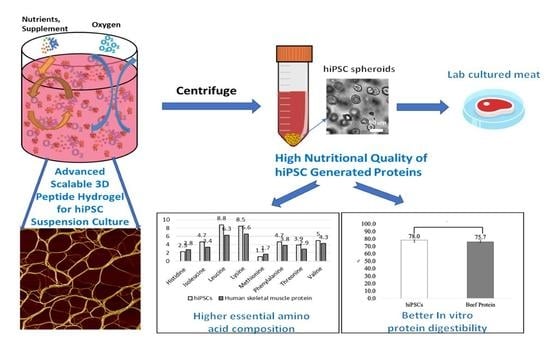
High Nutritional Quality of Human-Induced Pluripotent Stem Cell-Generated Proteins through an Advanced Scalable Peptide Hydrogel 3D Suspension System
Abstract
1. Introduction
2. Materials and Methods
2.1. Materials
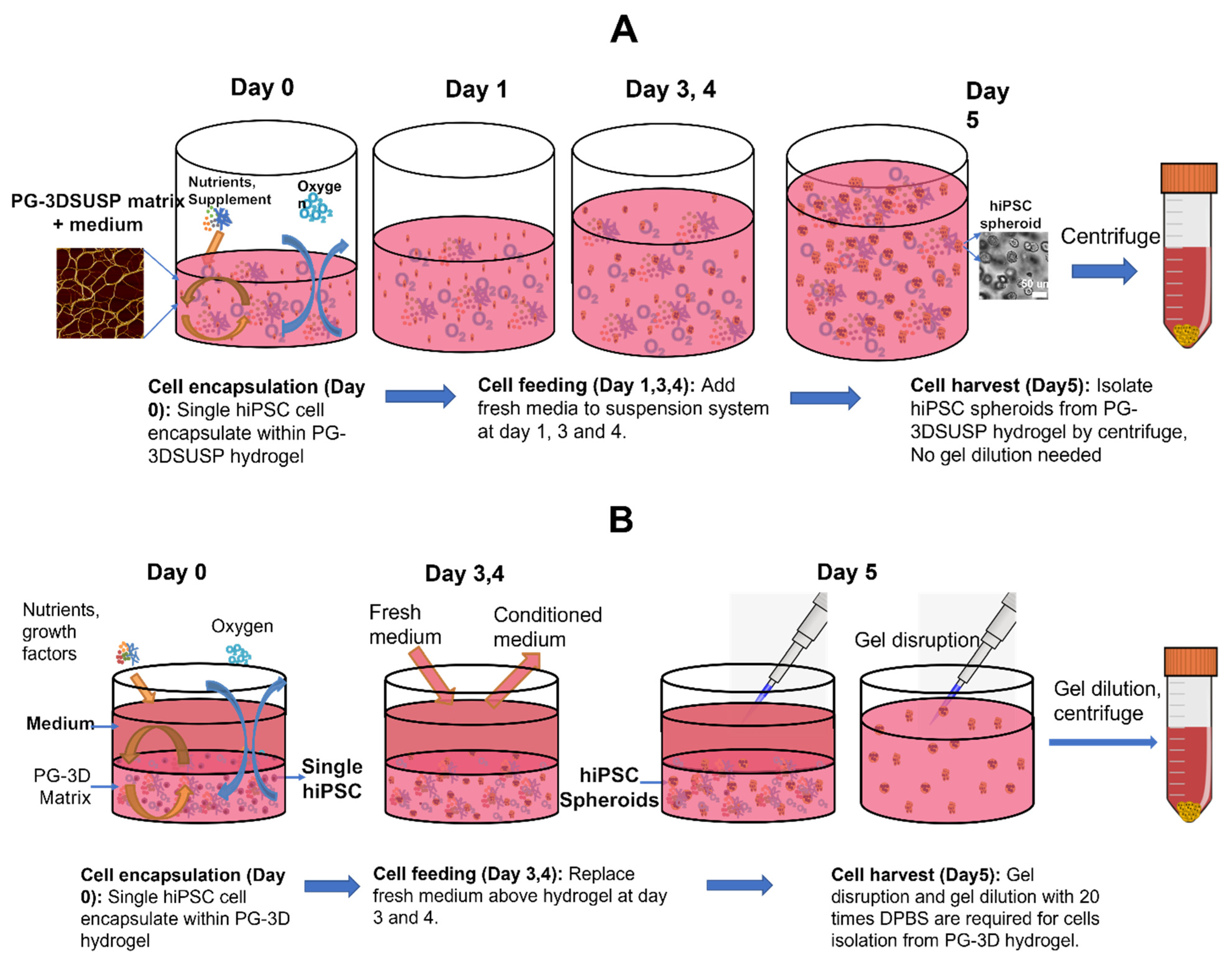
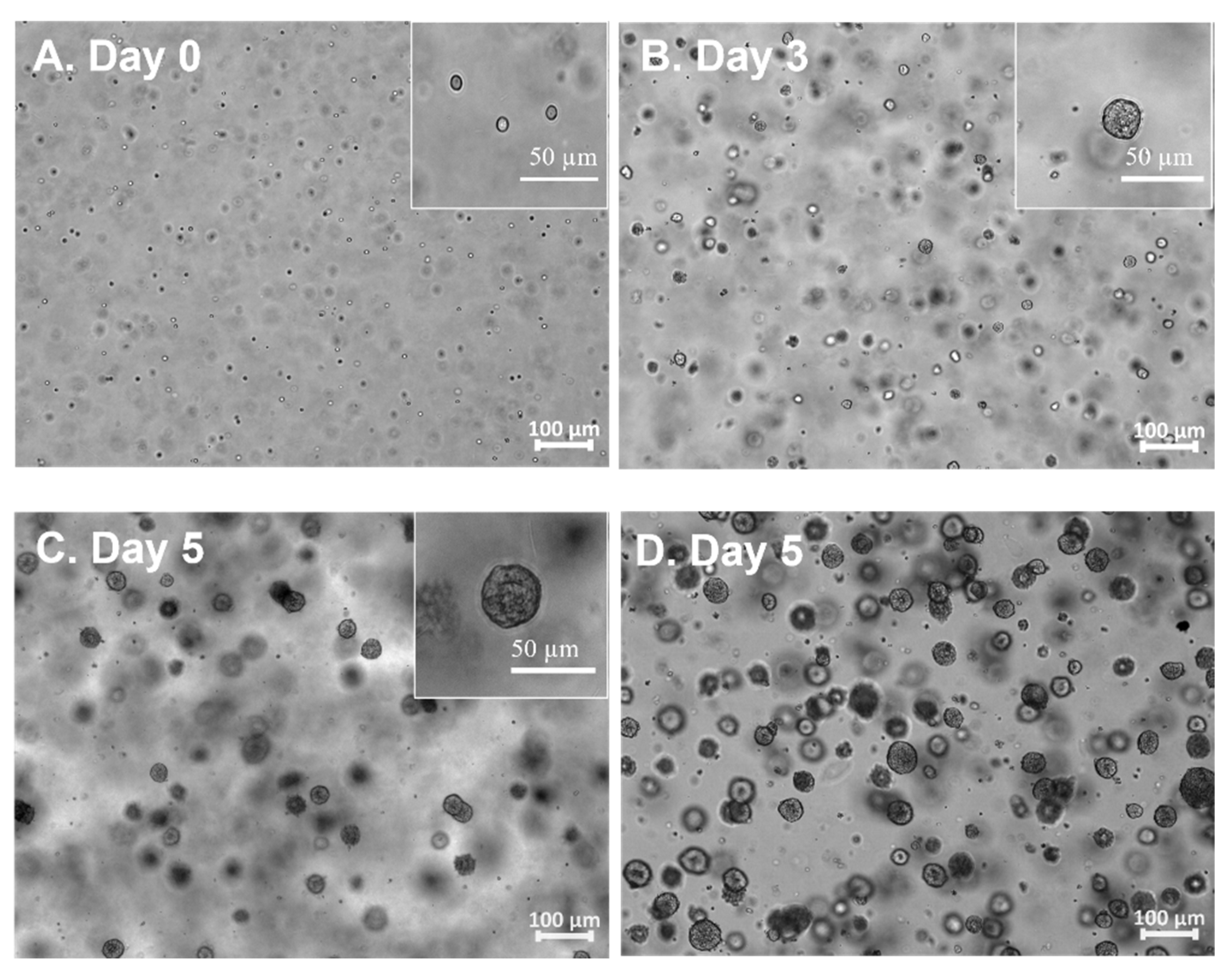
2.2. hiPSCs 3D Physiological Colony Culture in PGmatrix3D Suspension
2.3. hiPSC 3D Colonies Harvesting
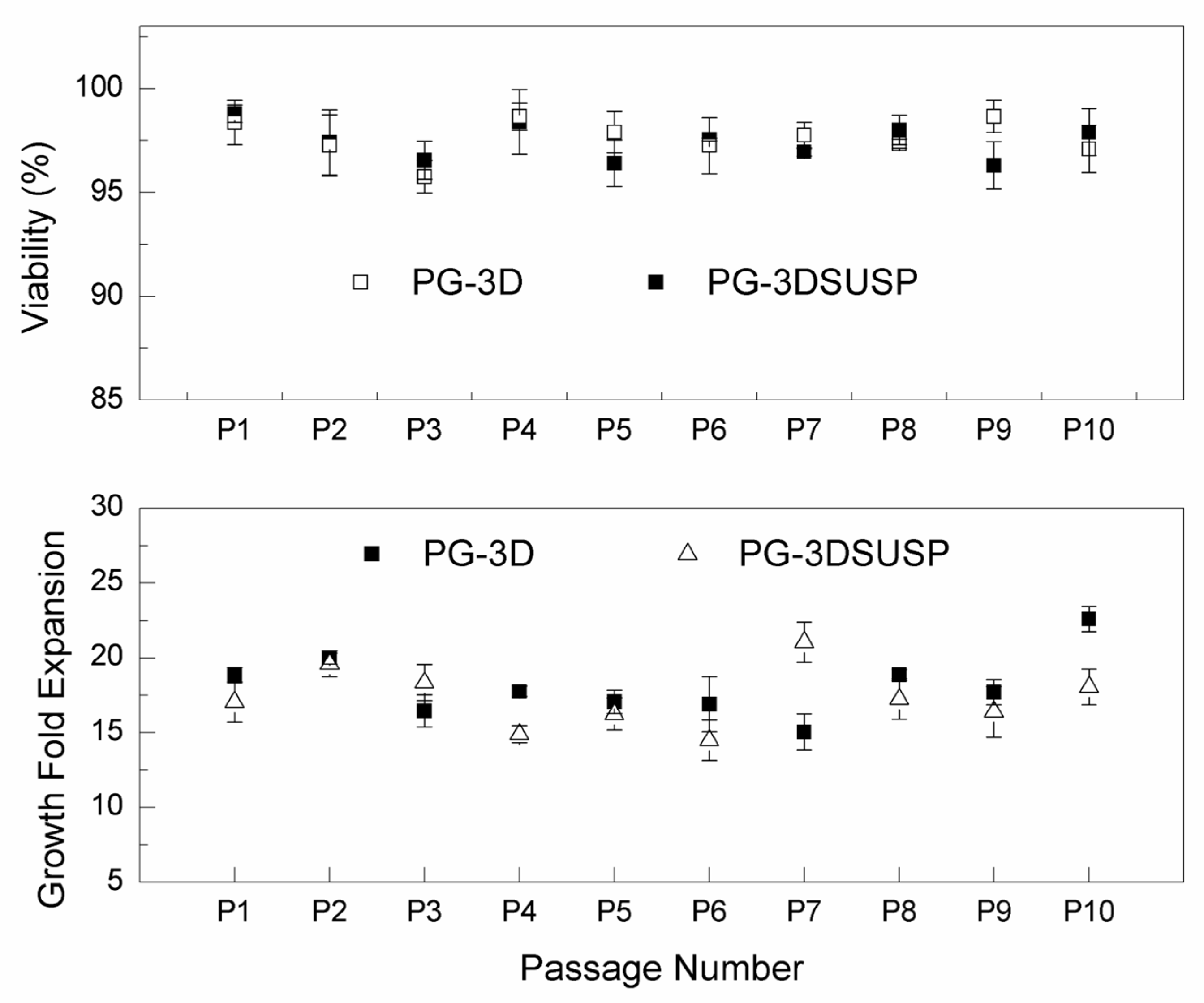
2.4. hiPSCs 3D Colonies Passage
2.5. hiPSC 3D Physiological Colony Culture in PGmatrix-hiPSC Culture
2.6. Real-Time Quantitative PCR (RT-qPCR)
2.7. Amino Acid Profile Analysis of hiPSC 3D Colonies
2.8. In Vitro Protein Digestion
2.9. Statistical Analysis
3. Results and Discussion
3.1. The hiPSC Production System
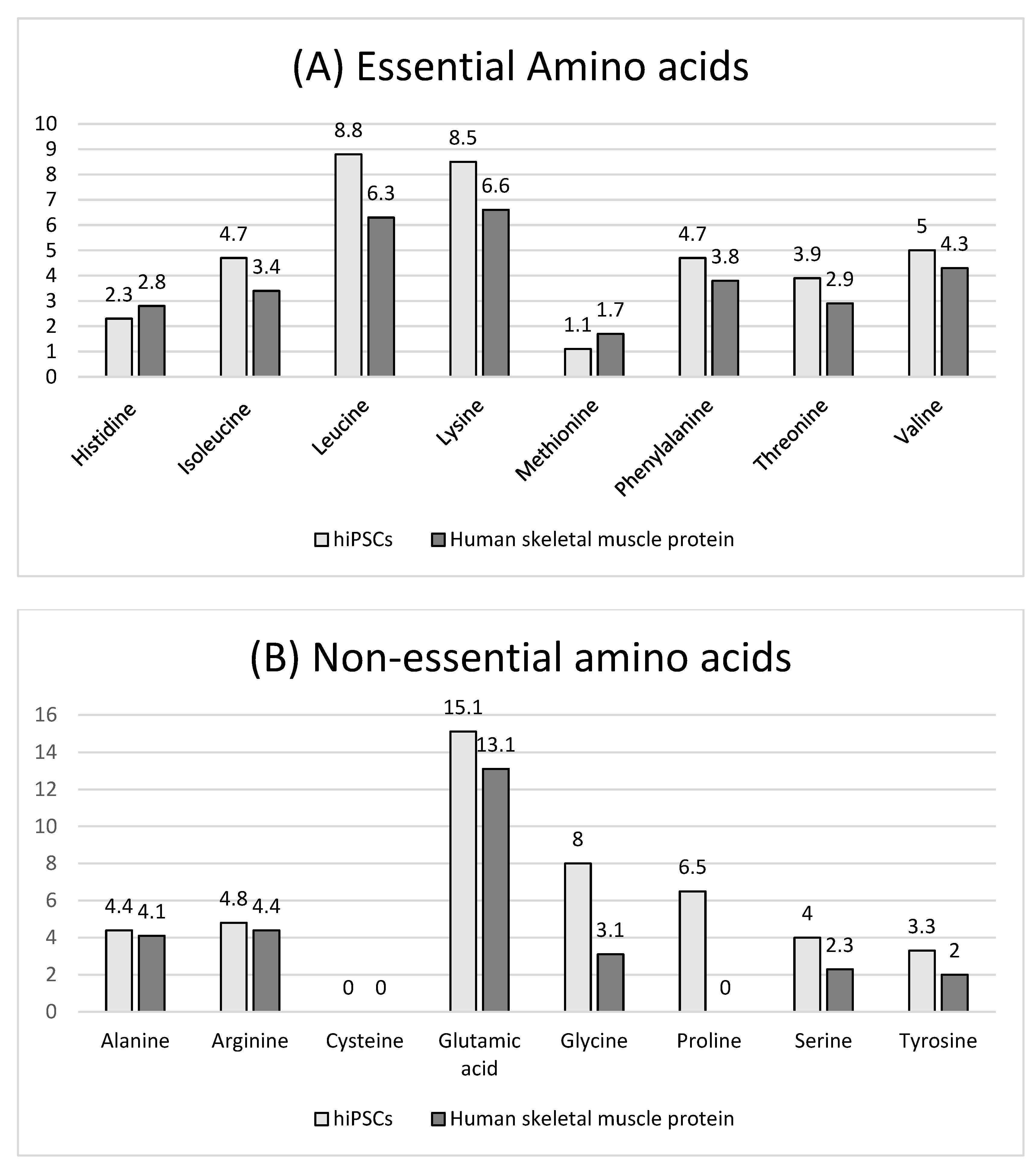
3.2. hiPSC Nutritional Properties
4. Conclusions
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Wu, G. Dietary protein intake and human health. Food Funct. 2016, 7, 1251–1265. [Google Scholar] [CrossRef] [PubMed]
- Davies, R.W.; Jakeman, P.M. Separating the Wheat from the Chaff: Nutritional Value of Plant Proteins and Their Potential Contribution to Human Health. Nutrients 2020, 12, 2410. [Google Scholar] [CrossRef] [PubMed]
- Atherton, P.J.; Smith, K.; Etheridge, T.; Rankin, D.; Rennie, M.J. Distinct anabolic signalling responses to amino acids in C2C12 skeletal muscle cells. Amino Acids 2010, 38, 1533–1539. [Google Scholar] [CrossRef] [PubMed]
- Gorissen, S.H.; Crombag, J.J.; Senden, J.M.; Waterval, W.A.; Bierau, J.; Verdijk, L.B.; van Loon, L.J. Protein content and amino acid composition of commercially available plant-based protein isolates. Amino Acids 2018, 50, 1685–1695. [Google Scholar] [CrossRef] [PubMed]
