Microbiome of Free-Living Amoebae (FLA) Isolated from Fresh Organic Produce: Potential Risk to Consumers?
Abstract
:1. Introduction
2. Materials and Methods
2.1. Samples and Processing
2.2. FLA Isolation
2.3. DNA Purification and Amplicon Sequencing
2.4. Data Analysis
2.5. Acanthamoeba spp. and Vermamoeba vermiformis qPCR Identification
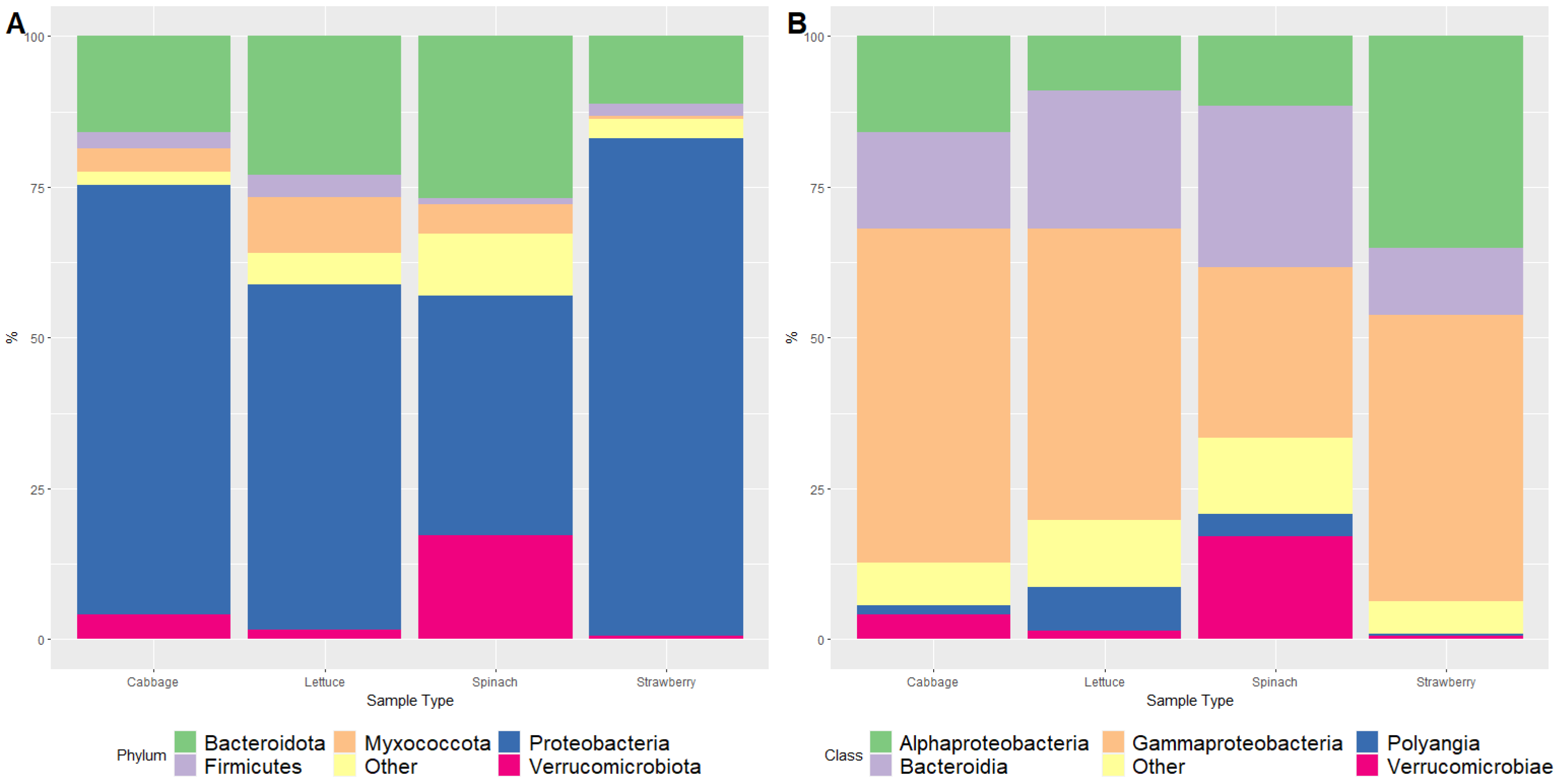
3. Results and Discussion
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Hughner, R.S.; McDonagh, P.; Prothero, A.; Shultz, C.J.; Stanton, J. Who are organic food consumers? A compilation and review of why people purchase organic food. J. Consum. Behav. 2007, 6, 94–110. [Google Scholar] [CrossRef]
- Merlini, V.V.; Pena, F.L.; da Cunha, D.T. Microbiological Quality of Organic and Conventional Leafy Vegetables. J. Food Qual. 2018, 2018, 4908316. [Google Scholar] [CrossRef]
- Kuan, C.H.; Rukayadi, Y.; Ahmad, S.H.; Wan Mohamed Radzi, C.W.J.; Thung, T.Y.; Premarathne, J.M.K.J.K.; Chang, W.S.; Loo, Y.Y.; Tan, C.W.; Ramzi, O.B.; et al. Comparison of the Microbiological Quality and Safety between Conventional and Organic Vegetables Sold in Malaysia. Front. Microbiol. 2017, 8, 1433. [Google Scholar] [CrossRef]
- Maffei, D.F.; Batalha, E.Y.; Landgraf, M.; Schaffner, D.W.; Franco, B.D. Microbiology of organic and conventionally grown fresh produce. Braz. J. Microbiol. 2016, 47 (Suppl. S1), 99–105. [Google Scholar] [CrossRef] [PubMed]
- MAPA (Ministerio de Agricultura, Pesca y Alimentación). Análisis de la Caracterización y Proyección de la Producción Ecológica en España en 2019; Secretaría General Técnica: Madrid, Spain, 2020; NIPO: 00321083X. [Google Scholar]
- Lynch, M.F.; Tauxe, R.V.; Hedberg, C.W. The growing burden of foodborne outbreaks due to contaminated fresh produce: Risks and opportunities. Epidemiol. Infect. 2009, 137, 307–315. [Google Scholar] [CrossRef]
- Alegbeleye, O.O.; Singleton, I.; Sant’ana, A.S. Sources and contamination routes of microbial pathogens to fresh produce during field cultivation: A review. Food Microbiol. 2018, 73, 177–208. [Google Scholar] [CrossRef] [PubMed]
- Rajwar, A.; Srivastava, P.; Sahgal, M. Microbiology of Fresh Produce: Route of Contamination, Detection Methods, and Remedy. Crit. Rev. Food Sci. Nutr. 2016, 56, 2383–2390. [Google Scholar] [CrossRef]
