Advances in Genetic Tools and Their Application in Streptococcus thermophilus
Abstract
:1. Introduction
2. Advantages of S. thermophilus as a Host
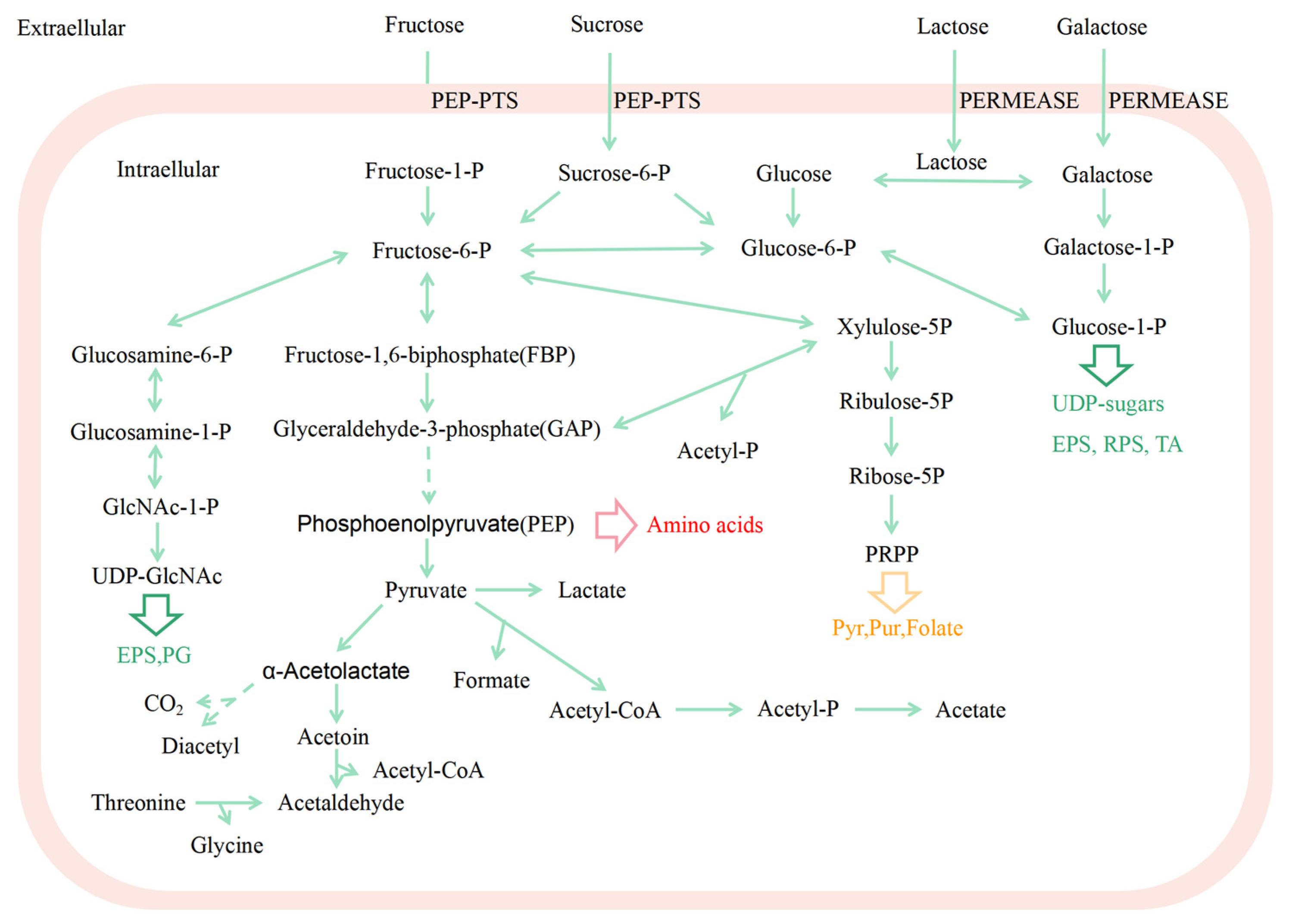
2.1. Clear Genetic Background and Physiological Characteristics
- 1.
- Sugar metabolism
- 2.
- Polysaccharide biosynthesis
- 3.
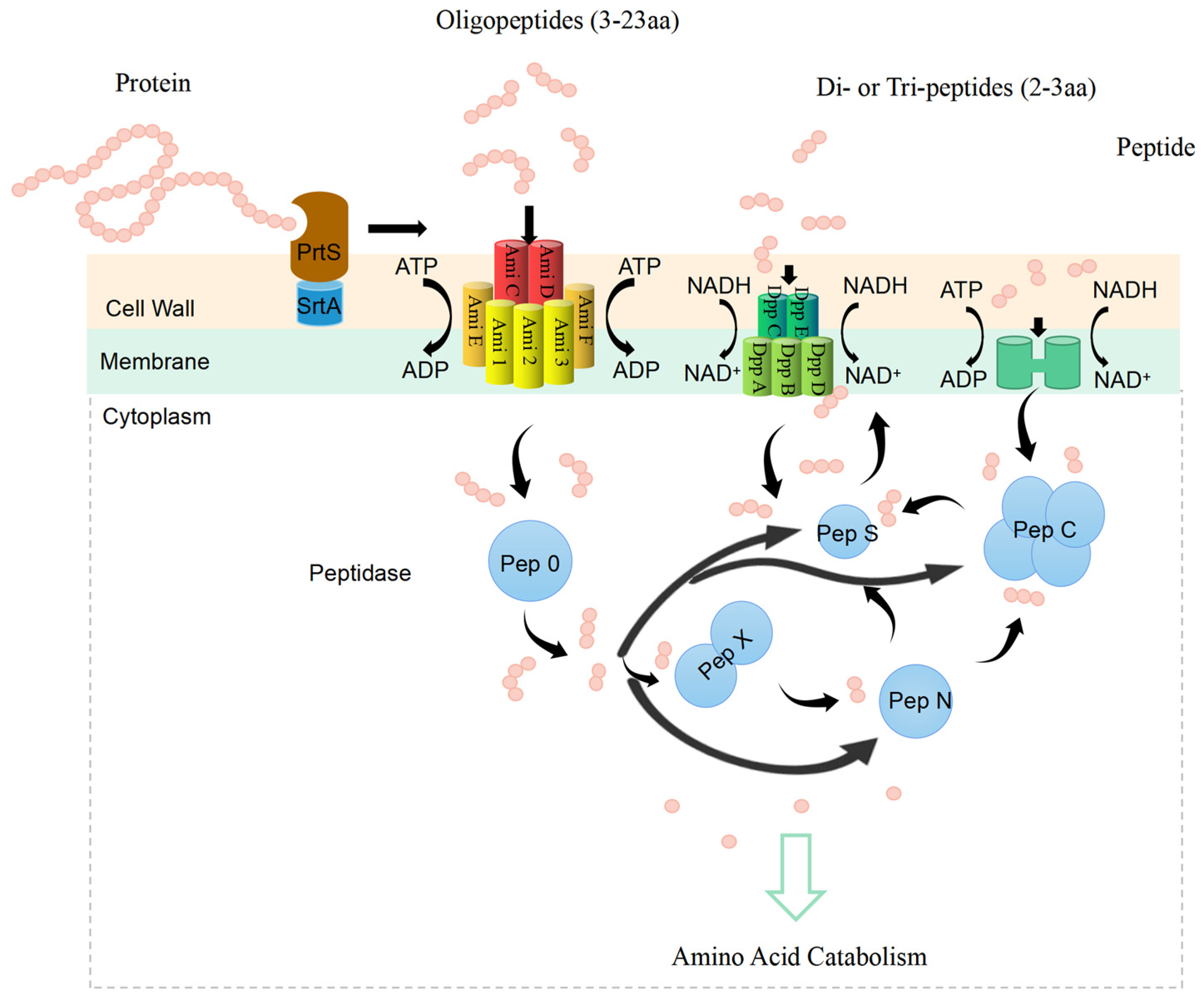
- Protein metabolism
2.2. Food-Grade Safety
3. DNA Components
3.1. Promoters
3.2. Terminator
- (i)
- L-shaped (canonical terminators): a trail of 10 bp that contains more than three uridylates follows a hairpin.
