Cronobacter spp. Isolated from Quick-Frozen Foods in China: Incidence, Genetic Characteristics, and Antibiotic Resistance
Abstract
:1. Introduction
2. Materials and Methods
2.1. Sampling
2.2. Quantitative Detection and Classification of Cronobacter spp.
2.3. O-Antigen Serotyping and Multilocus Sequence Typing
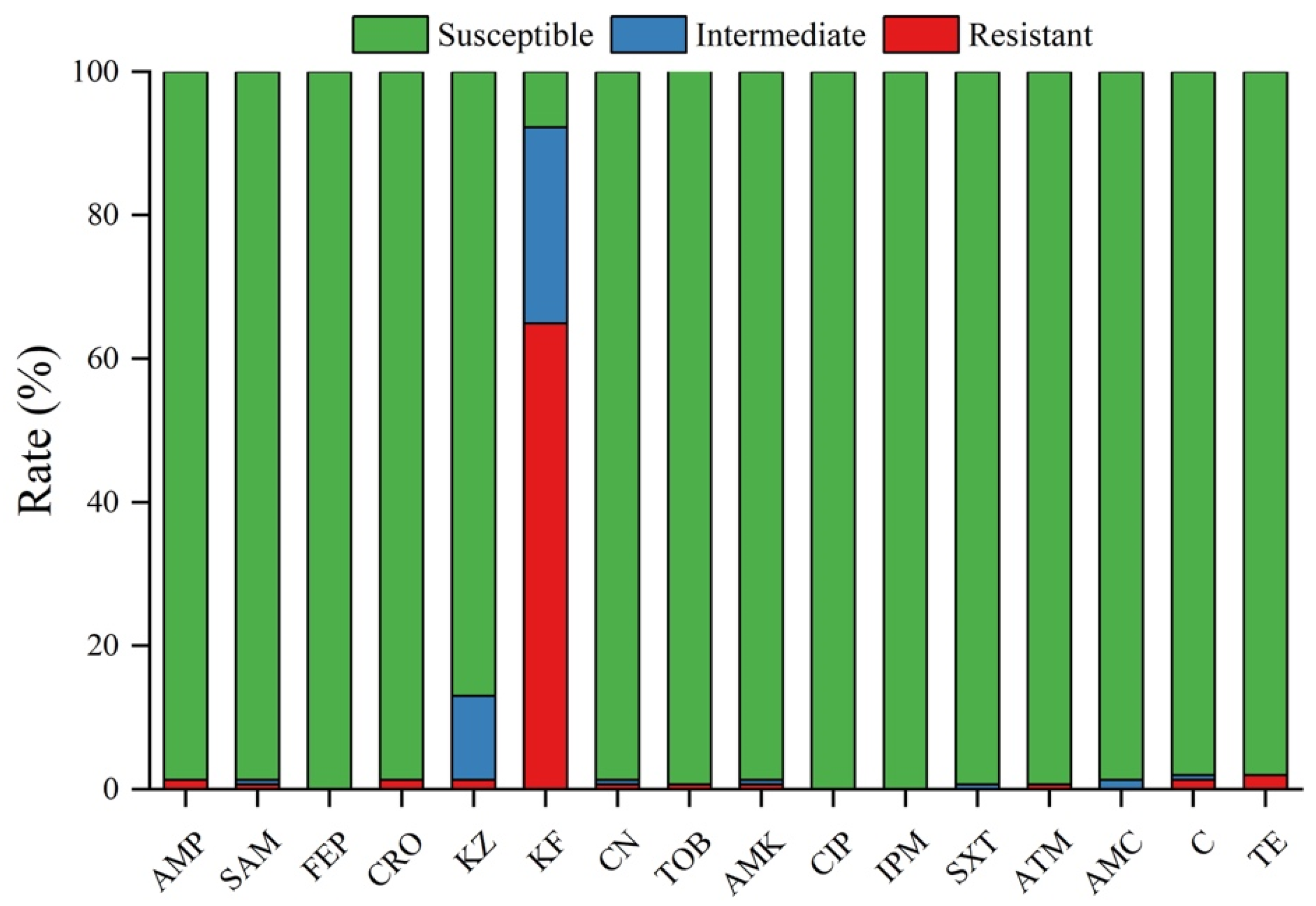
2.4. Antimicrobial Susceptibility Analysis
2.5. Statistical Analysis
3. Results
3.1. Contamination Level and Isolation of Cronobacter spp. in Quick-Frozen Foods
3.2. Antimicrobial Susceptibility Testing
3.3. Molecular Serotype and MLST Patterns of Cronobacter Isolates
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Farmer, J.J., III. My 40-year history with Cronobacter/Enterobacter sakazakii—Lessons learned, myths debunked, and recommendations. Front. Pediatr. 2015, 3, 84. [Google Scholar] [CrossRef] [Green Version]
- Forsythe, S.J.; Dickins, B.; Jolley, K.A. Cronobacter, the emergent bacterial pathogen Enterobacter sakazakii comes of age; MLST and whole genome sequence analysis. BMC Genomics 2014, 15, 1121. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Patrick, M.E.; Mahon, B.E.; Greene, S.A.; Rounds, J.; Cronquist, A.; Wymore, K.; Boothe, E.; Lathrop, S.; Palmer, A.; Bowen, A. Incidence of Cronobacter spp. infections, United States, 2003–2009. Emerg. Infect. Dis. 2014, 20, 1520–1523. [Google Scholar] [CrossRef] [PubMed]
- Lehner, A.; Tall, B.D.; Fanning, S.; Srikumar, S. Cronobacter spp.—opportunistic foodborne pathogens: An update on evolution, osmotic adaptation and pathogenesis. Curr. Clin. Microbiol. Rep. 2018, 5, 97–105. [Google Scholar] [CrossRef]
- Phair, K.; Pereira, S.G.; Kealey, C.; Fanning, S.; Brady, D.B. Insights into the mechanisms of Cronobacter sakazakii virulence. Microb. Pathog. 2022, 169, 105643. [Google Scholar] [CrossRef]
- Parra-Flores, J.; Aguirre, J.; Juneja, V.; Jackson, E.E.; Cruz-Córdova, A.; Silva-Sanchez, J.; Forsythe, S. Virulence and antibiotic resistance profiles of Cronobacter sakazakii and Enterobacter spp. involved in the diarrheic hemorrhagic outbreak in Mexico. Front. Microbiol. 2018, 9, 2206. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Townsend, S.M.; Hurrell, E.; Caubilla-Barron, J.; Loc-Carrillo, C.; Forsythe, S.J. Characterization of an extended-spectrum beta-lactamase Enterobacter hormaechei nosocomial outbreak, and other Enterobacter hormaechei misidentified as Cronobacter (Enterobacter) sakazakii. Microbiology 2008, 154, 3659–3667. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Forsythe, S.J. Updates on the Cronobacter genus. Annu. Rev. Food Sci. Technol. 2018, 9, 23–44. [Google Scholar] [CrossRef]
- Brandão, M.L.L.; Umeda, N.S.; Jackson, E.; Forsythe, S.J.; de Filippis, I. Isolation, molecular and phenotypic characterization, and antibiotic susceptibility of Cronobacter spp. from Brazilian retail foods. Food Microbiol. 2017, 63, 129–138. [Google Scholar] [CrossRef]
- Iversen, C.; Forsythe, S. Risk profile of Enterobacter sakazakii, an emergent pathogen associated with infant milk formula. Trends Food Sci. Technol. 2003, 14, 443–454. [Google Scholar] [CrossRef]
