Decade, Projects Bloomberg Intelligence|Press. 2021. Available online: https://www.bloomberg.com/company/press/plant-based-foods-market-to-hit-162-billion-in-next-decade-projects-bloomberg-intelligence/ (accessed on 28 March 2023).
- Bloomberg. Once-Hot Fake Meat Sees Sales Slide on Price and Being to “Woke”. 2022. Available online: https://news.bloomberglaw.com/capital-markets/once-hot-fake-meat-sees-sales-slide-on-price-and-being-too-woke (accessed on 28 March 2023).
- Reuters. Beyond Meat Sales under Thread as Plant-Based Boom Withers. 2022. Available online: https://www.reuters.com/business/retail-consumer/beyond-meat-sales-under-threat-plant-based-boom-withers-2022-08-03/ (accessed on 28 March 2023).
- Singhal, A.; Karaca, A.C.; Tyler, R.; Nickerson, M. Pulse Proteins: From Processing to Structure-Function Relationships. In Grain Legumes; InTech: Vienna, Austria, 2016. [Google Scholar]
- Methods of Testing Protein Functionality; Hall, G.M. (Ed.) Blackie Academic & Professional; Chapmann and Hall: London, UK, 1996. [Google Scholar]
- Zhang, Y.; Sharan, S.; Rinnan, Å.; Orlien, V. Survey on Methods for Investigating Protein Functionality and Related Molecular Characteristics. Foods 2021, 10, 2848. [Google Scholar] [CrossRef]
- Sathe, S.; Deshpande, S.; Salunkhe, K. Dry Beans of Phaseolus. A Review. Part 1. Chemical Composition: Proteins. C R C Crit. Rev. Food Sci. Nutr. 1984, 20, 1–46. [Google Scholar] [CrossRef]
- Shewry, P.R.; Napier, J.A.; Tatham, A.S. Seed Storage Proteins: Structures and Biosynthesis. Plant Cell 1995, 7, 945–956. [Google Scholar] [CrossRef] [Green Version]
- Rasheed, F.; Markgren, J.; Hedenqvist, M.; Johansson, E. Modeling to Understand Plant Protein Structure-Function Relationships—Implications for Seed Storage Proteins. Molecules 2020, 25, 873. [Google Scholar] [CrossRef] [Green Version]
- Pojić, M.; Mišan, A.; Tiwari, B. Eco-Innovative Technologies for Extraction of Proteins for Human Consumption from Renewable Protein Sources of Plant Origin. Trends Food Sci. Technol. 2018, 75, 93–104. [Google Scholar] [CrossRef]
- Del Mar Contreras, M.; Lama-Muñoz, A.; Manuel Gutiérrez-Pérez, J.; Espínola, F.; Moya, M.; Castro, E. Protein Extraction from Agri-Food Residues for Integration in Biorefinery: Potential Techniques and Current Status. Bioresour. Technol. 2019, 280, 459–477. [Google Scholar] [CrossRef] [PubMed]
- Vitale, A.; Hinz, G. Sorting of Proteins to Storage Vacuoles: How Many Mechanisms? Trends Plant Sci. 2005, 10, 316–323. [Google Scholar] [CrossRef] [PubMed]
- Warsame, A.O.; O’Sullivan, D.M.; Tosi, P. Seed Storage Proteins of Faba Bean (Vicia Faba L): Current Status and Prospects for Genetic Improvement. J. Agric. Food Chem. 2018, 66, 12617–12626. [Google Scholar] [CrossRef]
- Venkateswara Rao, M.; Sunil, C.K.; Rawson, A.; Chidanand, D.V.; Venkatachlapathy, N. Modifying the Plant Proteins Techno-Functionalities by Novel Physical Processing Technologies: A Review. Crit. Rev. Food Sci. Nutr. 2021, 1–22. [Google Scholar] [CrossRef]
- Short, E.C.; Kinchla, A.J.; Nolden, A.A. Plant-Based Cheeses: A Systematic Review of Sensory Evaluation Studies and Strategies to Increase Consumer Acceptance. Foods 2021, 10, 725. [Google Scholar] [CrossRef] [PubMed]
- Schutyser, M.A.I.; van der Goot, A.J. The Potential of Dry Fractionation Processes for Sustainable Plant Protein Production. Trends Food Sci. Technol. 2011, 22, 154–164. [Google Scholar] [CrossRef]
- MarketsAndMarkets. Food Enzymes Market Share, Size|2021–2026. 2021. Available online: https://www.marketsandmarkets.com/Market-Reports/food-enzymes-market-800.html (accessed on 12 April 2023).
- Amin, A.; Petersen, I.L.; Malmberg, C.; Orlien, V. Perspective on the Effect of Protein Extraction Method on the Antinutritional Factor (ANF) Content in Seeds. ACS Food Sci. Technol. 2022, 2, 604–612. [Google Scholar] [CrossRef]
- Sowbhagya, H.B.; Chitra, V.N. Enzyme-Assisted Extraction of Flavorings and Colorants from Plant Materials. Crit. Rev. Food Sci. Nutr. 2010, 50, 146–161. [Google Scholar] [CrossRef] [PubMed]
- Saldanha do Carmo, C.; Silventoinen, P.; Nordgård, C.T.; Poudroux, C.; Dessev, T.; Zobel, H.; Holtekjølen, A.K.; Draget, K.I.; Holopainen-Mantila, U.; Knutsen, S.H.; et al. Is Dehulling of Peas and Faba Beans Necessary Prior to Dry Fractionation for the Production of Protein- and Starch-Rich Fractions? Impact on Physical Properties, Chemical Composition and Techno-Functional Properties. J. Food Eng. 2020, 278, 109937. [Google Scholar] [CrossRef]
- Talmadge, K.W.; Keegstra, K.; Bauer, W.D.; Albersheim, P. The Structure of Plant Cell Walls. Plant Physiol. 1973, 51, 158–173. [Google Scholar] [CrossRef] [PubMed]
- Cheng, X.; Bi, L.; Zhao, Z.; Chen, Y. Advances in Enzyme Assisted Extraction of Natural Products. In Proceedings of the 3rd International Conference on Material, Mechanical and Manufacturing Engineering (IC3ME 2015), Guangzhou, China, 27–28 June 2015. [Google Scholar]
- Abu-Goukh, A.B.A.; Bashir, H.A. Changes in Pectic Enzymes and Cellulase Activity during Guava Fruit Ripening. Food Chem. 2003, 83, 213–218. [Google Scholar] [CrossRef]
- Andriotis, V.M.E.; Rejzek, M.; Barclay, E.; Rugen, M.D.; Field, R.A.; Smith, A.M. Cell Wall Degradation Is Required for Normal Starch Mobilisation in Barley Endosperm. Sci. Rep. 2016, 6, 33215. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Marathe, S.J.; Jadhav, S.B.; Bankar, S.B.; Kumari Dubey, K.; Singhal, R.S. Improvements in the Extraction of Bioactive Compounds by Enzymes. Curr. Opin. Food Sci. 2019, 25, 62–72. [Google Scholar] [CrossRef] [Green Version]
- Hanmoungjai, P.; Pyle, D.L.; Niranjan, K. Enzyme-Assisted Water-Extraction of Oil and Protein from Rice Bran. J. Chem. Technol. Biotechnol. 2002, 77, 771–776. [Google Scholar] [CrossRef]
- Sari, Y.W.; Mulder, W.J.; Sanders, J.P.M.; Bruins, M.E. Towards Plant Protein Refinery: Review on Protein Extraction Using Alkali and Potential Enzymatic Assistance. Biotechnol. J. 2015, 10, 1138–1157. [Google Scholar] [CrossRef]