- Amorós, I.; Alonso, J.L.; Cuesta, G. Cryptosporidium oocysts and Giardia cysts on salad products irrigated with contaminated water. J. Food Prot. 2010, 73, 1138–1140. [Google Scholar] [CrossRef]
- Bornier, F.; Zas, E.; Potheret, D.; Laaberki, M.-H.; Coupat-Goutaland, B.; Charpentier, X. Environmental Free-Living Amoebae Can Predate on Diverse Antibiotic-Resistant Human Pathogens. Appl. Environ. Microbiol. 2021, 87, e0074721. [Google Scholar] [CrossRef]
- Samba-Louaka, A.; Robino, E.; Cochard, T.; Branger, M.; Delafont, V.; Aucher, W.; Wambeke, W.; Bannantine, J.P.; Biet, F.; Héchard, Y. Environmental Mycobacterium avium subsp. Paratuberculosis Hosted by Free-Living Amoebae. Front. Cell. Infect. Microbiol. 2018, 8, 28. [Google Scholar] [CrossRef]
- Barker, J.; Brown, M.R.W. Trojan Horses of the microbial world: Protozoa and the survival of bacterial pathogens in the environment. Microbiology 1994, 140, 1253–1259. [Google Scholar] [CrossRef] [PubMed]
- Moreno-Mesonero, L.; Hortelano, I.; Moreno, Y.; Ferrús, M.A. Evidence of viable Helicobacter pylori and other bacteria of public health interest associated with free-living amoebae in lettuce samples by next generation sequencing and other molecular techniques. Int. J. Food Microbiol. 2020, 318, 108477. [Google Scholar] [CrossRef] [PubMed]
- Chavatte, N.; Lambrecht, E.; Van Damme, I.; Sabbe, K.; Houf, K. Abundance, diversity and community composition of free-living protozoa on vegetable sprouts. Food Microbiol. 2016, 55, 55–63. [Google Scholar] [CrossRef] [PubMed]
- Vaerewijck, M.J.; Sabbe, K.; Baré, J.; Houf, K. Occurrence and diversity of free-living protozoa on butterhead lettuce. Int. J. Food Microbiol. 2011, 147, 105–111. [Google Scholar] [CrossRef] [PubMed]
- Janda, W.M. Amoeba-Resistant Bacteria: Their Role in Human Infections. Clin. Microbiol. Newsl. 2010, 32, 177–184. [Google Scholar] [CrossRef]
- Delafont, V.; Brouke, A.; Bouchon, D.; Moulin, L.; Héchard, Y. Microbiome of free-living amoebae isolated from drinking water. Water Res. 2013, 47, 6958–6965. [Google Scholar] [CrossRef]
- Moreno-Mesonero, L.; Ferrús, M.A.; Moreno, Y. Determination of the bacterial microbiome of free-living amoebae isolated from wastewater by 16S rRNA amplicon-based sequencing. Environ. Res. 2020, 190, 109987. [Google Scholar] [CrossRef]
- Vaerewijck, M.J.M.; Baré, J.; Lambrecht, E.; Sabbe, K.; Houf, K. Interactions of Foodborne Pathogens with Free-living Protozoa: Potential Consequences for Food Safety. Compr. Rev. Food Sci. Food Saf. 2014, 13, 924–944. [Google Scholar] [CrossRef]