- (ii)
- I-shaped: where the path after the hairpin contains three uridines;
- (iii)
- U-shaped: multiple hairpin structures working together, with around 50 nt between them;
- (iv)
- X-shaped: convergent structures of the right and left strands that serve as terminators for the convergently transcribed genes; and
- (v)
- V-shaped: two hairpins, the second of which begins right after the first.
4. Plasmid Vectors
4.1. Non-Food Grade Expression Vectors
4.2. Food-Grade Expression Vectors
4.2.1. Dominant Selection Marker
4.2.2. Complementary Selection Markers
4.3. Endogenous Plasmid Vectors
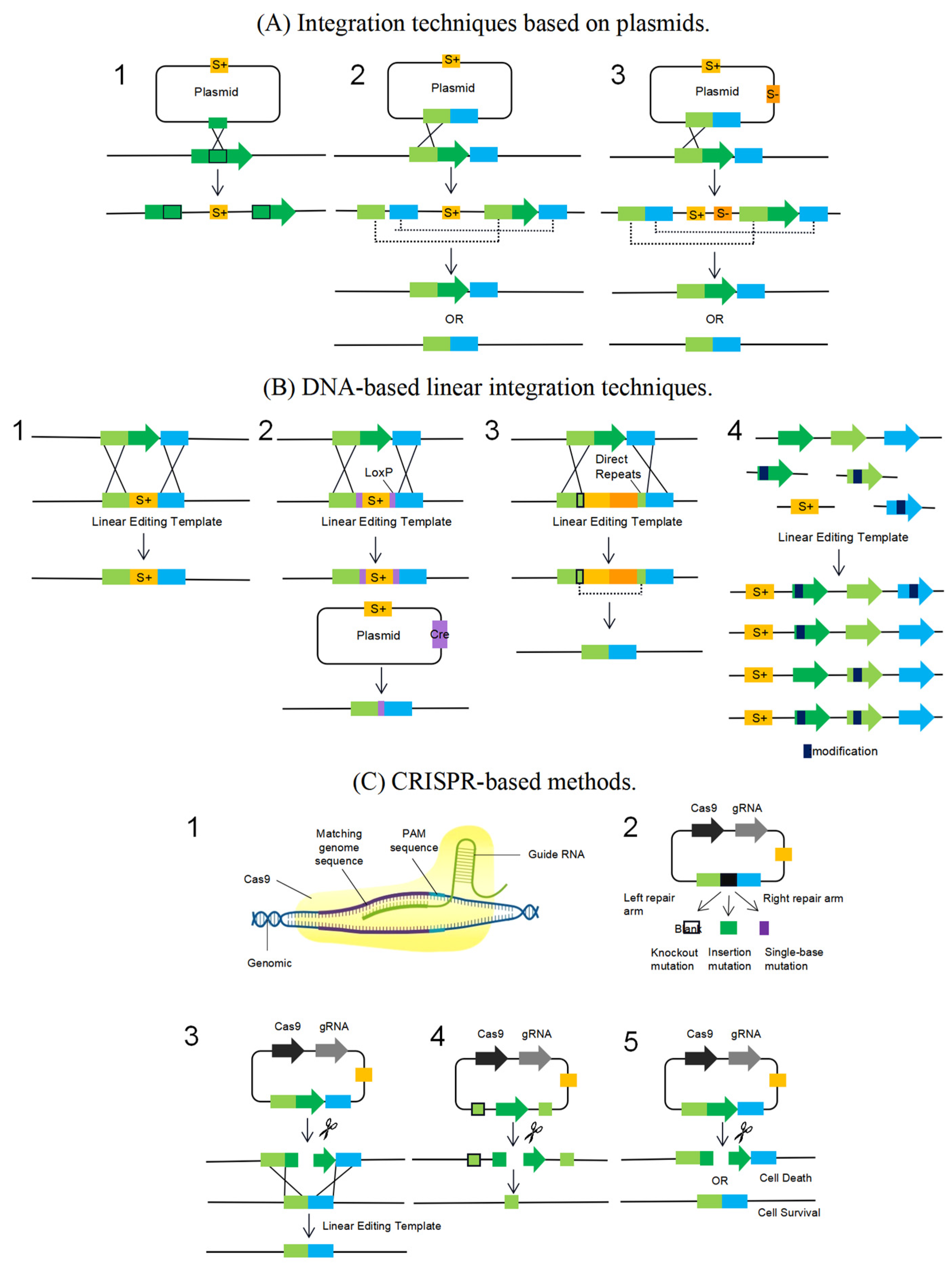
5. Genome Editing Tools
6. Application of Engineered Strains of S. thermophilus
7. Summary and Outlook
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Iyer, R.; Tomar, S.K.; Maheswari, T.; Singh, R. Streptococcus thermophilus strains: Multifunctional lactic acid bacteria. Int. Dairy J. 2010, 20, 133–141. [Google Scholar] [CrossRef]
- Prajapati, J.B.; Nathani, N.M.; Patel, A.K.; Senan, S.; Joshi, C.G. Genomic analysis of dairy starter culture Streptococcus thermophilus MTCC 5461. J. Microbiol. Biotechnol. 2013, 23, 459–466. [Google Scholar] [CrossRef] [PubMed]
- Alexandraki, V.; Kazou, M.; Pot, B.; Tsakalidou, E.; Papadimitriou, K. Complete Genome Sequence of the Yogurt Isolate Lactobacillus delbrueckii subsp. bulgaricus ACA-DC 87. Genome Announc. 2017, 5, e00868-17. [Google Scholar] [PubMed]
- Alexandraki, V.; Kazou, M.; Blom, J.; Pot, B.; Papadimitriou, K.; Tsakalidou, E. Comparative Genomics of Streptococcus thermophilus Support Important Traits Concerning the Evolution, Biology and Technological Properties of the Species. Front. Microbiol. 2019, 10, 2916. [Google Scholar] [CrossRef]
- Bolotin, A.; Quinquis, B.; Renault, P.; Sorokin, A.; Ehrlich, S.D.; Kulakauskas, S.; Lapidus, A.; Goltsman, E.; Mazur, M.; Pusch, G.D.; et al. Complete sequence and comparative genome analysis of the dairy bacterium Streptococcus thermophilus. Nat. Biotechnol. 2004, 22, 1554–1558. [Google Scholar] [CrossRef]
- Delorme, C.; Bartholini, C.; Bolotine, A.; Ehrlich, S.D.; Renault, P. Emergence of a Cell Wall Protease in the Streptococcus thermophilus Population. Appl. Environ. Microbiol. 2009, 76, 451–460. [Google Scholar] [CrossRef]
- Yu, J.; Sun, Z.; Liu, W.; Xi, X.; Song, Y.; Xu, H.; Lv, Q.; Bao, Q.; Menghe, B.; Sun, T. Multilocus sequence typing of Streptococcus thermophilus from naturally fermented dairy foods in China and Mongolia. BMC Microbiol. 2015, 15, 236. [Google Scholar] [CrossRef]
- Hols, P.; Hancy, F.; Fontaine, L.; Grossiord, B.; Prozzi, D.; Leblond-Bourget, N.; Decaris, B.; Bolotin, A.; Delorme, C.; Dusko Ehrlich, S.; et al. New insights in the molecular biology and physiology of Streptococcus thermophilus revealed by comparative genomics. FEMS Microbiol. Rev. 2005, 29, 435–463. [Google Scholar]
- Li, W.; Bian, X.; Evivie, S.E.; Huo, G.C. Comparative Analysis of Clustered Regularly Interspaced Short Palindromic Repeats (CRISPR) of Streptococcus thermophilus St-I and its Bacteriophage-Insensitive Mutants (BIM) Derivatives. Curr. Microbiol. 2016, 73, 393–400. [Google Scholar] [CrossRef]
- Mora, D.; Maguin, E.; Masiero, M.; Parini, C.; Ricci, G.; Manachini, P.L.; Daffonchio, D. Characterization of urease genes cluster of Streptococcus thermophilus. J. Appl. Microbiol. 2004, 96, 209–219. [Google Scholar] [CrossRef]
- Couvigny, B.; Thérial, C.; Gautier, C.; Renault, P.; Briandet, R.; Guédon, E. Streptococcus thermophilus Biofilm Formation: A Remnant Trait of Ancestral Commensal Life? PLoS ONE 2015, 10, e0128099. [Google Scholar] [CrossRef] [PubMed]
- van den Bogaard, P.T.; Hols, P.; Kuipers, O.P.; Kleerebezem, M.; de Vos, W.M. Sugar utilisation and conservation of the gal-lac gene cluster in Streptococcus thermophilus. Syst. Appl. Microbiol. 2004, 27, 10–17. [Google Scholar] [CrossRef] [PubMed]
- Poolman, B. Energy transduction in lactic acid bacteria. FEMS Microbiol. Rev. 1993, 12, 125–147. [Google Scholar] [CrossRef] [PubMed]
- O’Leary, V.S.; Woychik, J.H. Utilization of Lactose, Glucose, and Galactose by a Mixed Culture of Streptococcus thermophilus and Lactobacillus bulgaricus in Milk Treated with Lactase Enzyme. Appl. Environ. Microbiol. 1976, 32, 89–94. [Google Scholar] [CrossRef]
- Mora, D.; Fortina, M.G.; Parini, C.; Ricci, G.; Gatti, M.; Giraffa, G.; Manachini, P.L. Genetic diversity and technological properties of Streptococcus thermophilus strains isolated from dairy products. J. Appl. Microbiol. 2002, 93, 278–287. [Google Scholar] [CrossRef]