- Sari, Y.W.; Bruins, M.E.; Sanders, J.P.M. Enzyme Assisted Protein Extraction from Rapeseed, Soybean, and Microalgae Meals. Ind. Crops Prod. 2013, 43, 78–83. [Google Scholar] [CrossRef]
- Al-Zuhair, S.; Ashraf, S.; Hisaindee, S.; al Darmaki, N.; Battah, S.; Svistunenko, D.; Reeder, B.; Stanway, G.; Chaudhary, A. Enzymatic Pre-Treatment of Microalgae Cells for Enhanced Extraction of Proteins. Eng. Life Sci. 2017, 17, 175–185. [Google Scholar] [CrossRef] [Green Version]
- Vergara-Barberán, M.; Lerma-García, M.J.; Herrero-Martínez, J.M.; Simó-Alfonso, E.F. Use of an Enzyme-Assisted Method to Improve Protein Extraction from Olive Leaves. Food Chem. 2015, 169, 28–33. [Google Scholar] [CrossRef]
- Perović, M.N.; Knežević Jugović, Z.D.; Antov, M.G. Improved Recovery of Protein from Soy Grit by Enzyme-Assisted Alkaline Extraction. J. Food Eng. 2020, 276, 109894. [Google Scholar] [CrossRef]
- Prandi, B.; Zurlini, C.; Maria, C.I.; Cutroneo, S.; di Massimo, M.; Bondi, M.; Brutti, A.; Sforza, S.; Tedeschi, T. Targeting the Nutritional Value of Proteins from Legumes By-Products Through Mild Extraction Technologies. Front. Nutr. 2021, 8, 695793. [Google Scholar] [CrossRef]
- Perović, M.N.; Pajin, B.S.; Antov, M.G. The Effect of Enzymatic Pretreatment of Chickpea on Functional Properties and Antioxidant Activity of Alkaline Protein Isolate. Food Chem. 2022, 374, 131809. [Google Scholar] [CrossRef]
- Rommi, K.; Hakala, T.K.; Holopainen, U.; Nordlund, E.; Poutanen, K.; Lantto, R. Effect of Enzyme-Aided Cell Wall Disintegration on Protein Extractability from Intact and Dehulled Rapeseed (Brassica rapa L. and Brassica napus L.) Press Cakes. J. Agric. Food Chem. 2014, 62, 7989–7997. [Google Scholar] [CrossRef] [PubMed]
- Miranda, C.G.; Speranza, P.; Kurozawa, L.E.; Kawazoe Sato, A.C. Lentil Protein: Impact of Different Extraction Methods on Structural and Functional Properties. Heliyon 2022, 8, e11775. [Google Scholar] [CrossRef]
- Tirgarian, B.; Farmani, J.; Milani, J.M. Enzyme-Assisted Aqueous Extraction of Oil and Protein Hydrolysate from Sesame Seed. J. Food Meas. Charact. 2019, 13, 2118–2129. [Google Scholar] [CrossRef]
- Jiang, Y.; Zhou, X.; Zheng, Y.; Wang, D.; Deng, Y.; Zhao, Y. Impact of Ultrasonication/Shear Emulsifying/Microwave-Assisted Enzymatic Extraction on Rheological, Structural, and Functional Properties of Akebia trifoliata (Thunb.) Koidz. Seed Protein Isolates. Food Hydrocoll. 2021, 112, 106355. [Google Scholar] [CrossRef]
- Naseri, A.; Marinho, G.S.; Holdt, S.L.; Bartela, J.M.; Jacobsen, C. Enzyme-Assisted Extraction and Characterization of Protein from Red Seaweed Palmaria palmata. Algal Res. 2020, 47, 101849. [Google Scholar] [CrossRef]
- André, A. Rennets and Coagulants. In Encyclopedia of Dairy Sciences, 2nd ed.; Fuquay, J.W., Ed.; Elsevier Ltd.: Amsterdam, The Netherlands, 2011; pp. 574–578. [Google Scholar]
- Vogelsang-O’Dwyer, M.; Sahin, A.W.; Arendt, E.K.; Zannini, E. Enzymatic Hydrolysis of Pulse Proteins as a Tool to Improve Techno-Functional Properties. Foods 2022, 11, 1307. [Google Scholar] [CrossRef] [PubMed]
- Yi, D.; Lin, Q.; Johns, P.W. Estimation of Degree of Hydrolysis of Protein Hydrolysates by Size Exclusion Chromatography. Food Anal. Methods 2021, 14, 805–813. [Google Scholar] [CrossRef]
- Sharif, H.R.; Williams, P.A.; Sharif, M.K.; Abbas, S.; Majeed, H.; Masamba, K.G.; Safdar, W.; Zhong, F. Current Progress in the Utilization of Native and Modified Legume Proteins as Emulsifiers and Encapsulants—A Review. Food Hydrocoll. 2018, 76, 2–16. [Google Scholar] [CrossRef]
- Tapal, A.; Tiku, P.K. Nutritional and Nutraceutical Improvement by Enzymatic Modification of Food Proteins. In Enzymes in Food Biotechnology; Elsevier: Amsterdam, The Netherlands, 2019; pp. 471–481. ISBN 9780128132807. [Google Scholar]
- Tavano, O.L. Protein Hydrolysis Using Proteases: An Important Tool for Food Biotechnology. J. Mol. Catal. B Enzym. 2013, 90, 1–11. [Google Scholar] [CrossRef]
- Panyam, D.; Kilara, A. Enhancing the Functionality of Food Proteins by Enzymatic Modification. Trends Food Sci. Technol. 1996, 7, 120–125. [Google Scholar] [CrossRef]
- García Arteaga, V.; Apéstegui Guardia, M.; Muranyi, I.; Eisner, P.; Schweiggert-Weisz, U. Effect of Enzymatic Hydrolysis on Molecular Weight Distribution, Techno-Functional Properties and Sensory Perception of Pea Protein Isolates. Innov. Food Sci. Emerg. Technol. 2020, 65, 102449. [Google Scholar] [CrossRef]
- Del Mar Yust, M.; Pedroche, J.; del Carmen Millán-Linares, M.; Alcaide-Hidalgo, J.M.; Millán, F. Improvement of Functional Properties of Chickpea Proteins by Hydrolysis with Immobilised Alcalase. Food Chem. 2010, 122, 1212–1217. [Google Scholar] [CrossRef]
- Zhao, G.; Liu, Y.; Zhao, M.; Ren, J.; Yang, B. Enzymatic Hydrolysis and Their Effects on Conformational and Functional Properties of Peanut Protein Isolate. Food Chem. 2011, 127, 1438–1443. [Google Scholar] [CrossRef]
- Conde, J.M.; Escobar, M.D.M.Y.; Pedroche Jiménez, J.J.; Rodríguez, F.M.; Rodríguez Patino, J.M. Effect of Enzymatic Treatment of Extracted Sunflower Proteins on Solubility, Amino Acid Composition, and Surface Activity. J. Agric. Food Chem. 2005, 53, 8038–8045. [Google Scholar] [CrossRef]
- Brückner-Gühmann, M.; Heiden-Hecht, T.; Sözer, N.; Drusch, S. Foaming Characteristics of Oat Protein and Modification by Partial Hydrolysis. Eur. Food Res. Technol. 2018, 244, 2095–2106. [Google Scholar] [CrossRef]
- Paraman, I.; Hettiarachchy, N.S.; Schaefer, C.; Beck, M.I. Hydrophobicity, Solubility, and Emulsifying Properties of Enzyme-Modified Rice Endosperm Protein. Cereal Chem. 2007, 84, 343–349. [Google Scholar] [CrossRef]
- Yuan, B.; Ren, J.; Zhao, M.; Luo, D.; Gu, L. Effects of Limited Enzymatic Hydrolysis with Pepsin and High-Pressure Homogenization on the Functional Properties of Soybean Protein Isolate. LWT 2012, 46, 453–459. [Google Scholar] [CrossRef]