- Gilbert, J.-A.; Bendsen, N.; Tremblay, A.; Astrup, A. Effect of proteins from different sources on body composition. Nutr. Metab. Cardiovasc. Dis. 2011, 21, B16–B31. [Google Scholar] [CrossRef] [PubMed]
- Hoffman, J.R.; Falvo, M.J. Protein—Which is best? J. Sports Sci. Med. 2004, 3, 118. [Google Scholar]
- Hertzler, S.R.; Lieblein-Boff, J.C.; Weiler, M.; Allgeier, C. Plant Proteins: Assessing Their Nutritional Quality and Effects on Health and Physical Function. Nutrients 2020, 12, 3704. [Google Scholar] [CrossRef]
- Libera, J.; Iłowiecka, K.; Stasiak, D. Consumption of processed red meat and its impact on human health: A review. Int. J. Food Sci. Technol. 2021, 56, 6115–6123. [Google Scholar] [CrossRef]
- Deer, R.R.; Volpi, E. Protein intake and muscle function in older adults. Curr. Opin. Clin. Nutr. Metab. Care 2015, 18, 248–253. [Google Scholar] [CrossRef]
- Jinno, S.; Yamazaki, K.; Nakamura, Y.; Kinouchi, T. Daily protein and energy intakes of infants fed a commercial infant formula with a reduced protein concentration of 2.2 g/100 kcal: An impact of feeding interval on energy intake. Biosci. Biotechnol. Biochem. 2020, 84, 1259–1264. [Google Scholar] [CrossRef]
- Delimaris, I. Adverse Effects Associated with Protein Intake above the Recommended Dietary Allowance for Adults. Int. Sch. Res. Not. 2013, 2013, 126929. [Google Scholar] [CrossRef]
- Hector, A.J.; Phillips, S.M. Protein Recommendations for Weight Loss in Elite Athletes: A Focus on Body Composition and Performance. Int. J. Sport Nutr. Exerc. Metab. 2018, 28, 170–177. [Google Scholar] [CrossRef]
- Li, Y. Feeding the Future: Plant-Based Meat for Global Food Security and Environmental Sustainability. Cereal Foods World 2020, 65. [Google Scholar] [CrossRef]