- Schmid, M.; Iversen, C.; Gontia, I.; Stephan, R.; Hofmann, A.; Hartmann, A.; Jha, B.; Eberl, L.; Riedel, K.; Lehner, A. Evidence for a plant-associated natural habitat for Cronobacter spp. Res. Microbiol. 2009, 160, 608–614. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ling, N.; Li, C.S.; Zhang, J.M.; Wu, Q.P.; Zeng, H.Y.; He, W.; Ye, Y.; Wang, J.; Ding, Y.; Chen, M.; et al. Prevalence and molecular and antimicrobial characteristics of Cronobacter spp. isolated from raw vegetables in China. Front. Microbiol. 2018, 9, 1149. [Google Scholar] [CrossRef] [PubMed]
- Garbowska, M.; Berthold-Pluta, A.; Stasiak-Różańska, L. Microbiological quality of selected spices and herbs including presence of Cronobacter spp. Food Microbiol. 2015, 49, 1–5. [Google Scholar] [CrossRef] [PubMed]
- Csorba, C.; Pajić, M.; Blagojević, B.; Forsythe, S.; Radinović, M.; Velebit, B. Prevalence, characterization, and antibiotic susceptibility of Cronobacter spp. in a milk powder processing environment: The first reported case in Serbia. Food Sci. Nutr. 2022, 10, 554–563. [Google Scholar] [CrossRef]
- Chauhan, R.; Tall, B.D.; Gopinath, G.; Azmi, W.; Goel, G. Environmental risk factors associated with the survival, persistence, and thermal tolerance of Cronobacter sakazakii during the manufacture of powdered infant formula. Crit. Rev. Food Sci. Nutr. 2022, 1, 1–16. [Google Scholar] [CrossRef]
- Wei, J.P.; Zhang, Y.X.; Wang, X.; Chen, H.; Yuan, Y.H.; Yue, T.L. Distribution of cold-resistant bacteria in quick-frozen dumpling and its inhibition by different antibacterial agents. J. Food Process. Preserv. 2022, 49, e14710. [Google Scholar] [CrossRef]
- Møretrø, T.; Langsrud, S. Residential bacteria on surfaces in the food industry and their implications for food safety and quality. Compr. Rev. Food Sci. Food Saf. 2017, 16, 1022–1041. [Google Scholar] [CrossRef]
- Stellato, G.; Utter, D.R.; Voorhis, A.; De Angelis, M.D.; Eren, A.M.; Ercolini, D. A few Pseudomonas oligotypes dominate in the meat and dairy processing environment. Front. Microbiol. 2017, 8, 264. [Google Scholar] [CrossRef] [Green Version]
- Wu, S.; Wu, Q.P.; Zhang, J.M.; Chen, M.T.; Yan, Z.A.; Hu, H. Listeria monocytogenes prevalence and characteristics in retail raw foods in China. PLoS ONE 2015, 10, e0136682. [Google Scholar] [CrossRef]
- Wu, S.; Huang, J.; Wu, Q.P.; Zhang, J.M.; Zhang, F.; Yang, X.; Wu, H.; Zeng, H.; Chen, M.; Ding, Y.; et al. Staphylococcus aureus isolated from retail meat and meat products in China: Incidence, antibiotic resistance and genetic diversity. Front. Microbiol. 2018, 9, 2767. [Google Scholar] [CrossRef]
- Guo, H.; Yu, P.F.; Yu, S.B.; Wang, J.; Zhang, J.H.; Zhang, Y.; Liao, X.; Wu, S.; Ye, Q.; Yang, X.; et al. Incidence, toxin gene profiling, antimicrobial susceptibility, and genetic diversity of Bacillus cereus isolated from quick-frozen food in China. LWT Food Sci. Technol. 2021, 140, 110824. [Google Scholar] [CrossRef]