- Tang, C.H.; Wang, X.S.; Yang, X.Q. Enzymatic Hydrolysis of Hemp (Cannabis sativa L.) Protein Isolate by Various Proteases and Antioxidant Properties of the Resulting Hydrolysates. Food Chem. 2009, 114, 1484–1490. [Google Scholar] [CrossRef]
- Celus, I.; Brijs, K.; Delcour, J.A. Enzymatic Hydrolysis of Brewers’ Spent Grain Proteins and Technofunctional Properties of the Resulting Hydrolysates. J. Agric. Food Chem. 2007, 55, 8703–8710. [Google Scholar] [CrossRef]
- Wang, J.; Wang, T.; Yu, G.; Li, X.; Liu, H.; Liu, T.; Zhu, J. Effect of Enzymatic Hydrolysis on the Physicochemical and Emulsification Properties of Rice Bran Albumin and Globulin Fractions. LWT 2022, 156, 113005. [Google Scholar] [CrossRef]
- Galves, C.; Galli, G.; Miranda, C.G.; Kurozawa, L.E. Improving the Emulsifying Property of Potato Protein by Hydrolysis: An Application as Encapsulating Agent with Maltodextrin. Innov. Food Sci. Emerg. Technol. 2021, 70, 102696. [Google Scholar] [CrossRef]
- Mokni Ghribi, A.; Maklouf Gafsi, I.; Sila, A.; Blecker, C.; Danthine, S.; Attia, H.; Bougatef, A.; Besbes, S. Effects of Enzymatic Hydrolysis on Conformational and Functional Properties of Chickpea Protein Isolate. Food Chem. 2015, 187, 322–330. [Google Scholar] [CrossRef]
- Martínez, K.D.; Carrera Sánchez, C.; Rodríguez Patino, J.M.; Pilosof, A.M.R. Interfacial and Foaming Properties of Soy Protein and Their Hydrolysates. Food Hydrocoll. 2009, 23, 2149–2157. [Google Scholar] [CrossRef]
- Rodríguez Patino, J.M.; Miñones Conde, J.; Linares, H.M.; Pedroche Jiménez, J.J.; Carrera Sánchez, C.; Pizones, V.; Rodríguez, F.M. Interfacial and Foaming Properties of Enzyme-Induced Hydrolysis of Sunflower Protein Isolate. Food Hydrocoll. 2007, 21, 782–793. [Google Scholar] [CrossRef]
- Barac, M.; Cabrilo, S.; Stanojevic, S.; Pesic, M.; Pavlicevic, M.; Zlatkovic, B.; Jankovic, M. Functional Properties of Protein Hydrolysates from Pea (Pisum sativum, L.) Seeds. Int. J. Food Sci. Technol. 2012, 47, 1457–1467. [Google Scholar] [CrossRef]
- Fuke, Y.; Sekiguchi, M.; Matsuoka, H. Nature of Stem Bromelain Treatments on the Aggregation and Gelation of Soybean Proteins. J. Food Sci. 1985, 50, 1283–1288. [Google Scholar] [CrossRef]
- Chen, D.; Campanella, O.H. Limited Enzymatic Hydrolysis Induced Pea Protein Gelation at Low Protein Concentration with Less Heat Requirement. Food Hydrocoll. 2022, 128, 107547. [Google Scholar] [CrossRef]
- Nieto-Nieto, T.V.; Wang, Y.X.; Ozimek, L.; Chen, L. Effects of Partial Hydrolysis on Structure and Gelling Properties of Oat Globular Proteins. Food Res. Int. 2014, 55, 418–425. [Google Scholar] [CrossRef]
- Nisov, A.; Ercili-Cura, D.; Nordlund, E. Limited Hydrolysis of Rice Endosperm Protein for Improved Techno-Functional Properties. Food Chem. 2020, 302, 125274. [Google Scholar] [CrossRef]
- Hrckova, M.; Rusnakova, M.; Zemanovic, J. Enzymatic Hydrolysis of Defatted Soy Flour by Three Different Proteases and Their Effect on the Functional Preperties of Resulting Protein Hydrolysates. Czech J. Food Sci. 2001, 20, 7–14. [Google Scholar] [CrossRef] [Green Version]
- Sanchez, A.C.; Burgos, J. Thermal Gelation of Trypsin Hydrolysates of Sunflower Proteins: Effect of PH, Protein Concentration, and Hydrolysis Degree. J. Agric. Food Chem. 1996, 44, 3773–3777. [Google Scholar] [CrossRef]
- Olsen, J.V.; Ong, S.E.; Mann, M. Trypsin Cleaves Exclusively C-Terminal to Arginine and Lysine Residues. Mol. Cell. Proteom. 2004, 3, 608–614. [Google Scholar] [CrossRef] [Green Version]
- Šlechtová, T.; Gilar, M.; Kalíková, K.; Tesařová, E. Insight into Trypsin Miscleavage: Comparison of Kinetic Constants of Problematic Peptide Sequences. Anal. Chem. 2015, 87, 7636–7643. [Google Scholar] [CrossRef]
- Rodriguez, J.; Gupta, N.; Smith, R.D.; Pevzner, P.A. Does Trypsin Cut before Proline? J. Proteome Res. 2008, 7, 300–305. [Google Scholar] [CrossRef] [PubMed]
- Hamuro, Y.; Coales, S.J.; Molnar, K.S.; Tuske, S.J.; Morrow, J.A. Specificity of Immobilized Porcine Pepsin in H/D Exchange Compatible Conditions. Rapid Commun. Mass Spectrom. 2008, 22, 1041–1046. [Google Scholar] [CrossRef]
- Ahn, J.; Cao, M.J.; Yu, Y.Q.; Engen, J.R. Accessing the Reproducibility and Specificity of Pepsin and Other Aspartic Proteases. Biochim. Biophys. Acta Proteins Proteom. 2013, 1834, 1222–1229. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zheng, J.; Strutzenberg, T.S.; Reich, A.; Dharmarajan, V.; Pascal, B.D.; Crynen, G.C.; Novick, S.J.; Garcia-Ordonez, R.D.; Griffin, P.R. Comparative Analysis of Cleavage Specificities of Immobilized Porcine Pepsin and Nepenthesin II under Hydrogen/Deuterium Exchange Conditions. Anal. Chem. 2020, 92, 11018–11028. [Google Scholar] [CrossRef]
- Ambler, R.P. [21] Carboxypeptidases A and B. In Methods in Enzymology; Academic Press: Cambridge, MA, USA, 1972; Volume 25, pp. 262–272. [Google Scholar]
- Appel, W. Carboxypeptidases. In Methods of Enzymatic Analysis; Bergmeyer, H.U., Ed.; Academic Press-Elsevier: Cambridge, MA, USA, 1974; Volume 2, pp. 986–988. [Google Scholar]
- Christianson, D.W.; Lipscomb, W.N. Carboxypeptidase A. Acc. Chem. Res. 1989, 22, 62–69. [Google Scholar] [CrossRef]
- Tschesche, H. Carboxypeptidase C. Methods Enzym. 1977, 47, 73–84. [Google Scholar]
- Nandan, A.; Nampoothiri, K.M. Therapeutic and Biotechnological Applications of Substrate Specific Microbial Aminopeptidases. Appl. Microbiol. Biotechnol. 2020, 104, 5243–5257. [Google Scholar] [CrossRef]
- Sanz, Y. Aminopeptidases; Polaina, J., MacCabe, A.P., Eds.; Springer: Berlin/Heidelberg, Germany, 2007. [Google Scholar]
- De Castro Leite Júnior, B.R.; de Oliveira Martins, F.; Trevizano, L.M.; da Capela, A.P.; de Melo Carlos Dias, T.; Pacheco, A.F.C.; Martins, E.M.F. Applications of Enzymes in Food Processing. In Research and Technological Advances in Food Science; Academic Press: Cambridge, MA, USA, 2022; pp. 175–194. ISBN 9780323859172. [Google Scholar]