- Siddiqui, R.; Khan, N.A. Biology and pathogenesis of Acanthamoeba. Parasit. Vectors 2012, 5, 6. [Google Scholar] [CrossRef]
- Visvesvara, G.S.; Moura, H.; Schuster, F.L. Pathogenic and opportunistic free-living amoebae: Acanthamoeba spp., Balamuthia mandrillaris, Naegleria fowleri, and Sappinia diploidea. FEMS Immunol. Med. Microbiol. 2007, 50, 1–26. [Google Scholar] [CrossRef]
- Moreno-Mesonero, L.; Moreno, Y.; Alonso, J.L.; Ferrús, M.A. DVC-FISH and PMA-qPCR techniques to assess the survival of Helicobacter pylori inside Acanthamoeba castellanii. Res. Microbiol. 2016, 167, 29–34. [Google Scholar] [CrossRef]
- Bolyen, E.; Rideout, J.R.; Dillon, M.R.; Bokulich, N.A.; Abnet, C.C.; Al-Ghalith, G.A.; Alexander, H.; Alm, E.J.; Arumugam, M.; Asnicar, F.; et al. Reproducible, Interactive, Scalable and Extensible Microbiome Data Science using QIIME 2. Nat. Biotechnol. 2019, 37, 852–857. [Google Scholar] [CrossRef]
- Callahan, B.J.; Mcmurdie, P.J.; Rosen, M.J.; Han, A.W.; Johnson, A.J.A.; Holmes, S.P. DADA2: High-resolution sample inference from Illumina amplicon data. Nat. Methods 2016, 13, 581–583. [Google Scholar] [CrossRef] [PubMed]
- Camacho, C.; Coulouris, G.; Avagyan, V.; Ma, N.; Papadopoulos, J.; Bealer, K.; Madden, T.L. BLAST+: Architecture and applications. BMC Bioinform. 2009, 10, 421. [Google Scholar] [CrossRef]
- Quast, C.; Pruesse, E.; Yilmaz, P.; Gerken, J.; Schweer, T.; Yarza, P.; Peplies, J.; Glöckner, F.O. The SILVA Ribosomal RNA Gene Database Project: Improved Data Processing and Web-Based Tools. Nucleic Acids Res. 2013, 41, D590–D596. [Google Scholar] [CrossRef]
- Faith, D.P. Conservation evaluation and phylogenetic diversity. Biol. Conserv. 1992, 61, 1–10. [Google Scholar] [CrossRef]
- Lozupone, C.A.; Hamady, M.; Kelley, S.T.; Knight, R. Quantitative and Qualitative β Diversity Measures Lead to Different Insights into Factors That Structure Microbial Communities. Appl. Environ. Microbiol. 2007, 73, 1576–1585. [Google Scholar] [CrossRef]
- Wickham, H. ggplot2: Elegant Graphics for Data Analysis. In Use R! Springer: New York, NY, USA, 2016. [Google Scholar] [CrossRef]
- R Core Team. R: A Language and Environment for Statistical Computing; R Foundation for Statistical Computing: Vienna, Austria, 2022; Available online: https://www.R-project.org/ (accessed on 17 June 2023).
