Metagenomic Insights into the Microbiome and Resistance Genes of Traditional Fermented Foods in Arabia
Abstract
1. Introduction
2. Materials and Methods
2.1. 16S Amplicon Sequencing and Bioinformatics Analysis
2.2. Shotgun Sequencing and Bioinformatics Analyses
3. Results
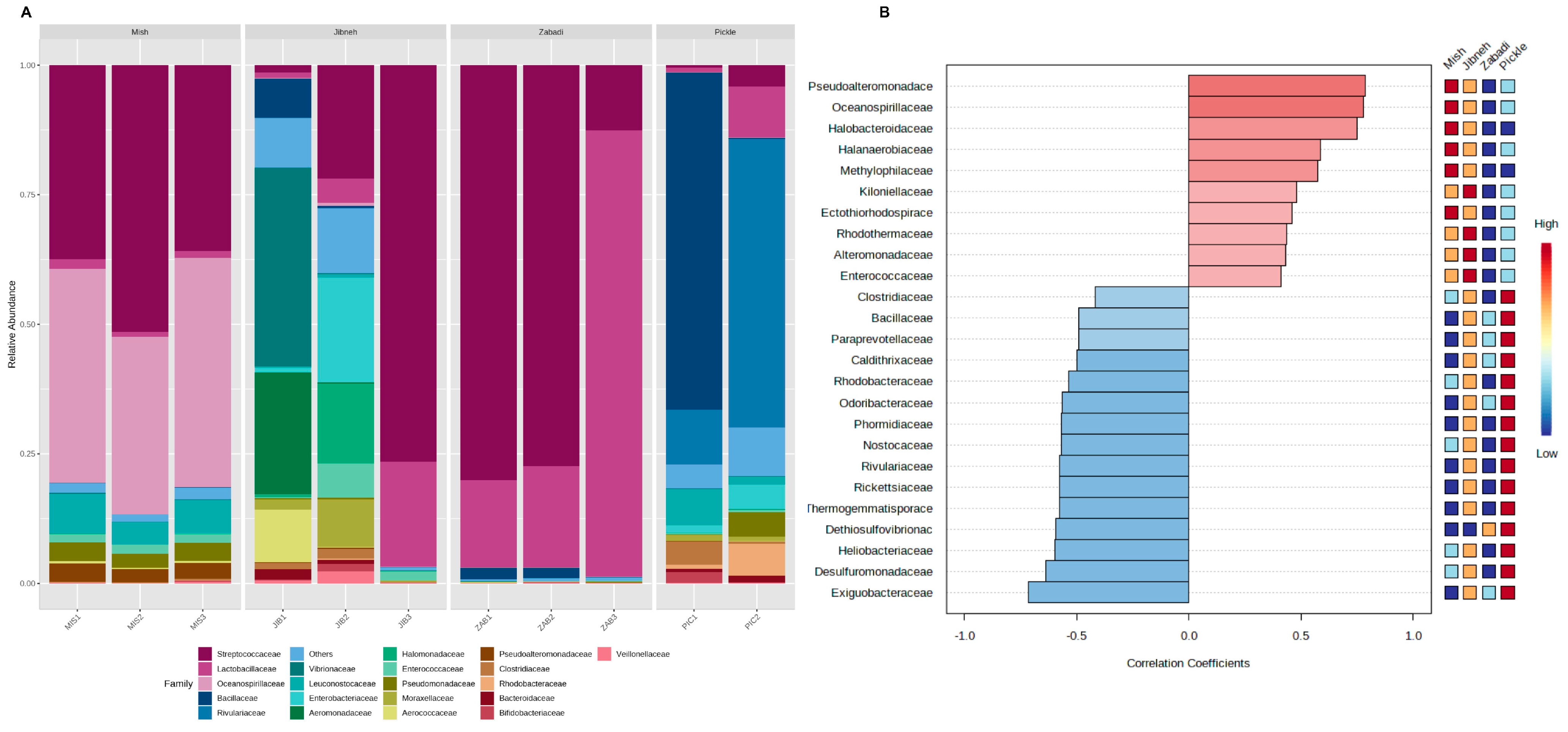
3.1. 16S Amplicon Sequencing Taxonomic Analysis
3.2. Taxonomy of Shotgun Metagenomic Sequencing
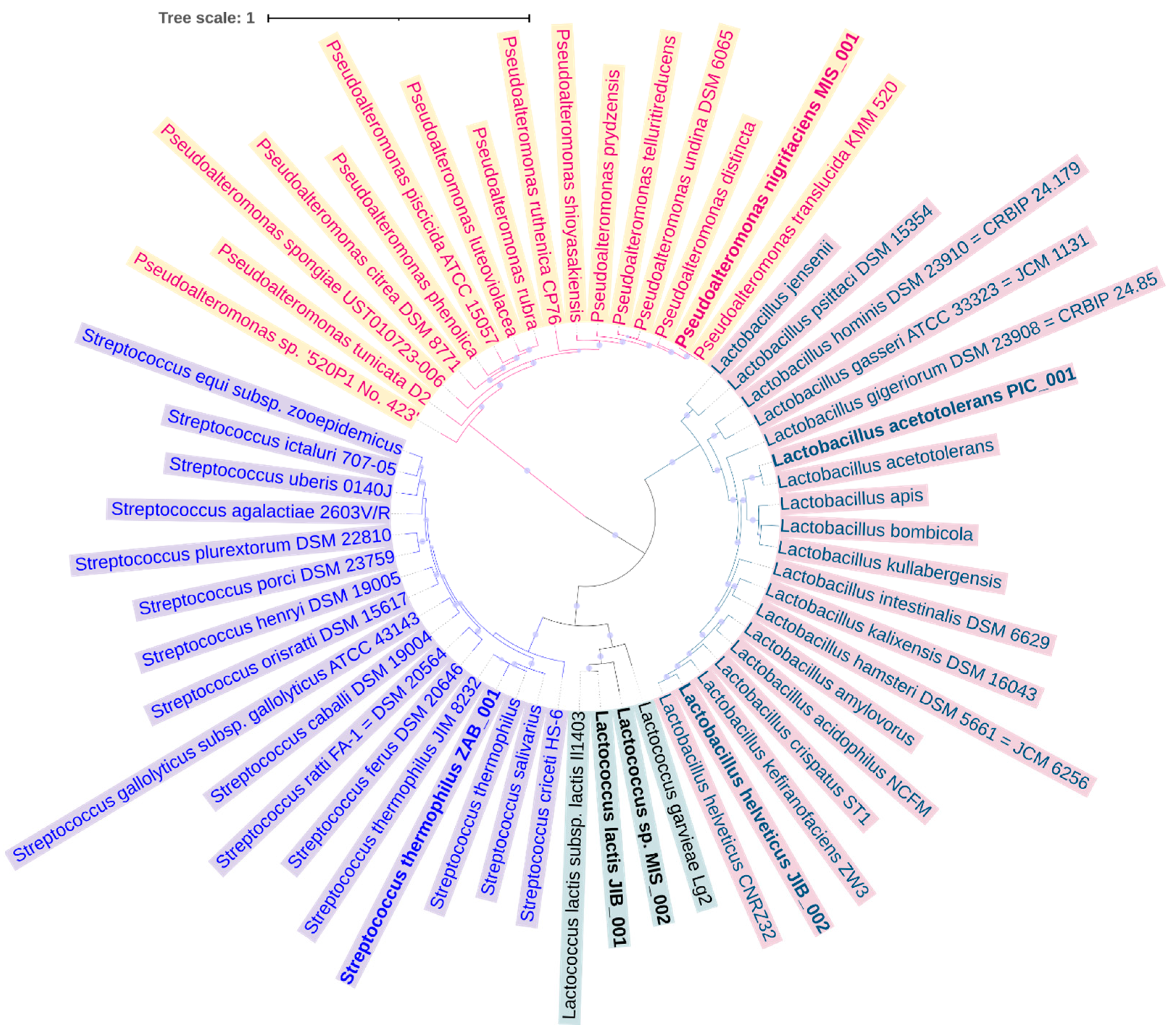
3.3. Binning and Genome Construction
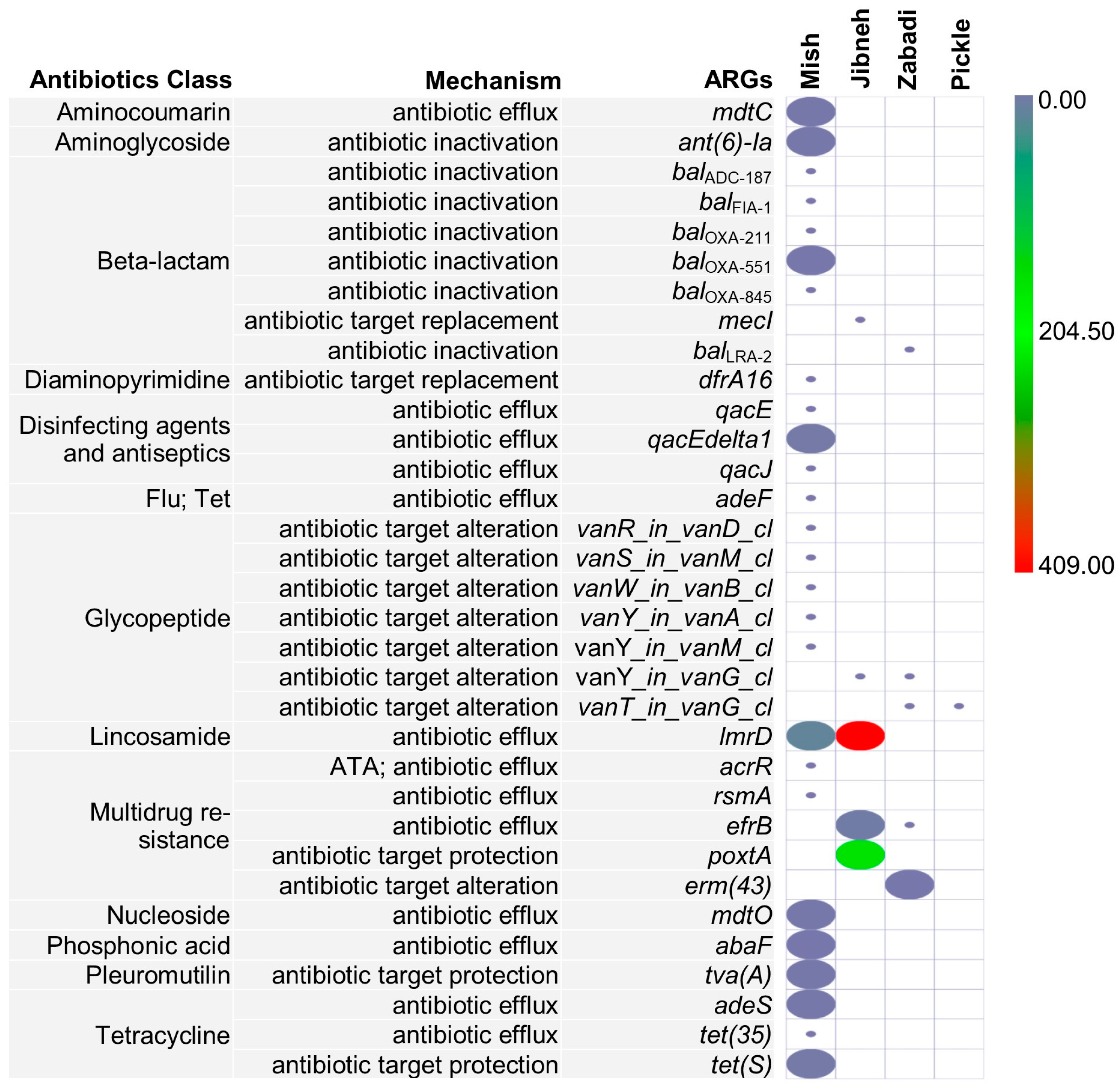
3.4. ARGs and Mobile Genetic Elements
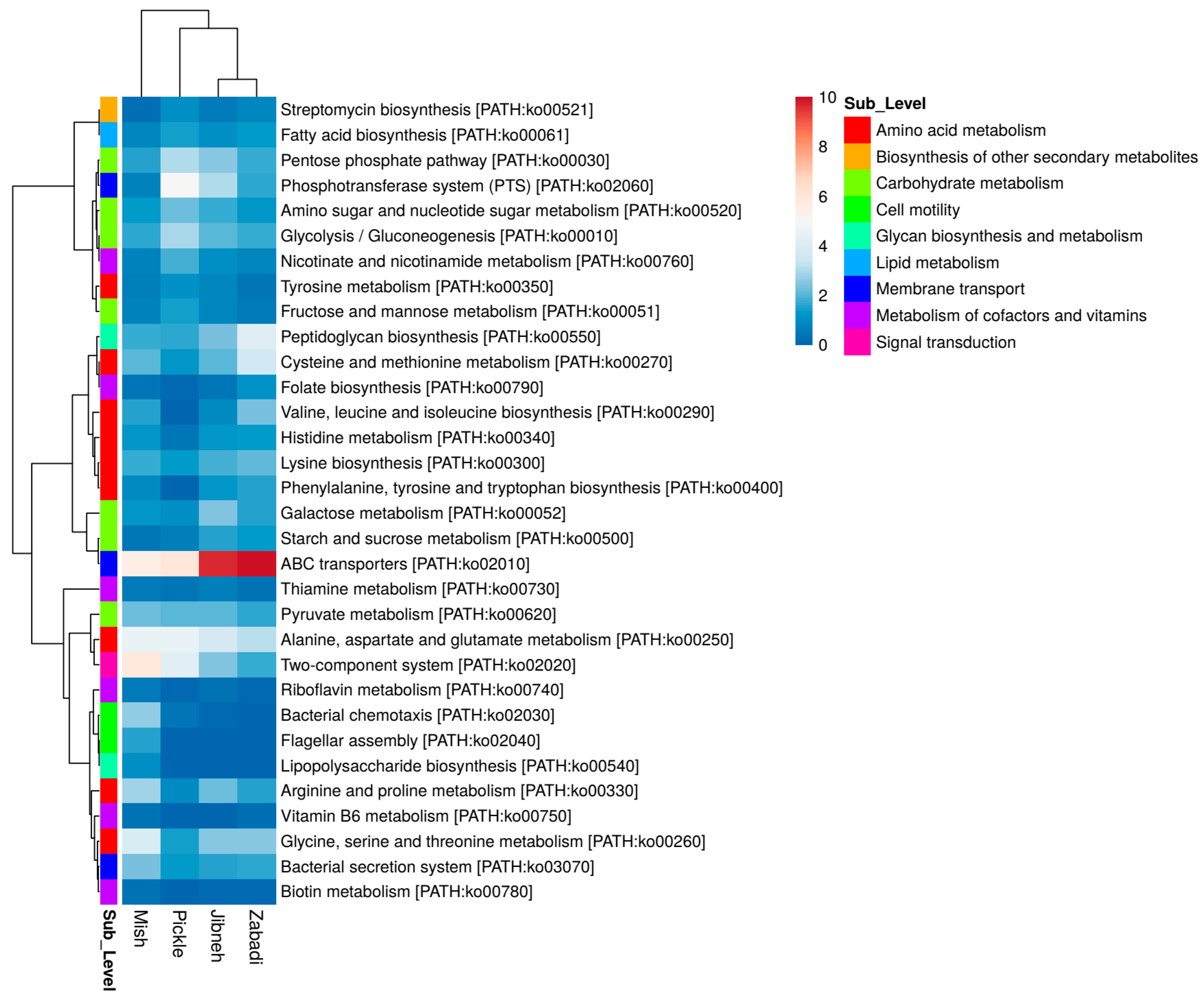
3.5. Functional Analysis of Fermented Foods Metagenomes
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Wilson, R.T. Traditional milk processing and value-added dairy products in selected Arab countries. Int. J. Dairy Technol. 2017, 70, 307–319. [Google Scholar] [CrossRef]
- Yasir, M.; Bibi, F.; Hashem, A.M.; Azhar, E.I. Comparative metagenomics and characterization of antimicrobial resistance genes in pasteurized and homemade fermented Arabian laban. Food Res. Int. 2020, 137, 109639. [Google Scholar] [CrossRef] [PubMed]
- Bin Masalam, M.S.; Bahieldin, A.; Alharbi, M.G.; Al-Masaudi, S.; Al-Jaouni, S.K.; Harakeh, S.M.; Al-Hindi, R.R. Isolation, Molecular Characterization and Probiotic Potential of Lactic Acid Bacteria in Saudi Raw and Fermented Milk. Evid. Based Complement. Altern. Med. 2018, 2018, 7970463. [Google Scholar] [CrossRef] [PubMed]
- Macori, G.; Cotter, P.D. Novel insights into the microbiology of fermented dairy foods. Curr. Opin. Biotechnol. 2018, 49, 172–178. [Google Scholar] [CrossRef]