- Wang, J.; Liu, M.; Wang, H.X.; Wu, Q.P.; Ding, Y.; Xu, T.X.; Ma, G.X.; Zhong, Y.; Zhang, J.; Chen, M.; et al. Occurrence, molecular characterization, and antimicrobial susceptibility of Yersinia enterocolitica isolated from retail food samples in China. LWT Food Sci. Technol. 2021, 150, 111876. [Google Scholar] [CrossRef]
- Wang, L.; Zhu, W.X.; Lu, G.G.; Wu, P.; Wei, Y.; Su, Y.; Jia, T.; Li, L.; Guo, X.; Huang, M.; et al. In silico species identification and serotyping for Cronobacter isolates by use of whole-genome sequencing data. Int. J. Food Microbiol. 2021, 358, 109405. [Google Scholar] [CrossRef]
- Li, H.; Fu, S.; Song, D.; Qin, X.; Zhang, W.; Man, C.; Yang, X.; Jiang, Y. Identification, typing and drug resistance of Cronobacter spp. in powdered infant formula and processing environment. Foods 2023, 12, 1084. [Google Scholar] [CrossRef] [PubMed]
- Zeng, H.Y.; Li, C.S.; Ling, N.; Zhang, J.M.; Chen, M.; Lei, T.; Wu, S.; Yang, X.; Luo, D.; Ding, Y.; et al. Prevalence, genetic analysis and CRISPR typing of Cronobacter spp. isolated from meat and meat products in China. Int. J. Food Microbiol. 2020, 321, 108549. [Google Scholar] [CrossRef] [PubMed]
- Li, C.S.; Zeng, H.Y.; Zhang, J.M.; Luo, D.D.; Chen, M.T.; Lei, T.; Yang, X.; Wu, H.; Cai, S.; Ye, Y.; et al. Cronobacter spp. isolated from aquatic products in China: Incidence, antibiotic resistance, molecular characteristic and CRISPR diversity. Int. J. Food Microbiol. 2020, 335, 108857. [Google Scholar] [CrossRef]
- Jaradat, Z.W.; Al-Mousa, W.A.; Elbetieha, A.M.; Ababneh, Q.O.; Al-Nabulsi, A.A.; Jang, H.; Gangiredla, J.; Patel, I.R.; Gopinath, G.R.; Tall, B.D. Virulence, antimicrobial susceptibility and phylogenetic analysis of Cronobacter sakazakii isolates of food origins from Jordan. J. Appl. Microbiol. 2022, 133, 2528–2546. [Google Scholar] [CrossRef] [PubMed]
- Lou, X.; Liu, T.; Zhang, W.; Yu, H.; Wang, H.; Song, S.; Chen, Q.; Fang, Z. The occurrence and distribution characteristics of Cronobacter in diverse cereal kernels, flour, and flour-based products. Food Microbiol. 2019, 84, 103269. [Google Scholar] [CrossRef]
- Das, S.K.; Kumar, S.H.; Nayak, B.B.; Lekshmi, M. Isolation and identification of Cronobacter spp. from fish and shellfish sold in retail markets. Curr. Microbiol. 2021, 78, 1973–1980. [Google Scholar] [CrossRef]
- Miranda, N.; Banerjee, P.; Simpson, S.; Kerdahi, K.; Sulaiman, I.M. Molecular surveillance of Cronobacter spp. isolated from a wide variety of foods from 44 different countries by sequence typing of 16S rRNA, rpoB and O-Antigen Genes. Foods 2017, 6, 36. [Google Scholar] [CrossRef] [Green Version]
- Fei, P.; Jiang, Y.C.; Gong, S.Y.; Li, R.; Jiang, Y.; Yuan, X.J.; Wang, Z.; Kang, H.; Ali, M.A. Occurrence, genotyping, and antibiotic susceptibility of Cronobacter spp. in drinking water and food samples from Northeast China. J. Food Prot. 2018, 81, 456–460. [Google Scholar] [CrossRef]
- Depardieu, F.; Podglajen, I.; Leclercq, R.; Collatz, E.; Courvalin, P. Modes and modulations of antibiotic resistance gene expression. Clin. Microbiol. Rev. 2007, 20, 79–114. [Google Scholar] [CrossRef] [Green Version]