- Post, M.J. Cultured meat from stem cells: Challenges and prospects. Meat Sci. 2012, 92, 297–301. [Google Scholar] [CrossRef]
- Wu, G.; Fanzo, J.; Miller, D.D.; Pingali, P.; Post, M.; Steiner, J.L.; Thalacker-Mercer, A.E. Production and supply of high-quality food protein for human consumption: Sustainability, challenges, and innovations. Ann. N. Y. Acad. Sci. 2014, 1321, 1–19. [Google Scholar] [CrossRef]
- Wikandari, R.; Manikharda Baldermann, S.; Ningrum, A.; Taherzadeh, M.J. Application of cell culture technology and genetic engineering for production of future foods and crop improvement to strengthen food security. Bioengineered 2021, 12, 11305–11330. [Google Scholar] [CrossRef] [PubMed]
- Kapałczyńska, M.; Kolenda, T.; Przybyła, W.; Zajączkowska, M.; Teresiak, A.; Filas, V.; Ibbs, M.; Bliźniak, R.; Łuczewski, L.; Lamperska, K. 2D and 3D cell cultures—A comparison of different types of cancer cell cultures. Arch. Med. Sci. 2018, 14, 910–919. [Google Scholar] [CrossRef] [PubMed]
- Stephens, N.; Di Silvio, L.; Dunsford, I.; Ellis, M.; Glencross, A.; Sexton, A. Bringing cultured meat to market: Technical, socio-political, and regulatory challenges in cellular agriculture. Trends Food Sci. Technol. 2018, 78, 155–166. [Google Scholar] [CrossRef] [PubMed]
- Bodiou, V.; Moutsatsou, P.; Post, M.J. Microcarriers for Upscaling Cultured Meat Production. Front. Nutr. 2020, 7, 10. [Google Scholar] [CrossRef]
- Zhu, Z.; Huangfu, D. Human pluripotent stem cells: An emerging model in developmental biology. Development 2013, 140, 705–717. [Google Scholar] [CrossRef]
- Li, Q.; Qi, G.; Liu, X.; Bai, J.; Zhao, J.; Tang, G.; Zhang, Y.S.; Chen-Tsai, R.; Zhang, M.; Wang, D.; et al. Universal Peptide Hydrogel for Scalable Physiological Formation and Bioprinting of 3D Spheroids from Human Induced Pluripotent Stem Cells. Adv. Funct. Mater. 2021, 31, 2104046. [Google Scholar] [CrossRef]