- Ramos, C.L.; Bressani, A.P.P.; Batista, N.N.; Martinez, S.J.; Dias, D.R.; Schwan, R.F. Indigenous fermented foods: Nutritional and safety aspects. Curr. Opin. Food Sci. 2023, 53, 101075. [Google Scholar] [CrossRef]
- Leech, J.; Cabrera-Rubio, R.; Walsh, A.M.; Macori, G.; Walsh, C.J.; Barton, W.; Finnegan, L.; Crispie, F.; O’Sullivan, O.; Claesson, M.J.; et al. Fermented-Food Metagenomics Reveals Substrate-Associated Differences in Taxonomy and Health-Associated and Antibiotic Resistance Determinants. mSystems 2020, 5, 10-1128. [Google Scholar] [CrossRef]
- Yasir, M.; Al-Zahrani, I.A.; Bibi, F.; Abd El Ghany, M.; Azhar, E.I. New insights of bacterial communities in fermented vegetables from shotgun metagenomics and identification of antibiotic resistance genes and probiotic bacteria. Food Res. Int. 2022, 157, 111190. [Google Scholar] [CrossRef]
- Hansen, E.B. Commercial bacterial starter cultures for fermented foods of the future. Int. J. Food Microbiol. 2002, 78, 119–131. [Google Scholar] [CrossRef]
- Escobar-Zepeda, A.; Sanchez-Flores, A.; Quirasco Baruch, M. Metagenomic analysis of a Mexican ripened cheese reveals a unique complex microbiota. Food Microbiol. 2016, 57, 116–127. [Google Scholar] [CrossRef]
- Ferrocino, I.; Bellio, A.; Giordano, M.; Macori, G.; Romano, A.; Rantsiou, K.; Decastelli, L.; Cocolin, L. Shotgun Metagenomics and Volatilome Profile of the Microbiota of Fermented Sausages. Appl. Environ. Microbiol. 2018, 84, e02120-17. [Google Scholar] [CrossRef]
- Liu, X.; Kuda, T.; Takahashi, H.; Kimura, B. Bacterial and fungal microbiota of spontaneously fermented Chinese products, Rubing milk cake and Yan-cai vegetable pickles. Food Microbiol. 2018, 72, 106–111. [Google Scholar] [CrossRef] [PubMed]