- Qvarnstrom, Y.; Visvesvara, G.S.; Sriram, R.; da Silva, A.J. Multiplex Real-Time PCR Assay for Simultaneous Detection of Acanthamoeba spp., Balamuthia mandrillaris, and Naegleria fowleri. J. Clin. Microbiol. 2006, 44, 3589–3595. [Google Scholar] [CrossRef] [PubMed]
- Gourabathini, P.; Brandl, M.T.; Redding, K.S.; Gunderson, J.H.; Berk, S.G. Interactions between Food-Borne Pathogens and Protozoa Isolated from Lettuce and Spinach. Appl. Environ. Microbiol. 2008, 74, 2518–2525. [Google Scholar] [CrossRef]
- Fatemi, M.; Niyyati, M.; Rouhani, S.; Karamati, S.A.; Mirjalali, H.; Karanis, P. Contamination of fresh vegetables in municipal stores with pathogenic Acanthamoeba genotypes; a public health concern. Int. J. Environ. Health Res. 2022, 33, 1010–1021. [Google Scholar] [CrossRef]
- Corsaro, D.; Wylezich, C.; Walochnik, J.; Venditti, D.; Michel, R. Molecular identification of bacterial endosymbionts of Sappinia strains. Parasitol. Res. 2017, 116, 549–558. [Google Scholar] [CrossRef] [PubMed]
- Garcia, A.; Goñi, P.; Cieloszyk, J.; Fernandez, M.T.; Calvo-Beguería, L.; Rubio, E.; Fillat, M.F.; Peleato, M.L.; Clavel, A. Identification of Free-Living Amoebae and Amoeba-Associated Bacteria from Reservoirs and Water Treatment Plants by Molecular Techniques. Environ. Sci. Technol. 2013, 47, 3132–3140. [Google Scholar] [CrossRef]
- Goñi, P.; Fernández, M.T.; Rubio, E. Identifying endosymbiont bacteria associated with free-living amoebae. Environ. Microbiol. 2014, 16, 339–349. [Google Scholar] [CrossRef] [PubMed]
- Balczun, C.; Scheid, P.L. Free-Living Amoebae as Hosts for and Vectors of Intracellular Microorganisms with Public Health Significance. Viruses 2017, 9, 65. [Google Scholar] [CrossRef] [PubMed]
- Samba-Louaka, A.; Delafont, V.; Rodier, M.-H.; Cateau, E.; Héchard, Y. Free-living amoebae and squatters in the wild: Ecological and molecular features. FEMS Microbiol. Rev. 2019, 43, 415–434. [Google Scholar] [CrossRef]
- Ratner, H. Topics in Clinical Microbiology Flavobacterium Meningosepticum. Infect. Control. Hosp. Epidemiol. 1984, 5, 237–241. [Google Scholar] [CrossRef]
- Duchaud, E.; Boussaha, M.; Loux, V.; Bernardet, J.-F.; Michel, C.; Kerouault, B.; Mondot, S.; Nicolas, P.; Bossy, R.; Caron, C.; et al. Complete genome sequence of the fish pathogen Flavobacterium psychrophilum. Nat. Biotechnol. 2007, 25, 763–769. [Google Scholar] [CrossRef]
- Magnet, A.; Peralta, R.H.S.; Gomes, T.S.; Izquierdo, F.; Fernandez-Vadillo, C.; Galvan, A.L.; Pozuelo, M.J.; Pelaz, C.; Fenoy, S.; Del Águila, C. Vectorial role of Acanthamoeba in Legionella propagation in water for human use. Sci. Total Environ. 2015, 505, 889–895. [Google Scholar] [CrossRef]
- García-Hernández, J.; Hernández, M.; Moreno, Y. Combination of Direct Viable Count and Fluorescent In Situ Hybridization (DVC-FISH) as a Potential Method for Identifying Viable Vibrio parahaemolyticus in Oysters and Mussels. Foods 2021, 10, 1502. [Google Scholar] [CrossRef]


| Acanthamoeba spp. | V. vermiformis | |
|---|---|---|
| Lettuce | 4/11 (36.4%) | 1/11 (9.1%) |
| Cabbage | 11/11 (100.0%) | 4/11 (36.4%) |
| Spinach | 4/11 (36.4%) | 4/11 (36.4%) |
| Strawberry | 7/7 (100.0%) | 1/7 (14.3%) |
| Total | 26/40 (65.0%) | 10/40 (25.0%) |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Soler, L.; Moreno, Y.; Moreno-Mesonero, L.; Amorós, I.; Alonso, J.L.; Ferrús, M.A. Microbiome of Free-Living Amoebae (FLA) Isolated from Fresh Organic Produce: Potential Risk to Consumers? Foods 2023, 12, 3102. https://doi.org/10.3390/foods12163102
Soler L, Moreno Y, Moreno-Mesonero L, Amorós I, Alonso JL, Ferrús MA. Microbiome of Free-Living Amoebae (FLA) Isolated from Fresh Organic Produce: Potential Risk to Consumers? Foods. 2023; 12(16):3102. https://doi.org/10.3390/foods12163102
Chicago/Turabian StyleSoler, Lara, Yolanda Moreno, Laura Moreno-Mesonero, Inmaculada Amorós, José Luís Alonso, and María Antonia Ferrús. 2023. "Microbiome of Free-Living Amoebae (FLA) Isolated from Fresh Organic Produce: Potential Risk to Consumers?" Foods 12, no. 16: 3102. https://doi.org/10.3390/foods12163102
APA StyleSoler, L., Moreno, Y., Moreno-Mesonero, L., Amorós, I., Alonso, J. L., & Ferrús, M. A. (2023). Microbiome of Free-Living Amoebae (FLA) Isolated from Fresh Organic Produce: Potential Risk to Consumers? Foods, 12(16), 3102. https://doi.org/10.3390/foods12163102