- Tacias-Pascacio, V.G.; Morellon-Sterling, R.; Siar, E.H.; Tavano, O.; Berenguer-Murcia, Á.; Fernandez-Lafuente, R. Use of Alcalase in the Production of Bioactive Peptides: A Review. Int. J. Biol. Macromol. 2020, 165, 2143–2196. [Google Scholar] [CrossRef] [PubMed]
- Adamson, N.J.; Reynolds, E.C. Characterization of Casein Phosphopeptides Prepared Using Alcalase: Determination of Enzyme Specificity. Enzym. Microb. Technol. 1996, 19, 202–207. [Google Scholar] [CrossRef]
- Doucet, D.; Otter, D.E.; Gauthier, S.F.; Foegeding, E.A. Enzyme-Induced Gelation of Extensively Hydrolyzed Whey Proteins by Alcalase: Peptide Identification and Determination of Enzyme Specificity. J. Agric. Food Chem. 2003, 51, 6300–6308. [Google Scholar] [CrossRef] [PubMed]
- McSweeney, P.L.H.; Olson, N.F.; Fox, P.F.; Healy, A.; Hϕjrup, P. Proteolytic Specificity of Plasmin on Bovine As1-Casein. Food Biotechnol. 2009, 7, 143–158. [Google Scholar] [CrossRef]
- Castellino, F.J. Plasmin. In Proteolytic Enzymes; Academic Press: Cambridge, MA, USA, 2013; Volume 3, pp. 2958–2968. ISBN 9780123822192. [Google Scholar]
- Hervio1, L.S.; Coombs2, G.S.; Bergstrom3, R.C.; Trivedi3, K.; Corey3, D.R.; Madison1, E.L. Negative Selectivity and the Evolution of Protease Cascades: The Specificity of Plasmin for Peptide and Protein Substrates. Chem. Biol. 2000, 7, 443–452. [Google Scholar] [CrossRef] [Green Version]
- Bruno, S.F.; Kudre, T.G.; Bhaskar, N. Effects of Different Pretreatments and Proteases on Recovery, Umami Taste Compound Contents and Antioxidant Potentials of Labeo Rohita Head Protein Hydrolysates. J. Food Sci. Technol. 2019, 56, 1966–1977. [Google Scholar] [CrossRef]
- Merz, M.; Eisele, T.; Berends, P.; Appel, D.; Rabe, S.; Blank, I.; Stressler, T.; Fischer, L. Flavourzyme, an Enzyme Preparation with Industrial Relevance: Automated Nine-Step Purification and Partial Characterization of Eight Enzymes. J. Agric. Food Chem. 2015, 63, 5682–5693. [Google Scholar] [CrossRef]
- Garcia-Mora, P.; Peñas, E.; Frias, J.; Martínez-Villaluenga, C. Savinase, the Most Suitable Enzyme for Releasing Peptides from Lentil (Lens culinaris Var. Castellana) Protein Concentrates with Multifunctional Properties. J. Agric. Food Chem. 2014, 62, 4166–4174. [Google Scholar] [CrossRef] [Green Version]
- Proctor, A.; Wang, Q.; Lawrence, D.S.; Allbritton, N.L. Selection and Optimization of Enzyme Reporters for Chemical Cytometry. In Methods in Enzymology; Academic Press: Cambridge, MA, USA, 2019; Volume 622, pp. 221–248. ISBN 9780128181195. [Google Scholar]
- Wilk, P.; Wątor, E.; Weiss, M.S. Prolidase—A Protein with Many Faces. Biochimie 2021, 183, 3–12. [Google Scholar] [CrossRef] [PubMed]
- Meshram, A.; Singhal, G.; Bhagyawant, S.S.; Srivastava, N. Plant-Derived Enzymes: A Treasure for Food Biotechnology. In Enzymes in Food Biotechnology; Academic Press: Cambridge, MA, USA, 2019; pp. 483–502. ISBN 9780128132807. [Google Scholar]
- Englund, P.T.; King, T.P.; Craig, L.C.; Walti, A. Studies on Ficin. I. Its Isolation and Characterization. Biochemistry 1968, 7, 163–175. [Google Scholar] [CrossRef] [PubMed]
- Morellon-Sterling, R.; El-Siar, H.; Tavano, O.L.; Berenguer-Murcia, Á.; Fernández-Lafuente, R. Ficin: A Protease Extract with Relevance in Biotechnology and Biocatalysis. Int. J. Biol. Macromol. 2020, 162, 394–404. [Google Scholar] [CrossRef] [PubMed]
- Storer, A.C.; Ménard, R. Papain. In Proteolytic Enzymes; Academic Press: Cambridge, MA, USA, 2013; Volume 2, pp. 1858–1861. ISBN 9780123822192. [Google Scholar]
- Harris, J.L.; Backes, B.J.; Leonetti, F.; Mahrus, S.; Ellman, J.A.; Craik, C.S. Rapid and General Profiling of Protease Specificity by Using Combinatorial Fluorogenic Substrate Libraries. Proc. Natl. Acad. Sci. USA 2000, 97, 7754–7759. [Google Scholar] [CrossRef] [Green Version]
- Hale, L.P.; Greer, P.K.; Trinh, C.T.; James, C.L. Proteinase Activity and Stability of Natural Bromelain Preparations. Int. Immunopharmacol. 2005, 5, 783–793. [Google Scholar] [CrossRef]
- Gerrard, J.A. Protein–Protein Crosslinking in Food: Methods, Consequences, Applications. Trends Food Sci. Technol. 2002, 13, 391–399. [Google Scholar] [CrossRef]
- Matheis, G.; Whitaker, J.R. A Review: Enzymatic Cross-Linking of Proteins Applicable to Foods. J. Food Biochem. 1987, 11, 309–327. [Google Scholar] [CrossRef]
- Heck, T.; Faccio, G.; Richter, M.; Thöny-Meyer, L. Enzyme-Catalyzed Protein Crosslinking. Appl. Microbiol. Biotechnol. 2013, 97, 461–475. [Google Scholar] [CrossRef] [Green Version]
- Moreno, H.M.; Pedrosa, M.M.; Tovar, C.A.; Borderías, A.J. Effect of Microbial Transglutaminase on the Production of Fish Myofibrillar and Vegetable Protein-Based Products; Academic Press: Cambridge, MA, USA, 2022; ISBN 9780323899291. [Google Scholar]
- Dube, M.; Schäfer, C.; Neidhart, S.; Carle, R. Texturisation and Modification of Vegetable Proteins for Food Applications Using Microbial Transglutaminase. Eur. Food Res. Technol. 2007, 225, 287–299. [Google Scholar] [CrossRef]
- Amirdivani, S.; Khorshidian, N.; Fidelis, M.; Granato, D.; Koushki, M.R.; Mohammadi, M.; Khoshtinat, K.; Mortazavian, A.M. Effects of Transglutaminase on Health Properties of Food Products. Curr. Opin. Food Sci. 2018, 22, 74–80. [Google Scholar] [CrossRef]
- Lerner, A.; Benzvi, C. Microbial Transglutaminase Is a Very Frequently Used Food Additive and Is a Potential Inducer of Autoimmune/Neurodegenerative Diseases. Toxics 2021, 9, 233. [Google Scholar] [CrossRef] [PubMed]
- Tang, C.H.; Luo, L.J.; Liu, F.; Chen, Z. Transglutaminase-Set Soy Globulin-Stabilized Emulsion Gels: Influence of Soy β-Conglycinin/Glycinin Ratio on Properties, Microstructure and Gelling Mechanism. Food Res. Int. 2013, 51, 804–812. [Google Scholar] [CrossRef]
- Nivala, O.; Nordlund, E.; Kruus, K.; Ercili-Cura, D. The Effect of Heat and Transglutaminase Treatment on Emulsifying and Gelling Properties of Faba Bean Protein Isolate. LWT 2021, 139, 110517. [Google Scholar] [CrossRef]
- Schäfer, C.; Zacherl, C.; Engel, K.H.; Neidhart, S.; Carle, R. Comparative Study of Gelation and Cross-Link Formation during Enzymatic Texturisation of Leguminous Proteins. Innov. Food Sci. Emerg. Technol. 2007, 8, 269–278. [Google Scholar] [CrossRef]