- Laws, A.; Gu, Y.; Marshall, V. Biosynthesis, characterisation, and design of bacterial exopolysaccharides from lactic acid bacteria. Biotechnol. Adv. 2001, 19, 597–625. [Google Scholar] [CrossRef]
- van Kranenburg, R.; Marugg, J.D.; Van Swam, I.I.; Willem, N.J.; de Vos, W.M. Molecular characterization of the plasmid-encoded eps gene cluster essential for exopolysaccharide biosynthesis in Lactococcus lactis. Mol. Microbiol. 1997, 24, 387–397. [Google Scholar] [CrossRef]
- Jolly, L.; Stingele, F. Molecular organization and functionality of exopolysaccharide gene clusters in lactic acid bacteria. Int. Dairy J. 2001, 11, 733–745. [Google Scholar] [CrossRef]
- Broadbent, J.R.; McMahon, D.J.; Welker, D.L.; Oberg, C.J.; Moineau, S. Biochemistry, genetics, and applications of exopolysaccharide production in Streptococcus thermophilus: A review. J. Dairy Sci. 2003, 86, 407–423. [Google Scholar] [CrossRef]
- Galia, W.; Perrin, C.; Genay, M.; Dary, A. Variability and molecular typing of Streptococcus thermophilus strains displaying different proteolytic and acidifying properties. Int. Dairy J. 2009, 19, 89–95. [Google Scholar] [CrossRef]
- Mora, D.; Ricci, G.; Guglielmetti, S.; Daffonchio, D.; Fortina, M.G. 16S-23S rRNA intergenic spacer region sequence variation in Streptococcus thermophilus and related dairy streptococci and development of a multiplex ITS-SSCP analysis for their identification. Microbiology 2003, 149 Pt 3, 807–813. [Google Scholar] [CrossRef]
- Bernaudat, F.; Frelet-Barrand, A.; Pochon, N.; Dementin, S.; Hivin, P.; Boutigny, S.; Rioux, J.B.; Salvi, D.; Seigneurin-Berny, D.; Richaud, P.; et al. Heterologous expression of membrane proteins: Choosing the appropriate host. PLoS ONE 2011, 6, e29191. [Google Scholar] [CrossRef]
- Westers, L.; Westers, H.; Quax, W.J. Bacillus subtilis as cell factory for pharmaceutical proteins: A biotechnological approach to optimize the host organism. Biochim. Biophys. Acta 2004, 1694, 299–310. [Google Scholar] [CrossRef]
- Morello, E.; Bermúdez-Humarán, L.G.; Llull, D.; Solé, V.; Miraglio, N.; Langella, P.; Poquet, I. Lactococcus lactis, an efficient cell factory for recombinant protein production and secretion. J. Mol. Microbiol. Biotechnol. 2008, 14, 48–58. [Google Scholar] [CrossRef] [PubMed]
- Zhou, M.; Theunissen, D.; Wels, M.; Siezen, R.J. LAB-Secretome: A genome-scale comparative analysis of the predicted extracellular and surface-associated proteins of Lactic Acid Bacteria. BMC Genom. 2010, 11, 651. [Google Scholar] [CrossRef] [PubMed]
- Wydau, S.; Dervyn, R.; Anba, J.; Dusko Ehrlich, S.; Maguin, E. Conservation of key elements of natural competence in Lactococcus lactis ssp. FEMS Microbiol. Lett. 2006, 257, 32–42. [Google Scholar] [CrossRef]
- Blomqvist, T.; Steinmoen, H.; Håvarstein, L.S. Natural genetic transformation: A novel tool for efficient genetic engineering of the dairy bacterium Streptococcus thermophilus. Appl. Environ. Microbiol. 2006, 72, 6751–6756. [Google Scholar] [CrossRef] [PubMed]
- Dandoy, D.; Fremaux, C.; de Frahan, M.H.; Horvath, P.; Boyaval, P.; Hols, P.; Fontaine, L. The fast milk acidifying phenotype of Streptococcus thermophilus can be acquired by natural transformation of the genomic island encoding the cell-envelope proteinase PrtS. Microb. Cell Factories 2011, 10 (Suppl. S1), S21. [Google Scholar] [CrossRef] [PubMed]
- Alper, H.; Fischer, C.; Nevoigt, E.; Stephanopoulos, G. Tuning genetic control through promoter engineering. Proc. Natl. Acad. Sci. USA 2005, 102, 12678–12683. [Google Scholar] [CrossRef]
- Solaiman, D.K.; Somkuti, G.A. Expression of Streptomyces melC and choA genes by a cloned Streptococcus thermophilus promoter STP2201. J. Ind. Microbiol. 1995, 15, 39–44. [Google Scholar] [CrossRef]
- Slos, P.; Bourquin, J.C.; Lemoine, Y.; Mercenier, A. Isolation and characterization of chromosomal promoters of Streptococcus salivarius subsp. thermophilus. Appl. Environ. Microbiol. 1991, 57, 1333–1339. [Google Scholar] [CrossRef]
- Rhimi, M.; Chouayekh, H.; Gouillouard, I.; Maguin, E.; Bejar, S. Production of D-tagatose, a low caloric sweetener during milk fermentation using L-arabinose isomerase. Bioresour. Technol. 2011, 102, 3309–3315. [Google Scholar] [CrossRef] [PubMed]
- Lecomte, X.; Gagnaire, V.; Briard-Bion, V.; Jardin, J.; Lortal, S.; Dary, A.; Genay, M. The naturally competent strain Streptococcus thermophilus LMD-9 as a new tool to anchor heterologous proteins on the cell surface. Microb. Cell Factories 2014, 13, 82. [Google Scholar] [CrossRef] [PubMed]