- Dimidi, E.; Cox, S.R.; Rossi, M.; Whelan, K. Fermented Foods: Definitions and Characteristics, Impact on the Gut Microbiota and Effects on Gastrointestinal Health and Disease. Nutrients 2019, 11, 1806. [Google Scholar] [CrossRef] [PubMed]
- Leeuwendaal, N.K.; Stanton, C.; O’Toole, P.W.; Beresford, T.P. Fermented Foods, Health and the Gut Microbiome. Nutrients 2022, 14, 1527. [Google Scholar] [CrossRef]
- Wolter, M.; Grant, E.T.; Boudaud, M.; Steimle, A.; Pereira, G.V.; Martens, E.C.; Desai, M.S. Leveraging diet to engineer the gut microbiome. Nat. Rev. Gastroenterol. Hepatol. 2021, 18, 885–902. [Google Scholar] [CrossRef] [PubMed]
- Clemente, J.C.; Manasson, J.; Scher, J.U. The role of the gut microbiome in systemic inflammatory disease. BMJ 2018, 360, j5145. [Google Scholar] [CrossRef] [PubMed]
- Khan, I.; Yasir, M.; Azhar, E.I.; Kumosani, T.; Barbour, E.K.; Bibi, F.; Kamal, M.A. Implication of gut microbiota in human health. CNS Neurol. Disord. Drug Targets 2014, 13, 1325–1333. [Google Scholar] [CrossRef]
- Kleerebezem, M.; Vaughan, E.E. Probiotic and gut lactobacilli and bifidobacteria: Molecular approaches to study diversity and activity. Annu. Rev. Microbiol. 2009, 63, 269–290. [Google Scholar] [CrossRef]
- LeBlanc, J.G.; Milani, C.; de Giori, G.S.; Sesma, F.; van Sinderen, D.; Ventura, M. Bacteria as vitamin suppliers to their host: A gut microbiota perspective. Curr. Opin. Biotechnol. 2013, 24, 160–168. [Google Scholar] [CrossRef]
- Veiga, P.; Pons, N.; Agrawal, A.; Oozeer, R.; Guyonnet, D.; Brazeilles, R.; Faurie, J.M.; van Hylckama Vlieg, J.E.; Houghton, L.A.; Whorwell, P.J.; et al. Changes of the human gut microbiome induced by a fermented milk product. Sci. Rep. 2014, 4, 6328. [Google Scholar] [CrossRef]