- Kuraishi, C.; Yamazaki, K.; Susa, Y. Transglutaminase: Its Utilization in The Food Industry. Food Rev. Int. 2007, 17, 221–246. [Google Scholar] [CrossRef]
- Matsuura, M.; Sasaki, M.; Sasaki, J.; Takeuchi, T. Process. for Producing Packed Tofu. U.S. Patent 6,042,851, 28 March 2000. [Google Scholar]
- Nonaka, M.; Soeda, T.; Yamagiwa, K.; Kowata, H.; Motogi, M.; Toiguchi, S. Process of Preparing Shelf-Stable “Tofu” at Normal Temperature for Long Term. U.S. Patent 5,055,310, 8 October 1991. [Google Scholar]
- Hu, X.; Zhao, M.; Sun, W.; Zhao, G.; Ren, J. Effects of Microfluidization Treatment and Transglutaminase Cross-Linking on Physicochemical, Functional, and Conformational Properties of Peanut Protein Isolate. J. Agric. Food Chem. 2011, 59, 8886–8894. [Google Scholar] [CrossRef]
- Tang, C.H.; Sun, X.; Yin, S.W.; Ma, C.Y. Transglutaminase-Induced Cross-Linking of Vicilin-Rich Kidney Protein Isolate: Influence on the Functional Properties and in Vitro Digestibility. Food Res. Int. 2008, 41, 941–947. [Google Scholar] [CrossRef]
- Porta, R.; Di Pierro, P.; Rossi-Marquez, G.; Mariniello, L.; Kadivar, M.; Arabestani, A. Microstructure and Properties of Bitter Vetch (Vicia Ervilia) Protein Films Reinforced by Microbial Transglutaminase. Food Hydrocoll. 2015, 50, 102–107. [Google Scholar] [CrossRef]
- Whitehurst, R.J.; Van Oort, M. Enzymes in Food Technology, 2nd ed.; John Wiley & Sons: Hoboken, NJ, USA, 2010. [Google Scholar]
- Yamaguchi, S.; Jeenes, D.J.; Archer, D.B. Protein-Glutaminase from Chryseobacterium Proteolyticum, an Enzyme That Deamidates Glutaminyl Residues in Proteins. Eur. J. Biochem. 2002, 268, 1410–1421. [Google Scholar] [CrossRef]
- Li, X.; Lin, C.; O’connor, P.B. Glutamine Deamidation: Differentiation of Glutamic Acid and γ-Glutamic Acid in Peptides by Electron Capture Dissociation. Anal. Chem. 2010, 82, 3606–3615. [Google Scholar] [CrossRef] [Green Version]
- Liu, X.; Wang, C.; Zhang, X.; Zhang, G.; Zhou, J.; Chen, J. Application Prospect of Protein-Glutaminase in the Development of Plant-Based Protein Foods. Foods 2022, 11, 440. [Google Scholar] [CrossRef]
- Hamada, J.S.; Swanson, P.B. Deamidation of Food Proteins to Improve Functionality. Crit. Rev. Food Sci. Nutr. 2009, 34, 283–292. [Google Scholar] [CrossRef]
- Yie, H.Y.; Yamaguchi, S.; Matsumura, Y. Effects of Enzymatic Deamidation by Protein-Glutaminase on Structure and Functional Properties of Wheat Gluten. J. Agric. Food Chem. 2006, 54, 6034–6040. [Google Scholar] [CrossRef]
- Suppavorasatit, I.; De Mejia, E.G.; Cadwallader, K.R. Optimization of the Enzymatic Deamidation of Soy Protein by Protein-Glutaminase and Its Effect on the Functional Properties of the Protein. J. Agric. Food Chem. 2011, 59, 11621–11628. [Google Scholar] [CrossRef] [PubMed]
- Jiang, Z.Q.; Sontag-Strohm, T.; Salovaara, H.; Sibakov, J.; Kanerva, P.; Loponen, J. Oat Protein Solubility and Emulsion Properties Improved by Enzymatic Deamidation. J. Cereal Sci. 2015, 64, 126–132. [Google Scholar] [CrossRef]
- Kunarayakul, S.; Thaiphanit, S.; Anprung, P.; Suppavorasatit, I. Optimization of Coconut Protein Deamidation Using Protein-Glutaminase and Its Effect on Solubility, Emulsification, and Foaming Properties of the Proteins. Food Hydrocoll. 2018, 79, 197–207. [Google Scholar] [CrossRef]
- Temthawee, W.; Panya, A.; Cadwallader, K.R.; Suppavorasatit, I. Flavor Binding Property of Coconut Protein Affected by Protein-Glutaminase: Vanillin-Coconut Protein Model. LWT 2020, 130, 109676. [Google Scholar] [CrossRef]
- Suppavorasatit, I.; Cadwallader, K.R. Effect of Enzymatic Deamidation of Soy Protein by Protein-Glutaminase on the Flavor-Binding Properties of the Protein under Aqueous Conditions. J. Agric. Food Chem. 2012, 60, 7817–7823. [Google Scholar] [CrossRef]
- Schlichtherle-Cerny, H.; Amadò, R. Analysis of Taste-Active Compounds in an Enzymatic Hydrolysate of Deamidated Wheat Gluten. J. Agric. Food Chem. 2002, 50, 1515–1522. [Google Scholar] [CrossRef]
- Liu, B.Y.; Zhu, K.X.; Guo, X.N.; Peng, W.; Zhou, H.M. Effect of Deamidation-Induced Modification on Umami and Bitter Taste of Wheat Gluten Hydrolysates. J. Sci. Food Agric. 2017, 97, 3181–3188. [Google Scholar] [CrossRef]
- McMahon, D.J.; Oommen, B.S. Supramolecular Structure of the Casein Micelle. J. Dairy Sci. 2008, 91, 1709–1721. [Google Scholar] [CrossRef] [Green Version]
- Kanaka, K.K.; Jeevan, C.; Chethan Raj, R.; Sagar, N.G.; Prasad, R.; Kotresh Prasad, C.; Shruthi, S. A Review on Ovalbumin Gene in Poultry. J. Entomol. Zool. Stud. 2018, 6, 1497–1503. [Google Scholar]
- Krishna, T.C.; Najda, A.; Bains, A.; Tosif, M.M.; Papliński, R.; Kapłan, M.; Chawla, P. Influence of Ultra-Heat Treatment on Properties of Milk Proteins. Polymers 2021, 13, 3164. [Google Scholar] [CrossRef] [PubMed]
- Dan den Akker, C.C.; Schleeger, M.; Bonn, M.; Koenderink, G.H. Structural Basis for the Polymorphism of β-Lactoglobulin Amyloid-Like Fibrils. In Bio-Nanoimaging: Protein Misfolding and Aggregation; Elsevier Inc.: Amsterdam, The Netherlands, 2013; pp. 333–343. ISBN 9780123944313. [Google Scholar]
- Hammann, F.; Schmid, M. Determination and Quantification of Molecular Interactions in Protein Films: A Review. Materials 2014, 7, 7975–7996. [Google Scholar] [CrossRef] [Green Version]
- Jain, A.; Kumar, A.; Salunke, D.M. Crystal Structure of the Vicilin from Solanum Melongena Reveals Existence of Different Anionic Ligands in Structurally Similar Pockets. Sci. Rep. 2016, 6, 23600. [Google Scholar] [CrossRef] [Green Version]
- Tang, C. Nanostructures of Soy Proteins for Encapsulation of Food Bioactive Ingredients. In Biopolymer Nanostructures for Food Encapsulation Purposes; Elsevier: Amsterdam, The Netherlands, 2019; pp. 247–285. ISBN 9780128156636. [Google Scholar]