- Kong, L.H.; Xiong, Z.Q.; Song, X.; Xia, Y.J.; Zhang, N.; Ai, L.Z. Characterization of a Panel of Strong Constitutive Promoters from Streptococcus thermophilus for Fine-Tuning Gene Expression. ACS Synth. Biol. 2019, 8, 1469–1472. [Google Scholar] [CrossRef]
- Chouayekh, H.; Serror, P.; Boudebbouze, S.; Maguin, E. Highly efficient production of the staphylococcal nuclease reporter in Lactobacillus bulgaricus governed by the promoter of the hlbA gene. FEMS Microbiol. Lett. 2009, 293, 232–239. [Google Scholar] [CrossRef]
- Renye, J.A., Jr.; Somkuti, G.A. Nisin-induced expression of pediocin in dairy lactic acid bacteria. J. Appl. Microbiol. 2010, 108, 2142–2151. [Google Scholar] [CrossRef] [PubMed]
- González-Márquez, H.; Perrin, C.; Bracquart, P.; Guimont, C.; Linden, G. A 16 kDa protein family overexpressed by Streptococcus thermophilus PB18 in acid environments. Microbiology 1997, 143 Pt 5, 1587–1594. [Google Scholar] [CrossRef]
- Petrova, P.M.; Gouliamova, D.E. Rapid screening of plasmid-encoded small hsp-genes in Streptococcus thermophilus. Curr. Microbiol. 2006, 53, 422–427. [Google Scholar] [CrossRef]
- Junjua, M.; Galia, W.; Gaci, N.; Uriot, O.; Genay, M.; Bachmann, H.; Kleerebezem, M.; Dary, A.; Roussel, Y. Development of the recombinase-based in vivo expression technology in Streptococcus thermophilus and validation using the lactose operon promoter. J. Appl. Microbiol. 2014, 116, 620–631. [Google Scholar] [CrossRef]
- Fontaine, L.; Goffin, P.; Dubout, H.; Delplace, B.; Baulard, A.; Lecat-Guillet, N.; Chambellon, E.; Gardan, R.; Hols, P. Mechanism of competence activation by the ComRS signalling system in streptococci. Mol. Microbiol. 2013, 87, 1113–1132. [Google Scholar] [CrossRef]
- Blount, B.A.; Weenink, T.; Ellis, T. Construction of synthetic regulatory networks in yeast. FEBS Lett. 2012, 586, 2112–2121. [Google Scholar] [CrossRef]
- Mitra, A.; Kesarwani, A.K.; Pal, D.; Nagaraja, V. WebGeSTer DB--a transcription terminator database. Nucleic Acids Res. 2011, 39, D129–D135. [Google Scholar] [CrossRef] [PubMed]
- Wong, W.Y.; Su, P.; Allison, G.E.; Liu, C.Q.; Dunn, N.W. A potential food-grade cloning vector for Streptococcus thermophilus that uses cadmium resistance as the selectable marker. Appl. Environ. Microbiol. 2003, 69, 5767–5771. [Google Scholar] [CrossRef] [PubMed]
- El Demerdash, H.A.; Heller, K.J.; Geis, A. Application of the shsp gene, encoding a small heat shock protein, as a food-grade selection marker for lactic acid bacteria. Appl. Environ. Microbiol. 2003, 69, 4408–4412. [Google Scholar] [CrossRef] [PubMed]
- Sasaki, Y.; Ito, Y.; Sasaki, T. ThyA as a selection marker in construction of food-grade host-vector and integration systems for Streptococcus thermophilus. Appl. Environ. Microbiol. 2004, 70, 1858–1864. [Google Scholar] [CrossRef]
- Labrie, S.; Bart, C.; Vadeboncoeur, C.; Moineau, S. Use of an alpha-galactosidase gene as a food-grade selection marker for Streptococcus thermophilus. J. Dairy Sci. 2005, 88, 2341–2347. [Google Scholar] [CrossRef]
- Somkuti, G.A.; Steinberg, D.H. Distribution and analysis of plasmids in Streptococcus thermophilus. J. Ind. Microbiol. 1986, 1, 157–163. [Google Scholar] [CrossRef]
- Solaiman, D.K.; Somkuti, G.A. Shuttle vectors developed from Streptococcus thermophilus native plasmid. Plasmid 1993, 30, 67–78. [Google Scholar] [CrossRef]
- Maguin, E.; Prévost, H.; Ehrlich, S.D.; Gruss, A. Efficient insertional mutagenesis in lactococci and other gram-positive bacteria. J. Bacteriol. 1996, 178, 931–935. [Google Scholar] [CrossRef]
- Solaiman, D.K.Y.; Somkuti, G.A. Construction of a green-fluorescent protein-based, insertion-inactivation shuttle vector for lactic acid bacteria and Escherichia coli. Biotechnol. Lett. 1997, 19, 1175–1179. [Google Scholar] [CrossRef]
- Solaiman, D.K.Y.; Somkuti, G.A. Characterization of pER371-based Streptococcus thermophilus-Escherichia coli shuttle vectors. Biotechnol. Lett. 1997, 19, 595–598. [Google Scholar] [CrossRef]
- Coderre, P.E.; Somkuti, G.A. Cloning and expression of the pediocin operon in Streptococcus thermophilus and other lactic fermentation bacteria. Curr. Microbiol. 1999, 39, 295–301. [Google Scholar] [CrossRef]
- Somkuti, G.A.; Steinberg, D.H. Promoter activity of the pER341-borne ST(Phsp) in heterologous gene expression in Escherichia coli and Streptococcus thermophilus(1). FEMS Microbiol. Lett. 1999, 179, 431–436. [Google Scholar] [CrossRef]