- Arslan, S.; Ertürk, H.G. Occurrence, virulence and antimicrobial susceptibility profiles of Cronobacter spp. from ready-to-eat foods. Curr. Microbiol. 2021, 78, 3403–3416. [Google Scholar] [CrossRef] [PubMed]
- Ling, N.; Jiang, X.; Forsythe, S.; Zhang, D.; Shen, Y.; Ding, Y.; Wang, J.; Zhang, J.; Wu, Q.; Ye, Y. Food safety risks and contributing factors of Cronobacter spp. Engineering 2022, 12, 128–138. [Google Scholar] [CrossRef]
- Uddin, T.M.; Chakraborty, A.J.; Khusro, A.; Zidan, R.M.; Mitra, S.; Emran, T.B.; Dhama, K.; Ripon, K.H.; Gajdács, K.; Sahibzada, M.U.K.; et al. Antibiotic resistance in microbes: History, mechanisms, therapeutic strategies and future prospects. J. Infect. Public Health 2021, 14, 1750–1766. [Google Scholar] [CrossRef] [PubMed]
- Verma, T.; Aggarwal, A.; Singh, S.; Sharma, S.; Sarma, S.J. Current challenges and advancements towards discovery and resistance of antibiotics. J. Mol. Struct. 2022, 122, 180–186. [Google Scholar] [CrossRef]
- Blažková, M.; Javůrková, B.; Vlach, J.; Göselová, S.; Karamonová, L.; Ogrodzki, P.; Forsythe, S.; Fukal, L. Diversity of O-antigens within the genus Cronobacter: From disorder to order. Appl. Environ. Microbiol. 2015, 81, 5574–5582. [Google Scholar] [CrossRef] [Green Version]
- Li, Y.H.; Yu, H.; Jiang, H.; Jiao, Y.; Zhang, Y.D.; Shao, J.H. Genetic diversity, antimicrobial susceptibility, and biofilm formation of Cronobacter spp. recovered from spices and cereals. Front. Microbiol. 2017, 8, 2567. [Google Scholar] [CrossRef] [Green Version]
- Fei, P.; Xing, M.; Feng, Y.G.; Liu, S.; Chang, Y.J.; Wang, Y.; Yu, Y.; Shi, E.; Zhang, Y.; Bian, X.; et al. Occurrence, molecular characterization, and antibiotic resistance of Cronobacter sakazakii in goat milk-based infant formula from Shanxi Province, China. Foodborne Pathog. Dis. 2021, 19, 304–310. [Google Scholar] [CrossRef]
- Alsonosi, A.; Hariri, S.; Kajsík, M.; Oriešková, M.; Hanulík, V.; Röderová, M.; Petrželová, J.; Kollárová, H.; Drahovská, H.; Forsythe, S.; et al. The speciation and genotyping of Cronobacter isolates from hospitalised patients. Eur. J. Clin. Microbiol. Infect. Dis. 2015, 34, 1979–1988. [Google Scholar] [CrossRef] [Green Version]
- Sonbol, H.; Joseph, S.; McAuley, C.M.; Craven, H.M.; Forsythe, S.J. Multilocus sequence typing of Cronobacter spp. from powdered infant formula and milk power production factories. Int. Dairy J. 2013, 30, 1–7. [Google Scholar] [CrossRef]
- Xu, X.K.; Li, C.S.; Wu, Q.P.; Zhang, J.M.; Huang, J.H.; Yang, G.Z. Prevalence, molecular characterization, and antibiotic susceptibility of Cronobacter spp. in Chinese ready-to-eat foods. Int. J. Food Microbiol. 2015, 204, 17–23. [Google Scholar] [CrossRef]
- Jackson, E.E.; Parra, F.J.; Fernandez, E.E.; Forsythe, S.J. Reevaluation of a suspected Cronobacter sakazakii outbreak in Mexico. J. Food Prot. 2015, 78, 1191–1196. [Google Scholar] [CrossRef]
- CLSI M100-S26; Performance Standards for Antimicrobial Susceptibility Testing. Twenty-Sixth Informational Supplement; Clinical and Laboratory Standards Institute [CLSI]: Wayne, PA, USA, 2018.