- Villa-Diaz, L.G.; Nandivada, H.; Ding, J.; Nogueira-De-Souza, N.C.; Krebsbach, P.H.; O’Shea, K.S.; Lahann, J.; Smith, G.D. Synthetic polymer coatings for long-term growth of human embryonic stem cells. Nat. Biotechnol. 2010, 28, 581–583. [Google Scholar] [CrossRef] [PubMed]
- Xu, J.; Qi, G.; Sui, C.; Wang, W.; Sun, X. 3D h9e peptide hydrogel: An advanced three-dimensional cell culture system for anticancer prescreening of chemopreventive phenolic agents. Toxicol. In Vitro 2019, 61, 104599. [Google Scholar] [CrossRef]
- Xu, J.; Qi, G.; Wang, W.; Sun, X.S. Advances in 3D peptide hydrogel models in cancer research. Sci. Food 2021, 5, 14. [Google Scholar] [CrossRef]
- Gerecht, S.; Burdick, J.A.; Ferreira, L.S.; Townsend, S.A.; Langer, R.; Vunjak-Novakovic, G. Hyaluronic acid hydrogel for controlled self-renewal and differentiation of human embryonic stem cells. Proc. Natl. Acad. Sci. USA 2007, 104, 11298–11303. [Google Scholar] [CrossRef]
- Lei, Y.; Schaffer, D.V. A fully defined and scalable 3D culture system for human pluripotent stem cell expansion and differentiation. Proc. Natl. Acad. Sci. USA 2013, 110, E5039–E5048. [Google Scholar] [CrossRef] [PubMed]
- Chayosumrit, M.; Tuch, B.; Sidhu, K. Alginate microcapsule for propagation and directed differentiation of hESCs to definitive endoderm. Biomaterials 2010, 31, 505–514. [Google Scholar] [CrossRef]
- Li, Z.; Leung, M.; Hopper, R.; Ellenbogen, R.; Zhang, M. Feeder-free self-renewal of human embryonic stem cells in 3D porous natural polymer scaffolds. Biomaterials 2010, 31, 404–412. [Google Scholar] [CrossRef]