- Turgeon, N.; Moineau, S. Isolation and characterization of a Streptococcus thermophilus plasmid closely related to the pMV158 family. Plasmid 2001, 45, 171–183. [Google Scholar] [CrossRef] [PubMed]
- Renye, J.A., Jr.; Somkuti, G.A. Insertion of a heterologous gene construct into a non-functional ORF of the Streptococcus thermophilus chromosome. Biotechnol. Lett. 2009, 31, 759–764. [Google Scholar] [CrossRef] [PubMed]
- Cui, Y.; Hu, T.; Qu, X.; Zhang, L.; Ding, Z.; Dong, A. Plasmids from Food Lactic Acid Bacteria: Diversity, Similarity, and New Developments. Int. J. Mol. Sci. 2015, 16, 13172–13202. [Google Scholar] [CrossRef]
- Somkuti, G.A.; Solaiman, D.K.; Steinberg, D.H. Structural and functional properties of the hsp16.4-bearing plasmid pER341 in Streptococcus thermophilus. Plasmid 1998, 40, 61–72. [Google Scholar] [CrossRef]
- O’Sullivan, T.; van Sinderen, D.; Fitzgerald, G. Structural and functional analysis of pCI65st, a 6.5 kb plasmid from Streptococcus thermophilus NDI-6. Microbiology 1999, 145 Pt 1, 127–134. [Google Scholar] [CrossRef]
- Su, P.; Jury, K.; Allison, G.E.; Wong, W.Y.; Kim, W.S.; Liu, C.Q.; Vancov, T.; Dunn, N.W. Cloning vectors for Streptococcus thermophilus derived from a native plasmid. FEMS Microbiol. Lett. 2002, 216, 43–47. [Google Scholar] [CrossRef]
- Geis, A.; El Demerdash, H.A.; Heller, K.J. Sequence analysis and characterization of plasmids from Streptococcus thermophilus. Plasmid 2003, 50, 53–69. [Google Scholar] [CrossRef]
- Petrova, P.; Miteva, V.; Ruiz-Masó, J.A.; del Solar, G. Structural and functional analysis of pt38, a 2.9kb plasmid of Streptococcus thermophilus yogurt strain. Plasmid 2003, 50, 176–189. [Google Scholar] [CrossRef] [PubMed]
- Solow, B.T.; Somkuti, G.A. Comparison of low-molecular-weight heat stress proteins encoded on plasmids in different strains of Streptococcus thermophilus. Curr. Microbiol. 2000, 41, 177–181. [Google Scholar] [CrossRef]
- Makarova, K.; Slesarev, A.; Wolf, Y.; Sorokin, A.; Mirkin, B.; Koonin, E.; Pavlov, A.; Pavlova, N.; Karamychev, V.; Polouchine, N.; et al. Comparative genomics of the lactic acid bacteria. Proc. Natl. Acad. Sci. USA 2006, 103, 15611–15616. [Google Scholar] [CrossRef] [PubMed]
- Girard, S.L.; Moineau, S. Analysis of two theta-replicating plasmids of Streptococcus thermophilus. Plasmid 2007, 58, 174–181. [Google Scholar] [CrossRef] [PubMed]
- Goh, Y.J.; Goin, C.; O’Flaherty, S.; Altermann, E.; Hutkins, R. Specialized adaptation of a lactic acid bacterium to the milk environment: The comparative genomics of Streptococcus thermophilus LMD-9. Microb. Cell Factories 2011, 10 (Suppl. S1), S22. [Google Scholar] [CrossRef]
- Turgeon, N.; Frenette, M.; Moineau, S. Characterization of a theta-replicating plasmid from Streptococcus thermophilus. Plasmid 2004, 51, 24–36. [Google Scholar] [CrossRef]
- Solaiman, D.K.; Somkuti, G.A. Characterization of a novel Streptococcus thermophilus rolling-circle plasmid used for vector construction. Appl. Microbiol. Biotechnol. 1998, 50, 174–180. [Google Scholar] [CrossRef]
- Somkuti, G.A.; Steinberg, D.H. Molecular organization of plasmid pER13 in Streptococcus thermophilus. Biotechnol. Lett. 2007, 29, 1991–1999. [Google Scholar] [CrossRef]
- Schoch, C.L.; Ciufo, S.; Domrachev, M.; Hotton, C.L.; Kannan, S.; Khovanskaya, R.; Leipe, D.; McVeigh, R.; O’Neill, K.; Robbertse, B.; et al. NCBI Taxonomy: A comprehensive update on curation, resources and tools. Database 2020, 2020, baaa062. [Google Scholar] [CrossRef]
- McDonnell, B.; Hanemaaijer, L.; Bottacini, F.; Kelleher, P.; Lavelle, K.; Sadovskaya, I.; Vinogradov, E.; Ver Loren van Themaat, E.; Kouwen, T.; Mahony, J.; et al. A cell wall-associated polysaccharide is required for bacteriophage adsorption to the Streptococcus thermophilus cell surface. Mol. Microbiol. 2020, 114, 31–45. [Google Scholar] [CrossRef]
- Yamauchi, R.; Maguin, E.; Horiuchi, H.; Hosokawa, M.; Sasaki, Y. The critical role of urease in yogurt fermentation with various combinations of Streptococcus thermophilus and Lactobacillus delbrueckii ssp. bulgaricus. J. Dairy Sci. 2019, 102, 1033–1043. [Google Scholar] [CrossRef] [PubMed]
- Defoor, E.; Kryger, M.B.; Martinussen, J. The orotate transporter encoded by oroP from Lactococcus lactis is required for orotate utilization and has utility as a food-grade selectable marker. Microbiology 2007, 153 Pt 11, 3645–3659. [Google Scholar] [CrossRef] [PubMed]