- Jiang, H.; Xiang, Y.; He, X.J.; Li, C.C.; Lin, F.; Shao, J.; Li, Y. Identification and antibiotic resistance of Cronobacter spp. isolated from dried edible mushrooms. J. Food Sci. 2022, 87, 3588–3598. [Google Scholar] [CrossRef] [PubMed]
- Carvalho, G.G.; Calarga, A.P.; Teodoro, J.R.; Queiroz, M.M.; Astudillo-Trujillo, C.A.; Levy, C.E.; Brocchi, M.; Kabuki, D.Y. Isolation, comparison of identification methods and antibiotic resistance of Cronobacter spp. in infant foods. Food Res. Int. 2020, 137, 109643. [Google Scholar] [CrossRef] [PubMed]
- Chon, J.W.; Song, K.Y.; Kim, S.Y.; Hyeon, J.Y.; Seo, K.H. Isolation and characterization of Cronobacter desiccated foods in Korea. J. Food Sci. 2012, 77, 354–358. [Google Scholar] [CrossRef] [PubMed]
- Moravkova, M.; Verbikova, V.; Huvarova, V.; Babak, V.; Cahlikova, H.; Karpiskova, R.; Kralik, P. Occurrence of Cronobacter spp. in ready-to-eat vegetable products, frozen vegetables, and sprouts examined using cultivation and real-time PCR methods. J. Food Sci. 2018, 83, 3054–3058. [Google Scholar] [CrossRef]
- Lou, X.Q.; Yu, H.; Wang, X.C.; Qi, J.J.; Zhang, W.; Wang, H.Q.; Si, G.J.; Song, S.; Huang, C.; Liu, T.; et al. Potential reservoirs and routes of Cronobacter transmission during cereal growing, processing and consumption. Food Microbiol. 2019, 79, 90–95. [Google Scholar] [CrossRef]
- Pluta, A.B.; Garbowska, M.; Stefańska, I.; Pluta, A. Microbiology quality of selected ready-to-eat leaf vegetables sprouts and non-pasteurized fresh fruit-vegetable juices including the presence of Cronobacter spp. Food Microbiol. 2017, 65, 221–230. [Google Scholar] [CrossRef]
- Santo, D.; Graça, A.; Nunes, C.; Quintas, C. Survival and growth of Cronobacter sakazakii on fresh-cut fruit and the effect of UV-C illumination and electrolyzed water in the reduction of its population. Int. J. Food Microbiol. 2016, 231, 10–15. [Google Scholar] [CrossRef]
- Ueda, S. The effects of temperature on the growth and heat resistance of Cronobacter spp. Biocontrol. Sci. 2017, 22, 125–129. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, C.; Zeng, H.; Zhang, J.; He, W.; Ling, N.; Chen, M.T.; Wu, S.; Lei, T.; Wu, H.; Ye, Y.; et al. Prevalence, antibiotic susceptibility, and molecular characterization of Cronobacter spp. isolated from edible mushrooms in China. Front. Microbiol. 2019, 10, 283. [Google Scholar] [CrossRef] [PubMed]
- Yang, X.J.; Huang, J.H.; Zhang, Y.; Liu, S.; Chen, L.; Xiao, C.; Zeng, H.; Wei, X.; Gu, Q.; Li, Y.; et al. Prevalence, abundance, serovars and antimicrobial resistance of Salmonella isolated from retail raw poultry meat in China. Sci. Total Environ. 2020, 713, 136385. [Google Scholar] [CrossRef]