- Dirar, H.A. The Indigenous Fermented Foods of the Sudan: A Study in African Food and Nutrition; CAB International: Wallingford, UK, 1993. [Google Scholar]
- El Zubeir, I.E.M.; Abdalla, W.M.; El Owni, O.A.O. Chemical composition of fermented milk (roub and mish) in Sudan. Food Control 2005, 16, 633–637. [Google Scholar] [CrossRef]
- Abd Elkader Mahmoud Abd El latif, R.; El Sherbini El-sayed Ali, M.; Abdelkhalek, A. Prevalence and Characterization of Some Pathogenic Bacteria in Fermented Milk Products and Mish Cheese in Dakahalia Governorate, Egypt. J. Adv. Vet. Res. 2022, 12, 446–450. [Google Scholar]
- Kazimierczak, K.A.; Scott, K.P. Antibiotics and resistance genes: Influencing the microbial ecosystem in the gut. Adv. Appl. Microbiol. 2007, 62, 269–292. [Google Scholar] [CrossRef] [PubMed]
- Khan, T.J.; Ahmed, Y.M.; Zamzami, M.A.; Mohamed, S.A.; Khan, I.; Baothman, O.A.S.; Mehanna, M.G.; Yasir, M. Effect of atorvastatin on the gut microbiota of high fat diet-induced hypercholesterolemic rats. Sci. Rep. 2018, 8, 662. [Google Scholar] [CrossRef] [PubMed]
- Wang, Q.; Garrity, G.M.; Tiedje, J.M.; Cole, J.R. Naive Bayesian classifier for rapid assignment of rRNA sequences into the new bacterial taxonomy. Appl. Environ. Microbiol. 2007, 73, 5261–5267. [Google Scholar] [CrossRef]
- Chong, J.; Liu, P.; Zhou, G.; Xia, J. Using MicrobiomeAnalyst for comprehensive statistical, functional, and meta-analysis of microbiome data. Nat. Protoc. 2020, 15, 799–821. [Google Scholar] [CrossRef]
- Arkin, A.P.; Cottingham, R.W.; Henry, C.S.; Harris, N.L.; Stevens, R.L.; Maslov, S.; Dehal, P.; Ware, D.; Perez, F.; Canon, S.; et al. KBase: The United States Department of Energy Systems Biology Knowledgebase. Nat. Biotechnol. 2018, 36, 566–569. [Google Scholar] [CrossRef]