- Tandang-Silvas, M.R.G.; Fukuda, T.; Fukuda, C.; Prak, K.; Cabanos, C.; Kimura, A.; Itoh, T.; Mikami, B.; Utsumi, S.; Maruyama, N. Conservation and Divergence on Plant Seed 11S Globulins Based on Crystal Structures. Biochim. Biophys. Acta Proteins Proteom. 2010, 1804, 1432–1442. [Google Scholar] [CrossRef] [PubMed]
- Adachi, M.; Kanamori, J.; Masuda, T.; Yagasaki, K.; Kitamura, K.; Mikami, B.; Utsumi, S. Crystal Structure of Soybean 11S Globulin: Glycinin A3B4 Homohexamer. Proc. Natl. Acad. Sci. USA 2003, 100, 7395–7400. [Google Scholar] [CrossRef] [Green Version]
- Luo, L.; Wang, Z.; Deng, Y.; Wei, Z.; Zhang, Y.; Tang, X.; Liu, G.; Zhou, P.; Zhao, Z.; Zhang, M.; et al. High-Pressure Homogenization: A Potential Technique for Transforming Insoluble Pea Protein Isolates into Soluble Aggregates. Food Chem. 2022, 397, 133684. [Google Scholar] [CrossRef]
- Dent, T.; Campanella, O.; Maleky, F. Enzymatic Hydrolysis of Soy and Chickpea Protein with Alcalase and Flavourzyme and Formation of Hydrogen Bond Mediated Insoluble Aggregates. Curr. Res. Food Sci. 2023, 6, 100487. [Google Scholar] [CrossRef]
- Shuai, X.; Gao, L.; Geng, Q.; Li, T.; He, X.; Chen, J.; Liu, C.; Dai, T. Effects of Moderate Enzymatic Hydrolysis on Structure and Functional Properties of Pea Protein. Foods 2022, 11, 2368. [Google Scholar] [CrossRef]
- Zheng, Z.; Li, J.; Liu, Y. Effects of Partial Hydrolysis on the Structural, Functional and Antioxidant Properties of Oat Protein Isolate. Food Funct. 2020, 11, 3144–3155. [Google Scholar] [CrossRef]
- De Leo, F.; Volpicella, M.; Licciulli, F.; Liuni, S.; Gallerani, R.; Ceci, L.R. PLANT-PIs: A Database for Plant Protease Inhibitors and Their Genes. Nucleic Acids Res. 2002, 30, 347–348. [Google Scholar] [CrossRef] [Green Version]
- Cid-Gallegos, M.S.; Corzo-Ríos, L.J.; Jiménez-Martínez, C.; Sánchez-Chino, X.M. Protease Inhibitors from Plants as Therapeutic Agents—A Review. Plant Foods Hum. Nutr. 2022, 77, 20–29. [Google Scholar] [CrossRef]
- Avilés-Gaxiola, S.; Chuck-Hernández, C.; Serna Saldívar, S.O. Inactivation Methods of Trypsin Inhibitor in Legumes: A Review. J. Food Sci. 2018, 83, 17–29. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Grossmann, L.; Zeeb, B.; Weiss, J. Diffusion Behavior of Microbial Transglutaminase to Induce Protein Crosslinking in Oil-in-Water Emulsions. J. Dispers. Sci. Technol. 2016, 37, 1745–1750. [Google Scholar] [CrossRef]
- Grossmann, L.; Wefers, D.; Bunzel, M.; Weiss, J.; Zeeb, B. Accessibility of Transglutaminase to Induce Protein Crosslinking in Gelled Food Matrices—Influence of Network Structure. LWT 2017, 75, 271–278. [Google Scholar] [CrossRef]
- Zeeb, B.; Grossmann, L.; Weiss, J. Accessibility of Transglutaminase to Induce Protein Crosslinking in Gelled Food Matrices—Impact of Membrane Structure. Food Biophys. 2016, 11, 176–183. [Google Scholar] [CrossRef]
- Zeeb, B.; McClements, D.J.; Weiss, J. Enzyme-Based Strategies for Structuring Foods for Improved Functionality. Annu. Rev. Food Sci. Technol. 2017, 8, 21–34. [Google Scholar] [CrossRef]
- Gruppi, A.; Dermiki, M.; Spigno, G.; Fitzgerald, R.J. Impact of Enzymatic Hydrolysis and Heat Inactivation on the Physicochemical Properties of Milk Protein Hydrolysates. Foods 2022, 11, 516. [Google Scholar] [CrossRef]
- Ong, L.; Li, X.; Ong, A.; Gras, S.L. New Insights into Cheese Microstructure. Annu. Rev. Food Sci. Technol. 2022, 13, 89–115. [Google Scholar] [CrossRef]
- Nadar, S.S.; Rao, P.; Rathod, V.K. Enzyme Assisted Extraction of Biomolecules as an Approach to Novel Extraction Technology: A Review. Food Res. Int. 2018, 108, 309–330. [Google Scholar] [CrossRef] [PubMed]
- Li, B.B.; Smith, B.; Hossain, M.M. Extraction of Phenolics from Citrus Peels: II. Enzyme-Assisted Extraction Method. Sep. Purif. Technol. 2006, 48, 189–196. [Google Scholar] [CrossRef]
- Tufvesson, P.; Fu, W.; Jensen, J.S.; Woodley, J.M. Process Considerations for the Scale-up and Implementation of Biocatalysis. Food Bioprod. Process. 2010, 88, 3–11. [Google Scholar] [CrossRef]
- Shiva; Rodríguez-Jasso, R.M.; López-Sandin, I.; Aguilar, M.A.; López-Badillo, C.M.; Ruiz, H.A. Intensification of Enzymatic Saccharification at High Solid Loading of Pretreated Agave Bagasse at Bioreactor Scale. J. Environ. Chem. Eng. 2023, 11, 109257. [Google Scholar] [CrossRef]
- Gouseti, O.; Jaime-Fonseca, M.R.; Fryer, P.J.; Mills, C.; Wickham, M.S.J.; Bakalis, S. Hydrocolloids in Human Digestion: Dynamic in-Vitro Assessment of the Effect of Food Formulation on Mass Transfer. Food Hydrocoll. 2014, 42, 378–385. [Google Scholar] [CrossRef] [Green Version]
- Jaime-Fonseca, M.R.; Gouseti, O.; Fryer, P.J.; Wickham, M.S.J.; Bakalis, S. Digestion of Starch in a Dynamic Small Intestinal Model. Eur. J. Nutr. 2015, 55, 2377–2388. [Google Scholar] [CrossRef]
- Burek, B.O.; Dawood, A.W.H.; Hollmann, F.; Liese, A.; Holtmann, D. Process Intensification as Game Changer in Enzyme Catalysis. Front. Catal. 2022, 2, 858706. [Google Scholar] [CrossRef]
- Liow, M.Y.; Gourich, W.; Chang, M.Y.; Loh, J.M.; Chan, E.S.; Song, C.P. Towards Rapid and Sustainable Synthesis of Biodiesel: A Review of Effective Parameters and Scale-up Potential of Intensification Technologies for Enzymatic Biodiesel Production. J. Ind. Eng. Chem. 2022, 114, 1–18. [Google Scholar] [CrossRef]
- Becker, L.; Sturm, J.; Eiden, F.; Holtmann, D. Analyzing and Understanding the Robustness of Bioprocesses. Trends Biotechnol. 2023. [Google Scholar] [CrossRef]
- Gaviria G, Y.S.; Zapata M, J.E. Optimization and Scale up of the Enzymatic Hydrolysis of Californian Red Worm Protein (Eisenia foetida). Heliyon 2023, 9, e16165. [Google Scholar] [CrossRef]
- Schmidt, C.M.; Nedele, A.K.; Hinrichs, J. Enzymatic Generation of Lactulose in Sweet and Acid Whey: Feasibility Study for the Scale up towards Robust Processing. Food Bioprod. Process. 2020, 119, 329–336. [Google Scholar] [CrossRef]
- Chatterjee, S.; Bambot, S.; Bormett, R.; Asher, S.; Russell, A.J. Enzymology Nonaqueous. Encycl. Mol. Biol. Mol. Med. 1996, 2, 249–258. [Google Scholar]