- Miyazaki, K. Molecular engineering of a PheS counterselection marker for improved operating efficiency in Escherichia coli. Biotechniques 2015, 58, 86–88. [Google Scholar] [CrossRef] [PubMed]
- Xin, Y.; Guo, T.; Mu, Y.; Kong, J. Development of a counterselectable seamless mutagenesis system in lactic acid bacteria. Microb. Cell Factories 2017, 16, 116. [Google Scholar] [CrossRef]
- Fontaine, L.; Boutry, C.; de Frahan, M.H.; Delplace, B.; Fremaux, C.; Horvath, P.; Boyaval, P.; Hols, P. A novel pheromone quorum-sensing system controls the development of natural competence in Streptococcus thermophilus and Streptococcus salivarius. J. Bacteriol. 2010, 192, 1444–1454. [Google Scholar] [CrossRef]
- Fontaine, L.; Dandoy, D.; Boutry, C.; Delplace, B.; de Frahan, M.H.; Fremaux, C.; Horvath, P.; Boyaval, P.; Hols, P. Development of a versatile procedure based on natural transformation for marker-free targeted genetic modification in Streptococcus thermophilus. Appl. Environ. Microbiol. 2010, 76, 7870–7877. [Google Scholar] [CrossRef]
- Zhang, S.; Zou, Z.; Kreth, J.; Merritt, J. Recombineering in Streptococcus mutans Using Direct Repeat-Mediated Cloning-Independent Markerless Mutagenesis (DR-CIMM). Front. Cell Infect. Microbiol. 2017, 7, 202. [Google Scholar] [CrossRef] [PubMed]
- Dalia, T.N.; Hayes, C.A.; Stolyar, S.; Marx, C.J.; McKinlay, J.B.; Dalia, A.B. Multiplex Genome Editing by Natural Transformation (MuGENT) for Synthetic Biology in Vibrio natriegens. ACS Synth. Biol. 2017, 6, 1650–1655. [Google Scholar] [CrossRef]
- Xu, K.; Ren, C.; Liu, Z.; Zhang, T.; Zhang, T.; Li, D.; Wang, L.; Yan, Q.; Guo, L.; Shen, J.; et al. Efficient genome engineering in eukaryotes using Cas9 from Streptococcus thermophilus. Cell Mol. Life Sci. 2015, 72, 383–399. [Google Scholar] [CrossRef]
- van der Els, S.; James, J.K.; Kleerebezem, M.; Bron, P.A. Versatile Cas9-Driven Subpopulation Selection Toolbox for Lactococcus lactis. Appl. Environ. Microbiol. 2018, 84, e02752-17. [Google Scholar] [CrossRef]
- Gong, T.; Tang, B.; Zhou, X.; Zeng, J.; Lu, M.; Guo, X.; Peng, X.; Lei, L.; Gong, B.; Li, Y. Genome editing in Streptococcus mutans through self-targeting CRISPR arrays. Mol. Oral. Microbiol. 2018, 33, 440–449. [Google Scholar] [CrossRef] [PubMed]
- Somkuti, G.A.; Solaiman, D.K.Y.; Steinberg, D.H. Cloning of a tyrosinase gene in Streptococcus thermophilus. Biotechnol. Lett. 1993, 15, 773–778. [Google Scholar] [CrossRef]
- Vaillancourt, K.; LeMay, J.D.; Lamoureux, M.; Frenette, M.; Moineau, S.; Vadeboncoeur, C. Characterization of a galactokinase-positive recombinant strain of Streptococcus thermophilus. Appl. Environ. Microbiol. 2004, 70, 4596–4603. [Google Scholar] [CrossRef] [PubMed]
- Chaves, A.C.; Fernandez, M.; Lerayer, A.L.; Mierau, I.; Kleerebezem, M.; Hugenholtz, J. Metabolic engineering of acetaldehyde production by Streptococcus thermophilus. Appl. Environ. Microbiol. 2002, 68, 5656–5662. [Google Scholar] [CrossRef]
- Renye, J.A., Jr.; Somkuti, G.A. Cloning of milk-derived bioactive peptides in Streptococcus thermophilus. Biotechnol. Lett. 2008, 30, 723–730. [Google Scholar] [CrossRef]
- Del Carmen, S.; de Moreno de LeBlanc, A.; Martin, R.; Chain, F.; Langella, P.; Bermúdez-Humarán, L.G.; LeBlanc, J.G. Genetically engineered immunomodulatory Streptococcus thermophilus strains producing antioxidant enzymes exhibit enhanced anti-inflammatory activities. Appl. Environ. Microbiol. 2014, 80, 869–877. [Google Scholar] [CrossRef]
- Feng, Y.; Li, W.; Wu, X.; He, L.; Ma, S. Rapid and efficient microwave-assisted sulfate modification of lentinan and its antioxidant and antiproliferative activities in vitro. Carbohydr. Polym. 2010, 82, 605–612. [Google Scholar] [CrossRef]
- Mikkonen, K.S.; Parikka, K.; Suuronen, J.-P.; Ghafar, A.; Serimaa, R.; Tenkanen, M. Enzymatic oxidation as a potential new route to produce polysaccharide aerogels. RSC Adv. 2014, 4, 11884–11892. [Google Scholar] [CrossRef]
- Zhao, B.; Zhang, J.; Yao, J.; Song, S.; Yin, Z.; Gao, Q. Selenylation modification can enhance antioxidant activity of Potentilla anserina L. polysaccharide. Int. J. Biol. Macromol. 2013, 58, 320–328. [Google Scholar] [CrossRef]
- Li, S.; Shah, N.P. Antioxidant and antibacterial activities of sulphated polysaccharides from Pleurotus eryngii and Streptococcus thermophilus ASCC 1275. Food Chem. 2014, 165, 262–270. [Google Scholar] [CrossRef]
- Krichen, F.; Karoud, W.; Sila, A.; Abdelmalek, B.E.; Ghorbel, R.; Ellouz-Chaabouni, S.; Bougatef, A. Extraction, characterization and antimicrobial activity of sulfated polysaccharides from fish skins. Int. J. Biol. Macromol. 2015, 75, 283–289. [Google Scholar] [CrossRef] [PubMed]