- Adil, M.M.; Rodrigues, G.M.; Kulkarni, R.U.; Rao, A.T.; Chernavsky, N.E.; Miller, E.W.; Schaffer, D.V. Efficient generation of hPSC-derived midbrain dopaminergic neurons in a fully defined, scalable, 3D biomaterial platform. Sci. Rep. 2017, 7, 40573. [Google Scholar] [CrossRef] [PubMed]
- Adil, M.M.; Vazin, T.; Ananthanarayanan, B.; Rodrigues, G.M.; Rao, A.T.; Kulkarni, R.U.; Miller, E.W.; Kumar, S.; Schaffer, D.V. Engineered hydrogels increase the post-transplantation survival of encapsulated hESC-derived midbrain dopaminergic neurons. Biomaterials 2017, 136, 1–11. [Google Scholar] [CrossRef]
- Li, Q. Synthetic Hydrogel-Based 3D Culture System for Maintenance of Human Induced Pluripotent Stem Cell. Ph.D. Dissertation, Kansas State University, Manhattan, KS, USA, 2017. [Google Scholar]
- Duval, K.; Grover, H.; Han, L.H.; Mou, Y.; Pegoraro, A.F.; Fredberg, J.; Chen, Z. Modeling Physiological Events in 2D vs. 3D Cell Culture. Physiology 2017, 32, 266–277. [Google Scholar] [CrossRef] [PubMed]
- Holmgren, G.; Ghosheh, N.; Zeng, X.; Bogestål, Y.; Sartipy, P.; Synnergren, J. Identification of stable reference genes in differentiating human pluripotent stem cells. Physiol. Genom. 2015, 47, 232–239. [Google Scholar] [CrossRef] [PubMed]
- Kambhampati, S.; Li, J.; Evans, B.S.; Allen, D.K. Accurate and efficient amino acid analysis for protein quantification using hydrophilic interaction chromatography coupled tandem mass spectrometry. Plant Methods 2019, 15, 46. [Google Scholar] [CrossRef] [PubMed]
- Hsu, H.W.; Vavak, D.L.; Satterlee, L.; Miller, G.A. A multienzyme technique for estimating protein digestibility. J. Food Sci. 1977, 42, 1269–1273. [Google Scholar] [CrossRef]
- Tinus, T.; Damour, M.; Van Riel, V.; Sopade, P.A. Particle size-starch–protein digestibility relationships in cowpea (Vigna unguiculata). J. Food Eng. 2012, 113, 254–264. [Google Scholar] [CrossRef]