- Picot-Allain, C.; Mahomoodally, M.F.; Ak, G.; Zengin, G. Conventional versus Green Extraction Techniques—A Comparative Perspective. Curr. Opin. Food Sci. 2021, 40, 144–156. [Google Scholar] [CrossRef]
- Smith, E.L.; Abbott, A.P.; Ryder, K.S. Deep Eutectic Solvents (DESs) and Their Applications. Chem. Rev. 2014, 114, 11060–11082. [Google Scholar] [CrossRef] [PubMed] [Green Version]
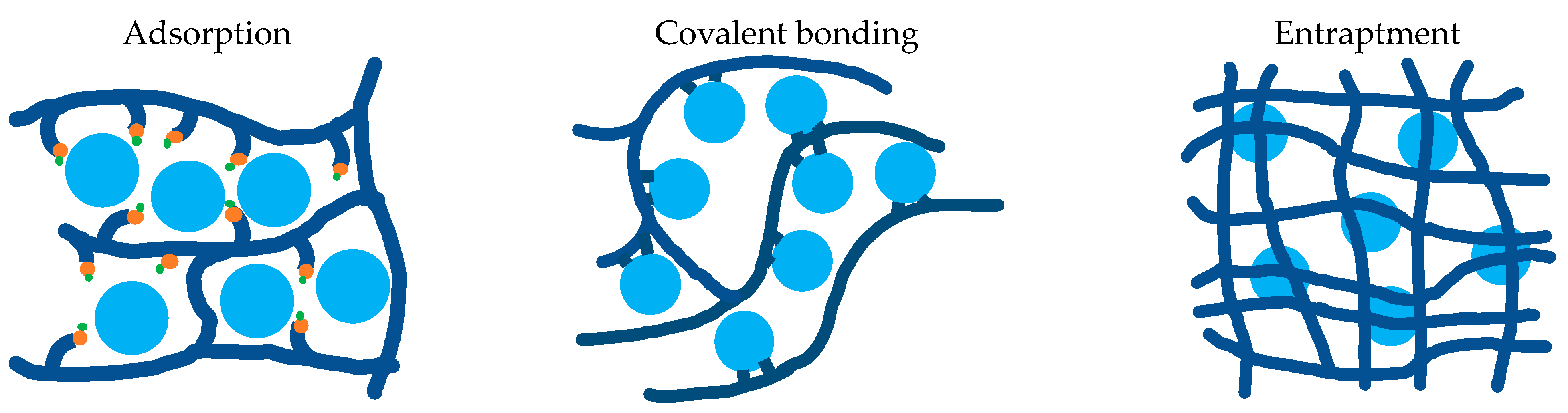
- Datta, S.; Christena, L.R.; Rajaram, Y.R.S. Enzyme Immobilization: An Overview on Techniques and Support Materials. 3 Biotech 2013, 3, 1–9. [Google Scholar] [CrossRef] [Green Version]
- Robinson, P.K. Enzymes: Principles and Biotechnological Applications. Essays Biochem. 2015, 59, 1–41. [Google Scholar] [CrossRef]
- Imam, H.T.; Marr, P.C.; Marr, A.C. Enzyme Entrapment, Biocatalyst Immobilization without Covalent Attachment. Green Chem. 2021, 23, 4980–5005. [Google Scholar] [CrossRef]
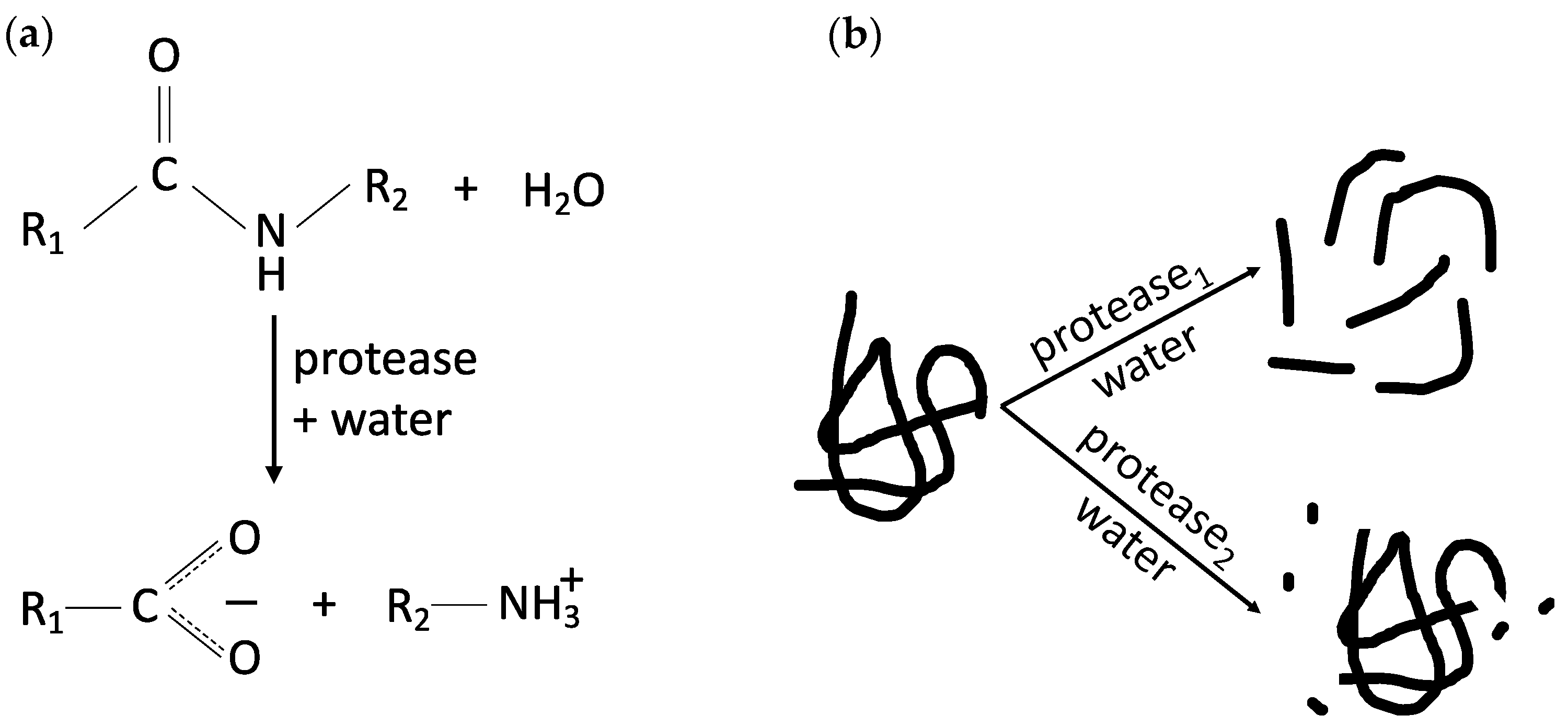




| Property Category | Example Functional Properties |
|---|---|
| Sensory | Color; flavor; smoothness; grittiness; mouthfeel |
| Hydration | Solubility; dispersibility; swelling; wettability; water absorption; water holding capacity; protein–water interactions |
| Surface properties | Emulsification; foaming; lipid-binding; surface hydrophobicity; amphiphilicity; surface charge; contact angle; oil holding capacity; film formation |
| Texture-related properties | Viscosity; elasticity; gelation; aggregation; extrudability; emulsification and foaming; adhesion; stickiness; chewiness; viscoelasticity; fiber formation ability |
| Protein Source | Enzyme (s) | Extraction Conditions | Yield | Quality | References |
|---|---|---|---|---|---|
| Defatted soybean cake | Cellulase, xylanase, pectinase | Mildly alkaline | 45% | Near-native; higher solubility and emulsifying properties when using enzymes in extraction | [40] |
| Pea | Papain | Mildly alkaline | 58% | Small peptides, amino acids; extensive proteolysis reduced emulsifying properties (also reported in Section 3.1) | [41] |
| Chickpea | Arabinofuranosidase or cocktail of cellulase and xylanase | Alkaline | 93% | Increased yield and functional properties of the protein isolate with both enzymatic treatments compared to alkaline extraction alone. | [42] |
| Rapeseed cake | Pectinex®, Depol®, Celluclast® | Neutral | Up to 74% | Not reported | [43] |
| Lentil | Viscozyme® | Mildly acidic for the enzymatic pre-treatment, then alkaline | 62% | Similar yield but higher purity and improved functional properties when using enzymes, compared to alkaline extraction alone. | [44] |
| Sesame | Neutrase®, Pectinex® | Not reported | 90% | Small peptides; extensive use of carbohydrases reduced purity as the product contained solubilised carbohydrates. | [45] |
| Akebia trifoliata | Cellulase | Alkaline | 20% | Higher purity and functional properties compared to alkaline extraction alone | [46] |
| Red seaweed | Alcalase®, Celluclast®, Shearzyme® | Mildly alkaline | 90% | Large, highly functional peptides under investigated conditions, despite using proteases | [47] |
| Property | Mechanisms through Which Hydrolysis May Increase It | Mechanisms through Which Hydrolysis May Reduce It | Examples (with References) |
|---|---|---|---|
| Solubility | Size reduction. Increase of ionizable groups. | Hydrophobic interactions of newly exposed groups. | Results vary considerably, but enzymatic hydrolysis appeared to increase solubility of chickpea [56], peanut [57], sunflower [58], oat [59], rice endosperm [60], and pea [55] protein at DH up to 23%. |
| Surface Hydrophobicity | Exposure of hidden hydrophobic groups to the surface. | Hydrophobic interactions of newly exposed groups, particularly at high DH. | Effect heavily depends on enzyme specificity and conditions. Can increase surface hydrophobicity of soy protein isolate [61]; hemp protein isolate [62]; brewers spent grain protein concentrate [63]. |
| Emulsification | Increased solubility. Increased surface hydrophobicity. Exposure of hidden hydrophobic groups that can adhere to the O/W interface. Increased amphiphilicity. Increased molecular flexibility and possibly disruption of the compact molecular structure [64,65]. | Extensive reduction in molecular size and hydrodynamic diameter at high DH. This may reduce potential of interfacial interactions and viscoelasticity of the resulting film. Reduced surface hydrophobicity at high DH. | Depending on conditions, limited hydrolysis (generally about up to 2–3% DH) overall increased emulsion capacity and stability in protein-stabilized O/W emulsions with rice bran albumin and globulin [64]; potato protein concentrate [65]; pea protein isolate [55]; chickpea protein isolate [66]. |
| Foaming | Similar to emulsification, factors that enhance surface interactions of the hydrolysates increase foamability. | Similar to emulsification, factors that decrease surface interactions of the hydrolysates reduce foamability. | Largely depending on conditions. At low DH, foaming properties increased for soy protein [67], sunflower protein isolate [68], pea protein isolate [55,69]. |
| Gelation | Factors that enhance protein–protein and reduce protein–water interactions. “Loosening” of the compact protein molecules. | Factors that enhance protein-water and reduce protein-protein interactions. Reduced molecular size. Reduced hydrophobicity. Increased surface charge. | Limited hydrolysis increased gelling properties of soybean proteins [70]; pea proteins [71]; peanut protein isolate [57]; oat protein [72]; rice endosperm protein [73]; defatted soy flour [74]; sunflower protein [75]. |
| Enzyme | Major Sources | Action Site | Product | References |
|---|---|---|---|---|
| Trypsin | Porcine or bovine intestine | Highly specific. Cleaves C-terminal to arginine (R) and lysine (K) residues. Less effective if acidic residue (glutamate (E) or aspartate (D)) is near the cleavage site. May cleave before proline (P). | Small peptides | [76,77,78] |