- Ye, Q.H.; Wu, Q.P.; Zhang, S.H.; Zhang, J.M.; Yang, G.; Wang, J.; Xue, L.; Chen, M. Characterization of extended-spectrum β-lactamase-producing Enterobacteriaceae from retail food in China. Front. Microbiol. 2018, 9, 1709. [Google Scholar] [CrossRef]
- Akineden, Ö.; Heinrich, V.; Gross, M.; Usleber, E. Reassessment of Cronobacter spp. originally isolated as Enterobacter sakazakii from infant food. Food Microbiol. 2017, 65, 44–50. [Google Scholar] [CrossRef] [PubMed]
- Scharinger, E.J.; Dietrich, R.; Wittwer, T.; Märtlbauer, E.; Schauer, K. Multiplexed lateral flow test for detection and differentiation of Cronobacter sakazakii serotypes O1 and O2. Front. Microbiol. 2017, 8, 1826. [Google Scholar] [CrossRef]
- Hariri, S.; Joseph, S.; Forsythe, S.J. Cronobacter sakazakii ST4 strains and neonatal meningitis, United States. Emerg. Infect. Dis. 2013, 19, 175–177. [Google Scholar] [CrossRef] [PubMed]


| Sample | No. of Samples | No. (%) of Positive Samples (p < 0.01) | No. of Positive Samples by Quantitative Methods by MPN/g Range | Positive Sample Contamination Level (MPN/g) | |||
|---|---|---|---|---|---|---|---|
| MPN < 10 | 10 ≤ MPN < 110 | 110 ≤ MPN | |||||
| Frozen flour products | 212 | 94 | 44.34% | 87 | 3 | 4 | 6.63 |
| Frozen poultry | 239 | 9 | 3.77% | 9 | 0 | 0 | 0.42 |
| Frozen meat | 125 | 5 | 4.00% | 5 | 0 | 0 | 0.22 |
| Total | 576 | 108 | 18.75% | 101 | 3 | 4 | 5.82 |
| Region | No. of Samples | No. of Positive Samples | Prevalence Rate (%) (p > 0.05) |
|---|---|---|---|
| South China | 207 | 35 | 16.90% |
| East China | 110 | 29 | 26.36% |
| Central China | 41 | 7 | 17.07% |
| North China | 55 | 9 | 16.36% |
| Northeast China | 42 | 7 | 16.67% |
| Northwest China | 66 | 14 | 21.21% |
| Southwest China | 55 | 7 | 12.73% |
| Isolates (Total) | Serotypes | Total | Type of Samples | MLST Allelic Type | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Frozen Flour Products | Frozen Poultry | Frozen Meat | atpD | fusA | glnS | gltB | gyrB | infB | pps | MLST Pattern (No. of Isolates) | |||
| C. sakazakii 109 | O1 | 44 | 43 | 0 | 1 | 15 | 15 | 80 | 126 | 124 | 56 | 159 | ST256 (4) |
| 15 | 67 | 49 | 9 | 14 | 19 | 15 | ST148 (8) | ||||||
| 10 | 17 | 30 | 59 | 57 | 66 | 83 | ST125 (2) | ||||||
| 3 | 38 | 120 | 154 | 150 | 15 | 140 | ST654 (1) | ||||||
| 11 | 8 | 7 | 45 | 8 | 15 | 10 | ST226 (5) | ||||||
| 1 | 1 | 1 | 1 | 1 | 1 | 1 | ST1 (6) | ||||||
| 15 | 8 | 39 | 36 | 38 | 38 | 47 | ST58 (5) | ||||||
| 3 | 8 | 52 | 13 | 21 | 65 | 73 | ST495 (1) | ||||||
| 3 | 11 | 13 | 18 | 11 | 88 | 104 | ST156 (1) | ||||||
| 71 | 10 | 68 | 18 | 94 | 92 | 121 | ST188 (1) | ||||||
| 44 | 69 | 80 | 147 | 96 | 93 | 123 | ST327 (1) | ||||||
| 15 | 186 | 251 | 298 | 209 | 246 | 362 | ST737 (1) | ||||||
| 3 | 8 | 13 | 15 | 22 | 20 | 21 | ST50 (2) | ||||||
| 3 | 8 | 3 | 3 | 18 | 46 | 127 | ST198 (1) | ||||||
| 3 | 10 | 9 | 275 | 247 | 20 | 51 | ST641 (1) | ||||||