- FAQ-JASP-Free and User-Friendly Statistical Software. Available online: jasp-stats.org (accessed on 26 September 2022).
- Liu, S.Q.; Tay, R.; Khan, M.; Ee, P.L.R.; Hedrick, J.L.; Yang, Y.Y. Synthetic hydrogels for controlled stem cell differentiation. Soft Matter 2010, 6, 67–81. [Google Scholar] [CrossRef]
- Nath, S.; Devi, G.R. Three-dimensional culture systems in cancer research: Focus on tumor spheroid model. Pharmacol. Ther. 2016, 163, 94–108. [Google Scholar] [CrossRef]
- Nicodemus, G.D.; Bryant, S.J. Cell encapsulation in biodegradable hydrogels for tissue engineering applications. Tissue Eng. Part B Rev. 2008, 14, 149–165. [Google Scholar] [CrossRef]
- Norton, L.E.; Layman, D.K.; Bunpo, P.; Anthony, T.G.; Brana, D.V.; Garlick, P.J. The leucine content of a complete meal directs peak activation but not duration of skeletal muscle protein synthesis and mammalian target of rapamycin signaling in rats. J. Nutr. 2009, 139, 1103–1109. [Google Scholar] [CrossRef]
- Saxton, R.A.; Knockenhauer, K.E.; Wolfson, R.L.; Chantranupong, L.; Pacold, M.E.; Wang, T.; Schwartz, T.U.; Sabatini, D.M. Structural basis for leucine sensing by the Sestrin2-mTORC1 pathway. Science 2016, 351, 53–58. [Google Scholar] [CrossRef]
- WHO. Protein and amino acid requirements in human nutrition. In World Health Organization Technical Report Series; WHO: Geneva, Switzerland, 2007; pp. 1–265. [Google Scholar]
- Shaghaghian, S.; McClements, D.J.; Khalesi, M.; Garcia-Vaquero, M.; Mirzapour-Kouhdasht, A. Digestibility and bioavailability of plant-based proteins intended for use in meat analogues: A review. Trends Food Sci. Technol. 2022, 129, 646–656. [Google Scholar] [CrossRef]