- Keegan, K.P.; Glass, E.M.; Meyer, F. MG-RAST, a Metagenomics Service for Analysis of Microbial Community Structure and Function. Methods Mol. Biol. 2016, 1399, 207–233. [Google Scholar] [CrossRef]
- Menzel, P.; Ng, K.L.; Krogh, A. Fast and sensitive taxonomic classification for metagenomics with Kaiju. Nat. Commun. 2016, 7, 11257. [Google Scholar] [CrossRef]
- Bolger, A.M.; Lohse, M.; Usadel, B. Trimmomatic: A flexible trimmer for Illumina sequence data. Bioinformatics 2014, 30, 2114–2120. [Google Scholar] [CrossRef]
- Meyer, F.; Paarmann, D.; D’Souza, M.; Olson, R.; Glass, E.M.; Kubal, M.; Paczian, T.; Rodriguez, A.; Stevens, R.; Wilke, A.; et al. The metagenomics RAST server—A public resource for the automatic phylogenetic and functional analysis of metagenomes. BMC Bioinform. 2008, 9, 386. [Google Scholar] [CrossRef]
- Kalantar, K.L.; Carvalho, T.; de Bourcy, C.F.A.; Dimitrov, B.; Dingle, G.; Egger, R.; Han, J.; Holmes, O.B.; Juan, Y.F.; King, R.; et al. IDseq-An open source cloud-based pipeline and analysis service for metagenomic pathogen detection and monitoring. Gigascience 2020, 9, giaa111. [Google Scholar] [CrossRef] [PubMed]
- Aldrete-Tapia, A.; Escobar-Ramírez, M.C.; Tamplin, M.L.; Hernández-Iturriaga, M. High-throughput sequencing of microbial communities in Poro cheese, an artisanal Mexican cheese. Food Microbiol. 2014, 44, 136–141. [Google Scholar] [CrossRef] [PubMed]
- Quigley, L.; O’Sullivan, O.; Beresford, T.P.; Ross, R.P.; Fitzgerald, G.F.; Cotter, P.D. High-throughput sequencing for detection of subpopulations of bacteria not previously associated with artisanal cheeses. Appl. Environ. Microbiol. 2012, 78, 5717–5723. [Google Scholar] [CrossRef]