| 69 | 8 | 13 | 95 | 86 | 105 | 47 | ST219 (1) | ||||||
| 11 | 8 | 7 | 5 | 8 | 15 | 10 | ST8 (1) | ||||||
| 16 | 17 | 49 | 32 | 127 | 92 | 245 | ST710 (1) | ||||||
| 16 | 17 | 49 | 205 | 127 | 92 | 462 * | ST930 * (1) | ||||||
| O2 | 23 | 21 | 2 | 0 | 3 | 17 | 49 | 68 | 58 | 63 | 65 | ST117 (2) | |
| 16 | 18 | 120 | 119 | 88 | 73 | 18 | ST308 (4) | ||||||
| 16 | 8 | 13 | 40 | 15 | 15 | 10 | ST64 (3) | ||||||
| 48 | 17 | 10 | 69 | 71 | 5 | 81 | ST42 (1) | ||||||
| 16 | 1 | 19 | 19 | 26 | 5 | 26 | ST22 (1) | ||||||
| 3 | 12 | 16 | 5 | 16 | 20 | 14 | ST17 (1) | ||||||
| 20 | 18 | 16 | 10 | 3 | 20 | 27 | ST23 (2) | ||||||
| 3 | 3 | 3 | 5 | 3 | 3 | 3 | ST3 (3) | ||||||
| 5 | 1 | 3 | 3 | 5 | 5 | 4 | ST4 (1) | ||||||
| 16 | 1 | 13 | 39 | 21 | 5 | 21 | ST264 (1) | ||||||
| 3 | 8 | 37 | 22 | 29 | 36 | 32 | ST31 (1) | ||||||
| 15 | 14 | 15 | 13 | 22 | 5 | 16 | ST13 (2) | ||||||
| 10 | 37 | 9 | 3 | 302 | 56 | 126 | ST836 (1) | ||||||
| 16 | 37 | 9 | 1 | 263 | 92 | 140 | ST938 * (1) | ||||||
| 20 | 17 | 306 * | 373 * | 16 | 274 * | 456 * | ST916 * (1) | ||||||
| O3 | 13 | 11 | 2 | 0 | 11 | 8 | 7 | 5 | 8 | 15 | 10 | ST8 (3) | |
| 5 | 1 | 3 | 3 | 5 | 5 | 4 | ST4 (3) | ||||||
| 15 | 8 | 13 | 94 | 99 | 118 | 126 | ST249 (1) | ||||||
| 3 | 68 | 49 | 88 | 81 | 56 | 96 | ST143 (1) | ||||||
| 15 | 8 | 13 | 94 | 99 | 98 | 126 | ST199 (1) | ||||||
| 15 | 1 | 3 | 294 | 15 | 48 | 280 | ST706 (1) | ||||||
| 15 | 8 | 13 | 94 | 99 | 304 * | 458 * | ST920 * (1) | ||||||
| 34 | 36 | 10 | 44 | 43 | 48 | 51 | ST46 (1) | ||||||
| 16 | 36 | 30 | 58 | 325 * | 303 * | 18 | ST919 * (1) | ||||||
| O4 | 11 | 11 | 0 | 0 | 3 | 15 | 28 | 22 | 5 | 38 | 19 | ST40 (4) | |
| 15 | 14 | 15 | 13 | 22 | 5 | 16 | ST13 (2) | ||||||
| 18 | 17 | 10 | 12 | 18 | 24 | 18 | ST12 (4) | ||||||
| 15 | 8 | 13 | 94 | 99 | 118 | 126 | ST249 (1) | ||||||
| O7 | 16 | 14 | 2 | 0 | 55 | 14 | 59 | 70 | 70 | 70 | 80 | ST73 (2) | |
| 19 | 16 | 19 | 41 | 19 | 15 | 23 | ST83 (4) | ||||||
| 3 | 36 | 52 | 374 | 18 | 90 | 466 | ST921 (1) | ||||||
| 16 | 36 | 3 | 249 | 58 | 305 * | 4 | ST922 * (1) | ||||||
| 3 | 1 | 9 | 378 * | 329 * | 56 | 103 | ST937 * (1) | ||||||
| 3 | 36 | 74 | 97 | 86 | 20 | 107 | ST184 (1) | ||||||
| 3 | 36 | 74 | 97 | 86 | 306 * | 107 | ST924 * (1) | ||||||
| 16 | 36 | 3 | 249 | 58 | 36 | 4 | ST587 (1) | ||||||
| 55 | 1 | 68 | 32 | 191 | 36 | 252 | ST470 (1) | ||||||
| 15 | 1 | 3 | 32 | 5 | 36 | 190 | ST93 (3) | ||||||
| uncertain | 2 | 0 | 0 | 2 | 3 | 11 | 13 | 18 | 11 | 17 | 13 | ST21 (2) | |
| C.malonaticus 22 | O1 | 5 | 4 | 1 | 0 | 12 | 7 | 8 | 8 | 10 | 16 | 43 | ST60 (2) |
| 57 | 7 | 64 | 8 | 17 | 16 | 128 | ST211 (1) | ||||||
| 61 | 7 | 12 | 7 | 189 | 14 | 247 | ST461 (1) | ||||||
| 10 | 7 | 17 | 8 | 214 | 314 * | 290 | ST936 * (1) | ||||||