| Pepsin | Porcine gastric mucosa | Broad specificity, with overall preference to cleave after bulky aromatic residues (maybe favoring phenylalanine (F)), leucine (L), and possibly methionine (M). Cleavage after histidine (H), lysine (K), arginine (R), proline (P) usually not as favored, unless adjacent to residues such as leucine (L) or phenylalanine (F). | Small peptides | [79,80,81] |
| Carboxy-peptidase (CP) | CP-A from bovine pancreas; CP-B from bovine or porcine pancreas; CP-Y (yeast CP) from baker’s yeast. | CPs are exopeptidases that cleave the carboxy end of proteins and peptides, usually one residue at a time. Depending on their substrate preference they can be classified as CPs-A (prefer aromatic and large aliphatic sidechains, hydrolyze slowly glycine (G) and acidic residues, rarely proline (P) and basic residues); CPs-B (with narrower specificity than CPs-A and preference towards the basic residues arginine (R), lysine (K) and some action on neutral amino acids); and CPs-C (can release proline (P) and other amino acids). CP-Y has broad specificity, similar to CP-A but cleaves rapidly glycine (G) and leucine (L), and slowly phenylalanine (F). | Typically single amino acids | [82,83,84,85] |
| Amino-peptidase (AP) | Microbes and porcine kidney. | APs are exopeptidases that cleave the amino end of proteins and peptides. They can be classified to aminoacylpeptidases, dipeptidyl- and tripeptidyl- peptidases (i.e., releasing single amino acids, dipeptides, tripeptides, respectively), with a tetra-peptidase recently reported. If acting only on di- or tri- peptides, they are di- and tri-peptidases, respectively. Based on substrate specificity they are classified into 2 categories: broad and narrow. | Amino acids, di-peptides, tri-peptides, rarely tetra-peptides | [86,87] |
| Alcalase | Microbes | Has broad specificity. Reported to cleave bonds on the carboxyl side of glutamic acid (E), methionine (M), leucine (L), tyrosine (Y), lysine (K), and glutamine (Q), also at phenylalanine (F), tryptophan (W), alanine (A), serine (S). | Small peptides | [88,89,90,91] |
| Plasmin or fibrinolysin | From bovine plasma or microbes | Has similar specificity to trypsin, but less efficient. Cleaves after arginine (R) and lysine (K) residues. | Small peptides | [92,93,94] |
| Flavor-zyme® | Microbial (Aspergillus oryzae) | Broad specificity, mostly endo activity | Small peptides and amino acids | [95,96] |
| Protamex | Microbial (Bacillussp.) | Broad specificity. | Small peptides | [95,97] |
| Neutrase® | Microbial (Bacillus amyloliquefaciens) | Broad specificity | Small peptides | [95] |
| Corolase 7089 | Fungal neutral protease | Broad specificity | Small peptides | [97] |
| Pronase | Microbial (Streptomyces griseus) | Broad specificity | Amino acids and peptides | [98] |
| Prolidase | Microbial | Cleaves before proline (P) or hydroxylproline in dipeptides. | Amino acids | [99] |
| Ficin | Fig (Ficus carica) | Generally prefers to cleave after aromatic residues e.g., tyrosine (Y), phenylalanine (F); exact specificity depends on form. | Small peptides | [100,101,102] |
| Papain | Papaya (Carica papayaL.) | Has broad specificity, with reported preference to cleave bonds at arginine (R), lysine (K), and phenylalanine (F). | Small peptides | [52,88,103,104] |
| Bromelain | Fruit or stem of pineapple (Ananas comosusL.) | Broad specificity. | Small peptides | [88,100,105] |
| Observation | Protein Source (with Ref) |
|---|---|
| Increased gel strength and firmness; Some studies mention increased water holding capacity. | Faba bean protein isolate [114]; pea protein isolate [115]; soy protein [116]; soybean milk [117,118] |
| Decreased solubility and increased surface hydrophobicity | Peanut protein isolate [119]; vicilin-rich kidney protein isolate [120] |
| Reduced CO2 and O2 permeability in edible protein films (for food packaging applications) | Bitter vetch protein films [121] |
| Observation | Possible Mechanism | Protein Source (with References) |
|---|---|---|
| Increased solubility and emulsifying/foaming properties, particularly at neutral pH | Increase in protein charge and associated inter-molecular repulsions may increase solubility; higher solubility may increase foaming and emulsifying properties. | Wheat gluten [127]; soy [128]; oat [129]; coconut [130]. |
| Improved taste, for example through decreasing binding affinity of proteins to tastants (vanillin), which become free, or by decreasing bitter taste and enhancing umami | By reducing binding affinity of proteins to tastants, therefore increasing the “free” tastant concentration. | Coconut [131,132]; soy [131,132]; wheat gluten [133,134]; wheat gluten hydrolysates [133,134]. |
| Reduced allergenic potential | Conformational changes of the proteins, particularly for proteins high in glutamine residues that are susceptible to deamidation | Wheat gluten [127] |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Gouseti, O.; Larsen, M.E.; Amin, A.; Bakalis, S.; Petersen, I.L.; Lametsch, R.; Jensen, P.E. Applications of Enzyme Technology to Enhance Transition to Plant Proteins: A Review. Foods 2023, 12, 2518. https://doi.org/10.3390/foods12132518
Gouseti O, Larsen ME, Amin A, Bakalis S, Petersen IL, Lametsch R, Jensen PE. Applications of Enzyme Technology to Enhance Transition to Plant Proteins: A Review. Foods. 2023; 12(13):2518. https://doi.org/10.3390/foods12132518
Chicago/Turabian StyleGouseti, Ourania, Mads Emil Larsen, Ashwitha Amin, Serafim Bakalis, Iben Lykke Petersen, Rene Lametsch, and Poul Erik Jensen. 2023. "Applications of Enzyme Technology to Enhance Transition to Plant Proteins: A Review" Foods 12, no. 13: 2518. https://doi.org/10.3390/foods12132518
APA StyleGouseti, O., Larsen, M. E., Amin, A., Bakalis, S., Petersen, I. L., Lametsch, R., & Jensen, P. E. (2023). Applications of Enzyme Technology to Enhance Transition to Plant Proteins: A Review. Foods, 12(13), 2518. https://doi.org/10.3390/foods12132518