- Li, J.; Zheng, Y.; Xu, H.; Xi, X.; Hou, Q.; Feng, S.; Wuri, L.; Bian, Y.; Yu, Z.; Kwok, L.Y.; et al. Bacterial microbiota of Kazakhstan cheese revealed by single molecule real time (SMRT) sequencing and its comparison with Belgian, Kalmykian and Italian artisanal cheeses. BMC Microbiol. 2017, 17, 13. [Google Scholar] [CrossRef]
- Elyas, Y.Y.A.; Yousif, N.M.E.; Ahmed, I.A.M. Screening of lactic acid bacteria from Sudanese fermented foods for bacteriocin production. J. Microbiol. Biotechnol. Food Sci. 2015, 4, 373–378. [Google Scholar] [CrossRef]
- Ahmed, N. Evaluation of Microbiological Quality of Sudanese Fermented Dairy Product ‘Mish’ During Storage. Adv. J. Food Sci. Technol 2010, 2, 155–158. [Google Scholar]
- Abdalla, W.M.; Zubeir, I.E.M.E. Microbial Hazards Associated with Fermented Milk (Roub and Mish) Processing in Sudan. Int. J. Dairy Sci. 2006, 1, 21–26. [Google Scholar] [CrossRef]
- Samelis, J.; Doulgeraki, A.I.; Bikouli, V.; Pappas, D.; Kakouri, A. Microbiological and metagenomic characterization of a retail delicatessen galotyri-like fresh acid-curd cheese product. Fermentation 2021, 7, 67. [Google Scholar] [CrossRef]
- Kõiv, V.; Adamberg, K.; Adamberg, S.; Sumeri, I.; Kasvandik, S.; Kisand, V.; Maiväli, Ü.; Tenson, T. Microbiome of root vegetables-a source of gluten-degrading bacteria. Appl. Microbiol. Biotechnol. 2020, 104, 8871–8885. [Google Scholar] [CrossRef]
- Pakwan, C.; Chitov, T.; Chantawannakul, P.; Manasam, M.; Bovonsombut, S.; Disayathanoowat, T. Bacterial compositions of indigenous Lanna (Northern Thai) fermented foods and their potential functional properties. PLoS ONE 2020, 15, e0242560. [Google Scholar] [CrossRef]