| O2 | 11 | 10 | 0 | 1 | 10 | 7 | 6 | 7 | 9 | 14 | 9 | ST7 (6) | |
| 10 | 7 | 6 | 99 | 9 | 14 | 9 | ST201 (2) | ||||||
| 10 | 162 | 67 | 7 | 77 | 204 | 279 | ST567 (1) | ||||||
| 10 | 13 | 64 | 75 | 239 | 14 | 8 | ST917 * (2) | ||||||
| O3 | 5 | 4 | 1 | 0 | 61 | 7 | 67 | 7 | 77 | 81 | 95 | ST408 (1) | |
| 17 | 7 | 17 | 11 | 17 | 22 | 12 | ST11 (3) | ||||||
| 10 | 13 | 67 | 7 | 131 | 124 | 174 | ST300 (1) | ||||||
| uncertain | 1 | 0 | 0 | 1 | 13 | 7 | 247 | 293 | 77 | 127 | 355 | ST701 (1) | |
| C. dublinensis 19 | O1 | 12 | 12 | 0 | 0 | 112 | 41 | 198 | 251 | 211 | 154 | 109 | ST574 (1) |
| 94 | 100 | 113 | 133 | 211 | 130 | 282 | ST561 (1) | ||||||
| 39 | 43 | 45 | 49 | 49 | 52 | 57 | ST77 (3) | ||||||
| 70 | 20 | 192 | 230 | 84 | 192 | 271 | ST524 (1) | ||||||
| 94 | 100 | 263 | 133 | 211 | 310 * | 464 * | ST932 * (1) | ||||||
| 94 | 100 | 263 | 133 | 211 | 309 * | 463 * | ST931 * (1) | ||||||
| 151 | 20 | 70 | 376 * | 327 * | 145 | 36 | ST926 * (1) | ||||||
| 67 | 20 | 71 | 91 | 195 | 308 | 461 | ST929 * (1) | ||||||
| 67 | 20 | 226 | 92 | 85 | 86 | 460 * | ST928 * (1) | ||||||
| 67 | 20 | 307 * | 375 * | 326 * | 214 | 459 * | ST923 * (1) | ||||||
| O2 | 6 | 6 | 0 | 0 | 233 * | 48 | 34 | 309 | 315 | 27 | 457 * | ST918 * (1) | |
| 30 | 46 | 33 | 26 | 36 | 27 | 41 | ST80 (1) | ||||||
| 58 | 63 | 75 | 76 | 73 | 76 | 108 | ST167 (2) | ||||||
| 40 | 48 | 133 | 152 | 221 | 312 * | 187 * | ST934 * (1) | ||||||
| 58 | 63 | 308 * | 377 * | 73 | 307 * | 108 | ST927 * (1) | ||||||
| uncertain | 1 | 0 | 1 | 0 | 63 | 20 | 35 | 247 | 119 | 78 | 317 | ST606 (1) | |
| C.turicensis 4 | O3 | 4 | 4 | 0 | 0 | 68 | 100 | 309 * | 93 | 328 * | 311 * | 465 * | ST933 * (1) |
| 25 | 26 | 22 | 21 | 31 | 37 | 35 | ST35 (1) | ||||||
| 47 | 5 | 4 | 296 | 116 | 150 | 150 | ST714 (1) | ||||||
| 47 | 1 | 311 * | 93 | 60 | 313 * | 153 | ST935 * (1) | ||||||
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Li, Q.; Li, C.; Chen, L.; Cai, Z.; Wu, S.; Gu, Q.; Zhang, Y.; Wei, X.; Zhang, J.; Yang, X.; et al. Cronobacter spp. Isolated from Quick-Frozen Foods in China: Incidence, Genetic Characteristics, and Antibiotic Resistance. Foods 2023, 12, 3019. https://doi.org/10.3390/foods12163019
Li Q, Li C, Chen L, Cai Z, Wu S, Gu Q, Zhang Y, Wei X, Zhang J, Yang X, et al. Cronobacter spp. Isolated from Quick-Frozen Foods in China: Incidence, Genetic Characteristics, and Antibiotic Resistance. Foods. 2023; 12(16):3019. https://doi.org/10.3390/foods12163019
Chicago/Turabian StyleLi, Qi, Chengsi Li, Ling Chen, Zhihe Cai, Shi Wu, Qihui Gu, Youxiong Zhang, Xianhu Wei, Jumei Zhang, Xiaojuan Yang, and et al. 2023. "Cronobacter spp. Isolated from Quick-Frozen Foods in China: Incidence, Genetic Characteristics, and Antibiotic Resistance" Foods 12, no. 16: 3019. https://doi.org/10.3390/foods12163019