- Yuan, Y.; Macquarrie, D. Microwave assisted extraction of sulfated polysaccharides (fucoidan) from Ascophyllum nodosum and its antioxidant activity. Carbohydr. Polym. 2015, 129, 101–107. [Google Scholar] [CrossRef] [PubMed]
- Zhao, Y.L.; Yang, T.; Tong, Y.; Wang, J.; Luan, J.H.; Jiao, Z.B.; Chen, D.; Yang, Y.; Hu, A.; Liu, C.T.; et al. Heterogeneous precipitation behavior and stacking-fault-mediated deformation in a CoCrNi-based medium-entropy alloy. Acta Mater. 2017, 138, 72–82. [Google Scholar] [CrossRef]
| Promoters | Species of Origin | Main Features | References |
|---|---|---|---|
| STP2201 | S. thermophilus | Active in both S. thermophilus and E. coli | [30] |
| P25 | S. thermophilus | Active in both S. thermophilus and E. coli | [31] |
| PprtS | S. thermophilus | Functional in S. thermophilus | [33] |
| Native promoter library-1 | S. thermophilus | 28 promoters with different strengths, static regulation of gene expression level for metabolic engineering | [34] |
| PhlbA | L. delbrueckii subsp. bulgaricus | Strong and constitutive expression in S. thermophilus | [35] |
| PnisA | L. lactis | Inducible by nisin | [36] |
| Phsp16 | S. thermophilus | Inducible by acid shock | [37] |
| Pshsp | S. thermophilus | Inducible by heat shock | [38] |
| Plac | S. thermophilus | Inducible by lactose | [39] |
| Promoters with ComR-box | Streptococci species | Inducible by ComS17–24 | [40] |
| Features | S.thermophilus CNRZ1066 Chromosome, Complete Genome | S. thermophilus JIM 8232, Complete Genome | S. thermophilus LMD-9, Complete Genome | S. thermophilus LMG 18,311 Chromosome, Complete Genome | S. thermophilus ND03 Chromosome, Complete Genome |
|---|---|---|---|---|---|
| Genes | 2000 | 2230 | 1795 | 1973 | 1990 |
| Terminators | 754 | 807 | 721 | 738 | 770 |
| GreatestΔG | 604 | 642 | 569 | 593 | 621 |
| L | 598 | 631 | 566 | 587 | 595 |
| L GreatestΔG | 506 | 535 | 479 | 498 | 509 |
| I | 156 | 176 | 155 | 151 | 175 |
| I greatestΔG | 98 | 107 | 90 | 95 | 112 |
| X | 40 | 43 | 33 | 36 | 40 |
| X greatestΔG | 27 | 31 | 24 | 25 | 33 |
| U | 63 | 75 | 67 | 59 | 64 |
| V | 0 | 0 | 0 | 0 | 0 |
| References | [42] | [42] | [42] | [42] | [42] |
| Plasmid | Replicon (Selection Gene) | Size (kb) | Utilization | References |
|---|---|---|---|---|
| pGKV210 | pND919 (cadA, cadC) | 2.9 | Heavy metal resistance, cadmium resistance | [43] |
| pHRM1 | pSt08 (shsp) | 6.4 | Thermoresistance vector (shsp) | [44] |
| pBUL1 + pSY1 | pSintA1 (thyA) | 8.0 | Thymidylate auxotrophic complementary selection marker | [45] |
| pNZ123 | pRAF301 (aga) | 2.5 | Selection marker for α-galactosidase (aga encoding) | [46] |
| pER8 | 2.2 | Cryptic plasmids of S. thermophilus | [47] | |
| pER371 | 2.7 | |||
| pMEU5 and pMEU6 | pER8 (erm, bla) | 5.7 | Shuttle vector for E. coli and S. thermophilus and other LAB | [48] |
| pMEU9 and pMEU10 | pER8 (erm, bla, cat) | 6.9 | ||
| pG + host9 | pWV01 (Ts) (erm) | 3.7 | Thermosensitive vector for gene inactivation and random insertional mutagenesis | [49] |
| p5aGFP2201a | pMEU5a + pUCP8201 (bla, erm, gfp) | 6.4 | Shuttle vector for E. coli, S. thermophilus and other LAB | [50] |
| pMeu14′-1 | pER371 + pUER28b (bla, erm) | 5.3 | Shuttle vector for E. coli and S. thermophilus | [51] |
| pPC418 | p5aGFP2201a (erm, bla) | 7.9 | Pediocin expression in LAB | [52] |
| pPC318 | p5aGFP2201a (bla) | 9.1 | ||
| pG341a/b | pMEU5a (bla, erm) | 6.4 | Heterologous gene expression in S. thermophilus and E. coli | [53] |
| pSMQ172cat | pSMQ172 (cat) | 5.7 | Theta replication shuttle vector for E. coli, S. thermophilus, S. salivarius, and L. lactis | [54] |
| pINTRS | pWV01 (Ts) (erm) | 5.3 | Food-grade vector for chromosomic insertion of heterologous DNA or gene inactivation | [55] |
| Plasmid Name | Strain | Replication | Size (kb) | (G + C)% | Protein | Gene | Rep Protein | NCBI Accession | References |
|---|---|---|---|---|---|---|---|---|---|
| pSMQ172 | SMQ-172 | RCR | 4.23 | 38 | 4 | 5 | Rep 223 aa | NC_004958.1 | [54] |
| pSMQ-316 | θ | 6.71 | 37.7 | 5 | 5 | NC_010859.1 | [54] | ||
| pCI65st | NDI-6 | RCR | 6.5 | 34.5 | 5 | 5 | RepA 315 aa | AF027167.1 | [57] |
| pND103 | ST2-1 | 3.53 | 32.4 | 4 | 4 | NC_004747.1 | [58] | ||
| pSt0 | St0 | RCR | 8.1 | 37 | 6 | 6 | NC_025154 | [59] | |
| pSt04 | St04 | RCR | 3.1 | AJ242477 | [60] | ||||
| pSt08 | St08 | RCR | 7.51 | 9 | 1 | Rep 313 aa | AJ 239049 | [60] | |
| pSt106 | 5.283 | 36 | 1 | Rep 287 aa | AJ 242479 | [60] | |||