- Kothe, C.I.; Bolotin, A.; Kraiem, B.F.; Dridi, B.; Food Microbiome, T.; Renault, P. Unraveling the world of halophilic and halotolerant bacteria in cheese by combining cultural, genomic and metagenomic approaches. Int. J. Food Microbiol. 2021, 358, 109312. [Google Scholar] [CrossRef] [PubMed]
- Furtado, D.N.; Todorov, S.D.; Landgraf, M.; Destro, M.T.; Franco, B.D. Bacteriocinogenic Lactococcus lactis subsp. lactis DF04Mi isolated from goat milk: Evaluation of the probiotic potential. Braz. J. Microbiol. [Publ. Braz. Soc. Microbiol.] 2014, 45, 1047–1054. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Han, F.; Wu, G.; Zhang, Y.; Zheng, H.; Han, S.; Li, X.; Cai, W.; Liu, J.; Zhang, W.; Zhang, X.; et al. Streptococcus thermophilus Attenuates Inflammation in Septic Mice Mediated by Gut Microbiota. Front. Microbiol. 2020, 11, 598010. [Google Scholar] [CrossRef] [PubMed]
- Sugimura, T.; Jounai, K.; Ohshio, K.; Tanaka, T.; Suwa, M.; Fujiwara, D. Immunomodulatory effect of Lactococcus lactis JCM5805 on human plasmacytoid dendritic cells. Clin. Immunol. 2013, 149, 509–518. [Google Scholar] [CrossRef]
- Zielińska, D.; Długosz, E.; Zawistowska-Deniziak, A. Functional Properties of Food Origin Lactobacillus in the Gastrointestinal Ecosystem-In Vitro Study. Probiot. Antimicrob. Proteins 2019, 11, 820–829. [Google Scholar] [CrossRef]
- MIR. (Mordor Intelligence Report). Pickles and Pickle Products Market—Growth, Trends, COVID-19 Impact, and Forecasts (2022–2027). 2020. Available online: https://www.mordorintelligence.com/industry-reports/pickles-and-pickle-products-market (accessed on 17 February 2022).