- Butts, C.A.; Monro, J.A.; Moughan, P.J. In vitro determination of dietary protein and amino acid digestibility for humans. Br. J. Nutr. 2012, 108 (Suppl. 2), S282–S287. [Google Scholar] [CrossRef] [PubMed]
- Rieder, A.; Afseth, N.K.; Böcker, U.; Knutsen, S.H.; Kirkhus, B.; Mæhre, H.K.; Ballance, S.; Wubshet, S.G. Improved estimation of in vitro protein digestibility of different foods using size exclusion chromatography. Food Chem. 2021, 358, 129830. [Google Scholar] [CrossRef] [PubMed]
- Mendes, F.Q.; Oliveira, M.G.D.A.; Costa, N.M.B.; Pires, C.V.; Passos, F.R. Capability of in vitro digestibility methods to predict in vivo digestibility of vegetal and animal proteins. Arch. Latinoam. Nutr. 2016, 66, 5–16. [Google Scholar]
- Ketnawa, S.; Ogawa, Y. In vitro protein digestibility and biochemical characteristics of soaked, boiled and fermented soybeans. Sci. Rep. 2021, 11, 14257. [Google Scholar] [CrossRef]
- Sá, A.G.A.; Moreno, Y.M.F.; Carciofi, B.A.M. Food processing for the improvement of plant proteins digestibility. Crit. Rev. Food Sci. Nutr. 2020, 60, 3367–3386. [Google Scholar] [CrossRef]
- Lee, S.; Choi, Y.S.; Jo, K.; Yong, H.I.; Jeong, H.G.; Jung, S. Improvement of meat protein digestibility in infants and the elderly. Food Chem. 2021, 356, 129707. [Google Scholar] [CrossRef]

| Gene | Sequence |
|---|---|
| hTERT | Forward (5′ to 3′): GGAGCAAGTTGCAAAGCATTG |
| Reverse (3′ to 5′): TCCCACGACGTAGTCCATGTT | |
| NANOG | Forward (5′ to 3′): TGTGATTTGTGGGCCTGA |
| Reverse (3′ to 5′): GTGGGTTGTTTGCCTTTG | |
| OCT4 | Forward (5′ to 3′): AAAGAGAAAGCGAACCAG |
| Reverse (3′ to 5′): CCACATCCTTCTCGAGCC | |
| REX1 | Forward (5′ to 3′): GTTTCGTGTGTCCCTTTC |
| Reverse (3′ to 5′): CTTTCCCTCTTGTTCATTC | |
| UTF1 | Forward (5′ to 3′): CTCCCAGCGAACCAG |
| Reverse (3′ to 5′): GCGTCCGCAGACTTC | |
| hEID2 | Forward (5′ to 3′): GAAGCCTGCAGAGCAAGG |