| pJ34 | J34 | RCR | 3.38 | 1 | RepA 315 aa | AJ242475 | [60] | ||
| pSt22-2 | St22 | [60] | |||||||
| pER1-1 | RCR | 3.365 | 2 | 1 | RepA 314 aa | AJ 242476 | [60] | ||
| pER1-2 | 4.45 | 36.9 | 5 | 5 | NC_025196.1 | [60] | |||
| pt38 | ST2783 | 2.91 | 32.4 | 5 | 9 | Rep 311 aa | NC_005098.1 | [61] | |
| pER16 | 4.27 | 3 | Rep 315 aa | AF177166 | [61] | ||||
| pER35 | ST135 | 9.53 | 36.5 | 5 | 5 | RepA 315 aa | NC_000937.1 | [61] | |
| pER36 | ST136 | 3.5 | 34.4 | 2 | 2 | RepA 315 aa | NC_000938.1 | [62] | |
| pK1002C2 | K1002C2 | 3.38 | 35 | 2 | 2 | RepA 314 aa | NC_019231.1 | [63] | |
| pK2007C6 | K2007C6 | 2.98 | 35.1 | 2 | 2 | RepA 314 aa | NC_019232.1 | [63] | |
| pSMQ173b | SMQ-173 | RCR | 4.45 | 37 | 5 | 5 | Rep 146 aa | NC_005323.1 | [64] |
| pSMQ-308 | 8.14 | 37.8 | 6 | 6 | NC_005322.1 | [64] | |||
| pSTER_A | LMD-9 | RCR | 4.45 | 37 | 4 | 4 | NC_008500.1 | [65] | |
| pSTER_B | LMD-9 | RCR | 3.36 | 35.1 | 2 | 2 | Rep 314 aa | NC_008501.1 | [65] |
| pER341 | ST134 | RCR | 2.798 | 33.7 | 2 | AF019139.1 | [65] | ||
| pER371 | ST371 | 2.67 | 38.2 | 3 | 3 | Rep 247 aa | NC_004968.1 | [66] | |
| pER13 | ST113 | RCR | 4.14 | 38.4 | 4 | 4 | RepB 217 aa | NC_002776.1 | [67] |
| pSTHERMO | STH_CIRM_65 | 3.35 | 33.6 | 3 | 3 | Rep | NZ_LR822016.1 | [68] | |
| pST64987 | ST64987 | 7.98 | 37.5 | 7 | 7 | NZ_CP049054.1 | [69] | ||
| paSTHERMO | STH_CIRM_956 | 4.40 | 39.4 | 3 | 3 | Rep | NZ_LR822021.1 | [69] | |
| pbSTHERMO | STH_CIRM_956 | 2.16 | 36.8 | 1 | 1 | Rep | NZ_LR822022.1 | [69] | |
| pSTHERMO | STH_CIRM_998 | 4.40 | 39.3 | 3 | 3 | Rep | NZ_LR822028.1 | [69] | |
| pSTHERMO | STH_CIRM_336 | 3.82 | 33.4 | 2 | 2 | Rep | NZ_LR822018.1 | [69] | |
| pSTHERMO | STH_CIRM_1121 | 3.53 | 32.3 | 2 | 2 | Rep | NZ_LR822038.1 | [69] | |
| pSTHERMO | STH_CIRM_67 | 3.35 | 35.5 | 3 | 3 | Rep | NZ_LR824003.1 | [69] | |
| p.P3A | TK-P3A | 3.50 | 37.3 | 2 | 3 | NZ_CP045597.1 | [69] | ||
| p202_03 | MAG_rmk202_sterm | 14.14 | 36.1 | 15 | 17 | NZ_CP046135.1 | [69] | ||
| p1 | TH-4 | 3.36 | 35.1 | 2 | 3 | Rep | NZ_CP102539.1 | [69] | |
| p2 | TH-4 | 4.45 | 37 | 5 | 5 | NZ_CP102540.1 | [69] | ||
| paSTHERMO | STH_CIRM_368 | 4.45 | 39.4 | 3 | 3 | NZ_LR822024.1 | [70] |
| Bacterial Chassis | Peptide | Wild-Type/Gene Source | Expression Details | Purpose | Reference |
|---|---|---|---|---|---|
| S. thermophilus LMD9 | L-arabinose isomerase | Geobacillus stearothermophilus | The plasmid pMR4 | Diabetic patients | [32] |
| S. thermophilus ST128 | PepA-D or papA-D (pediocin operon) | Pediococcus acidilactici | NICE system, under PnisA control | Against Listeria in dairy food | [36] |
| S. thermophilus ST128 | Tyrosinase | S. antibioticus | Cloning vector pIL253 | The synthesis of tyrosinase protein by genetic transformants | [82] |
| S. thermophilus SMQ-301 | Galactokinase | S. salivarius | The plasmid pTRKL2TK, galK (galactokinase) and galM (mutarotase) | Galactose reduction in dairy products | [83] |
| S. thermophilus | Serine hydroxymethyltransferase (SHMT) | S. thermophilus NIZOB505 | Promoter PLacA, the glyA gene | Control and improvement of acetaldehyde production in fermented (dairy) products with S. thermophilus as starter culture | [84] |
| Streptococcus thermophilus ST128 | Bioactive peptides BL-11 and C-12 | Bos taurus | The ST2201 promoter | Bioactive peptides from milk proteins | [85] |
| Streptococcus thermophilus CRL 807 | Superoxide dismutase and catalase | L. casei | The plasmid pIL253-sodA, pIL253-mnkat | Antioxidant enzyme production to confer anti-inflammatory potential | [86] |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhao, R.; Chen, Z.; Liang, J.; Dou, J.; Guo, F.; Xu, Z.; Wang, T. Advances in Genetic Tools and Their Application in Streptococcus thermophilus. Foods 2023, 12, 3119. https://doi.org/10.3390/foods12163119
Zhao R, Chen Z, Liang J, Dou J, Guo F, Xu Z, Wang T. Advances in Genetic Tools and Their Application in Streptococcus thermophilus. Foods. 2023; 12(16):3119. https://doi.org/10.3390/foods12163119
Chicago/Turabian StyleZhao, Ruiting, Zouquan Chen, Jie Liang, Jiaxin Dou, Fangyu Guo, Zhenshang Xu, and Ting Wang. 2023. "Advances in Genetic Tools and Their Application in Streptococcus thermophilus" Foods 12, no. 16: 3119. https://doi.org/10.3390/foods12163119
APA StyleZhao, R., Chen, Z., Liang, J., Dou, J., Guo, F., Xu, Z., & Wang, T. (2023). Advances in Genetic Tools and Their Application in Streptococcus thermophilus. Foods, 12(16), 3119. https://doi.org/10.3390/foods12163119