- Toth, A.G.; Csabai, I.; Maroti, G.; Jerzsele, A.; Dubecz, A.; Patai, A.V.; Judge, M.F.; Nagy, S.A.; Makrai, L.; Banyai, K.; et al. A glimpse of antimicrobial resistance gene diversity in kefir and yoghurt. Sci. Rep. 2020, 10, 22458. [Google Scholar] [CrossRef]
- Ma, Q.; Fu, Y.; Sun, H.; Huang, Y.; Li, L.; Yu, Q.; Dinnyes, A.; Sun, Q. Antimicrobial resistance of Lactobacillus spp. from fermented foods and human gut. LWT 2017, 86, 201–208. [Google Scholar] [CrossRef]
- Hill, M.J. Intestinal flora and endogenous vitamin synthesis. Eur. J. Cancer Prev. Off. J. Eur. Cancer Prev. Organ. (ECP) 1997, 6 (Suppl. 1), S43–S45. [Google Scholar] [CrossRef]
- Giello, M.; La Storia, A.; De Filippis, F.; Ercolini, D.; Villani, F. Impact of Lactobacillus curvatus 54M16 on microbiota composition and growth of Listeria monocytogenes in fermented sausages. Food Microbiol. 2018, 72, 1–15. [Google Scholar] [CrossRef]
- Singh, N.; Gurav, A.; Sivaprakasam, S.; Brady, E.; Padia, R.; Shi, H.; Thangaraju, M.; Prasad, P.D.; Manicassamy, S.; Munn, D.H.; et al. Activation of Gpr109a, receptor for niacin and the commensal metabolite butyrate, suppresses colonic inflammation and carcinogenesis. Immunity 2014, 40, 128–139. [Google Scholar] [CrossRef]
- Zhang, J.; Wang, X.; Huo, D.; Li, W.; Hu, Q.; Xu, C.; Liu, S.; Li, C. Metagenomic approach reveals microbial diversity and predictive microbial metabolic pathways in Yucha, a traditional Li fermented food. Sci. Rep. 2016, 6, 32524. [Google Scholar] [CrossRef] [PubMed]
- Martens, J.H.; Barg, H.; Warren, M.J.; Jahn, D. Microbial production of vitamin B12. Appl. Microbiol. Biotechnol. 2002, 58, 275–285. [Google Scholar] [CrossRef]
- Noda, H.; Akasaka, N.; Ohsugi, M. Biotin production by bifidobacteria. J. Nutr. Sci. Vitaminol. 1994, 40, 181–188. [Google Scholar] [CrossRef]
- Pompei, A.; Cordisco, L.; Amaretti, A.; Zanoni, S.; Matteuzzi, D.; Rossi, M. Folate production by bifidobacteria as a potential probiotic property. Appl. Environ. Microbiol. 2007, 73, 179–185. [Google Scholar] [CrossRef] [PubMed]
- Wu, H.; Ito, K.; Shimoi, H. Identification and characterization of a novel biotin biosynthesis gene in Saccharomyces cerevisiae. Appl. Environ. Microbiol. 2005, 71, 6845–6855. [Google Scholar] [CrossRef] [PubMed]
- Ruzzo, E.K.; Capo-Chichi, J.M.; Ben-Zeev, B.; Chitayat, D.; Mao, H.; Pappas, A.L.; Hitomi, Y.; Lu, Y.F.; Yao, X.; Hamdan, F.F.; et al. Deficiency of asparagine synthetase causes congenital microcephaly and a progressive form of encephalopathy. Neuron 2013, 80, 429–441. [Google Scholar] [CrossRef]


Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Yasir, M.; Alkhaldy, A.A.; Soliman, S.A.; Turkistani, S.A.; Azhar, E.I. Metagenomic Insights into the Microbiome and Resistance Genes of Traditional Fermented Foods in Arabia. Foods 2023, 12, 3342. https://doi.org/10.3390/foods12183342
Yasir M, Alkhaldy AA, Soliman SA, Turkistani SA, Azhar EI. Metagenomic Insights into the Microbiome and Resistance Genes of Traditional Fermented Foods in Arabia. Foods. 2023; 12(18):3342. https://doi.org/10.3390/foods12183342
Chicago/Turabian StyleYasir, Muhammad, Areej A. Alkhaldy, Samah Abdullah Soliman, Safaa A. Turkistani, and Esam I. Azhar. 2023. "Metagenomic Insights into the Microbiome and Resistance Genes of Traditional Fermented Foods in Arabia" Foods 12, no. 18: 3342. https://doi.org/10.3390/foods12183342
APA StyleYasir, M., Alkhaldy, A. A., Soliman, S. A., Turkistani, S. A., & Azhar, E. I. (2023). Metagenomic Insights into the Microbiome and Resistance Genes of Traditional Fermented Foods in Arabia. Foods, 12(18), 3342. https://doi.org/10.3390/foods12183342