| Reverse (3′ to 5′): ATATCGAGGTCCACCCTGTG | |
| ZENF324B | Forward (5′ to 3′): GAGAATGGCCACGAGCTTT |
| Reverse (3′ to 5′): TTTACACTGTGGCAGGCATC | |
| hCAPN10 | Forward (5′ to 3′): GGAGGTGACCACAGATGACC |
| Reverse (3′ to 5′): GTAAGGGGAGCCAGAACACA |
| 8 | hiPSC Pluripotency-Related Gene Expression | ||||
|---|---|---|---|---|---|
| REX1 | OCT4 | NANOG | UTF1 | hTERT | |
| PG-3D | 1 | 1 | 1 | 1 | 1 |
| PG-3DSUSP | 0.88 | 1.99 | 1.46 | 1.09 | 1.55 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Xu, S.; Qi, G.; Durrett, T.P.; Li, Y.; Liu, X.; Bai, J.; Chen, M.-S.; Sun, X.; Wang, W. High Nutritional Quality of Human-Induced Pluripotent Stem Cell-Generated Proteins through an Advanced Scalable Peptide Hydrogel 3D Suspension System. Foods 2023, 12, 2713. https://doi.org/10.3390/foods12142713
Xu S, Qi G, Durrett TP, Li Y, Liu X, Bai J, Chen M-S, Sun X, Wang W. High Nutritional Quality of Human-Induced Pluripotent Stem Cell-Generated Proteins through an Advanced Scalable Peptide Hydrogel 3D Suspension System. Foods. 2023; 12(14):2713. https://doi.org/10.3390/foods12142713
Chicago/Turabian StyleXu, Shan, Guangyan Qi, Timothy P. Durrett, Yonghui Li, Xuming Liu, Jianfa Bai, Ming-Shun Chen, Xiuzhi (Susan) Sun, and Weiqun Wang. 2023. "High Nutritional Quality of Human-Induced Pluripotent Stem Cell-Generated Proteins through an Advanced Scalable Peptide Hydrogel 3D Suspension System" Foods 12, no. 14: 2713. https://doi.org/10.3390/foods12142713
APA StyleXu, S., Qi, G., Durrett, T. P., Li, Y., Liu, X., Bai, J., Chen, M.-S., Sun, X., & Wang, W. (2023). High Nutritional Quality of Human-Induced Pluripotent Stem Cell-Generated Proteins through an Advanced Scalable Peptide Hydrogel 3D Suspension System. Foods, 12(14), 2713. https://doi.org/10.3390/foods12142713