Tracking Microbial Diversity and Hygienic-Sanitary Status during Processing of Farmed Rainbow Trout (Oncorhynchus mykiss)
Abstract
:1. Introduction
2. Materials and Methods
2.1. Sampling and Experimental Design
2.2. Physicochemical Analysis
2.3. Microbial Analysis
2.4. Isolation and Molecular Identification
2.5. Statistical Analysis
3. Results and Discussion
3.1. Physicochemical Assessment
3.2. Microbiological Assessment
3.2.1. Water Samples
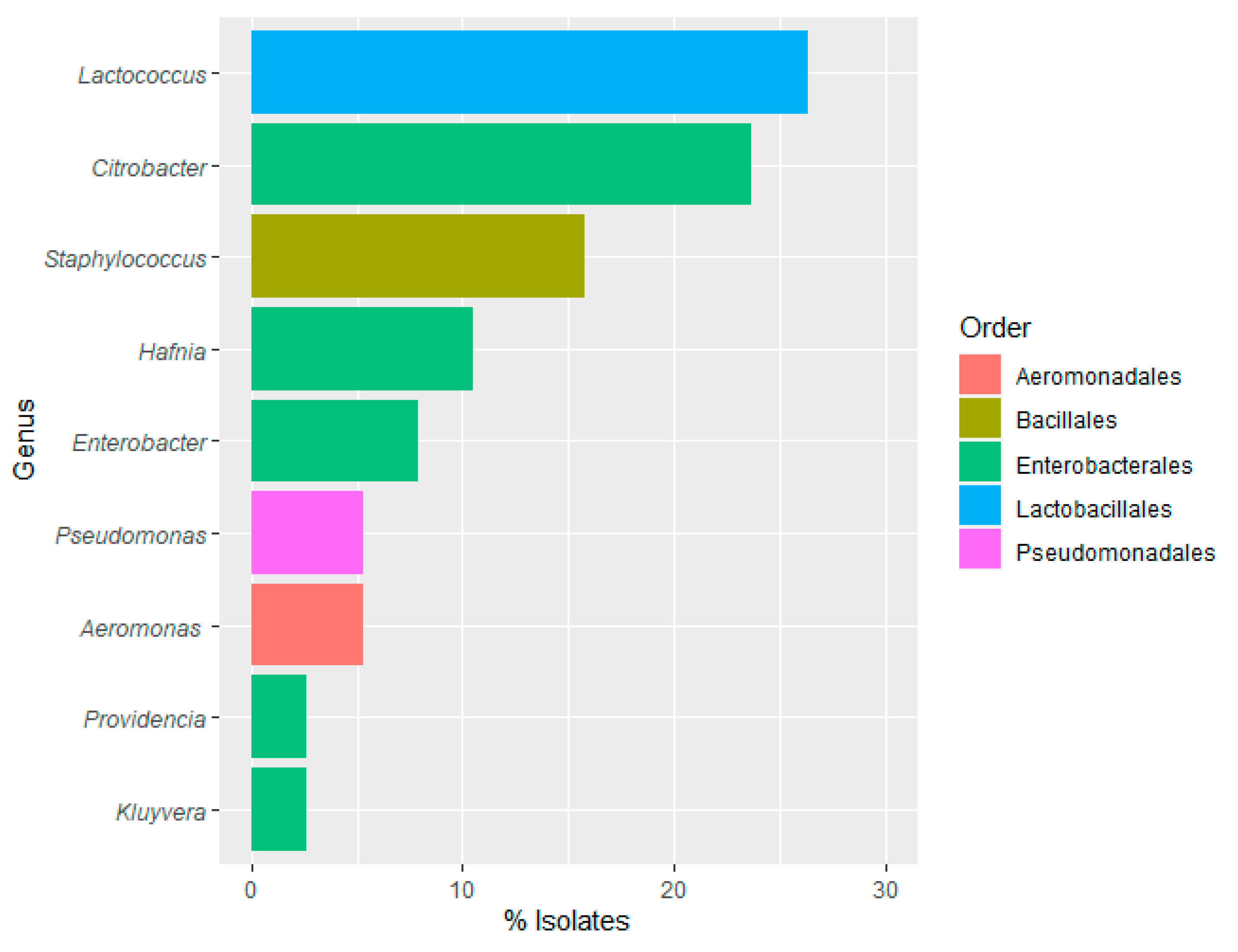
Bacterial Genus Presence in Water Samples
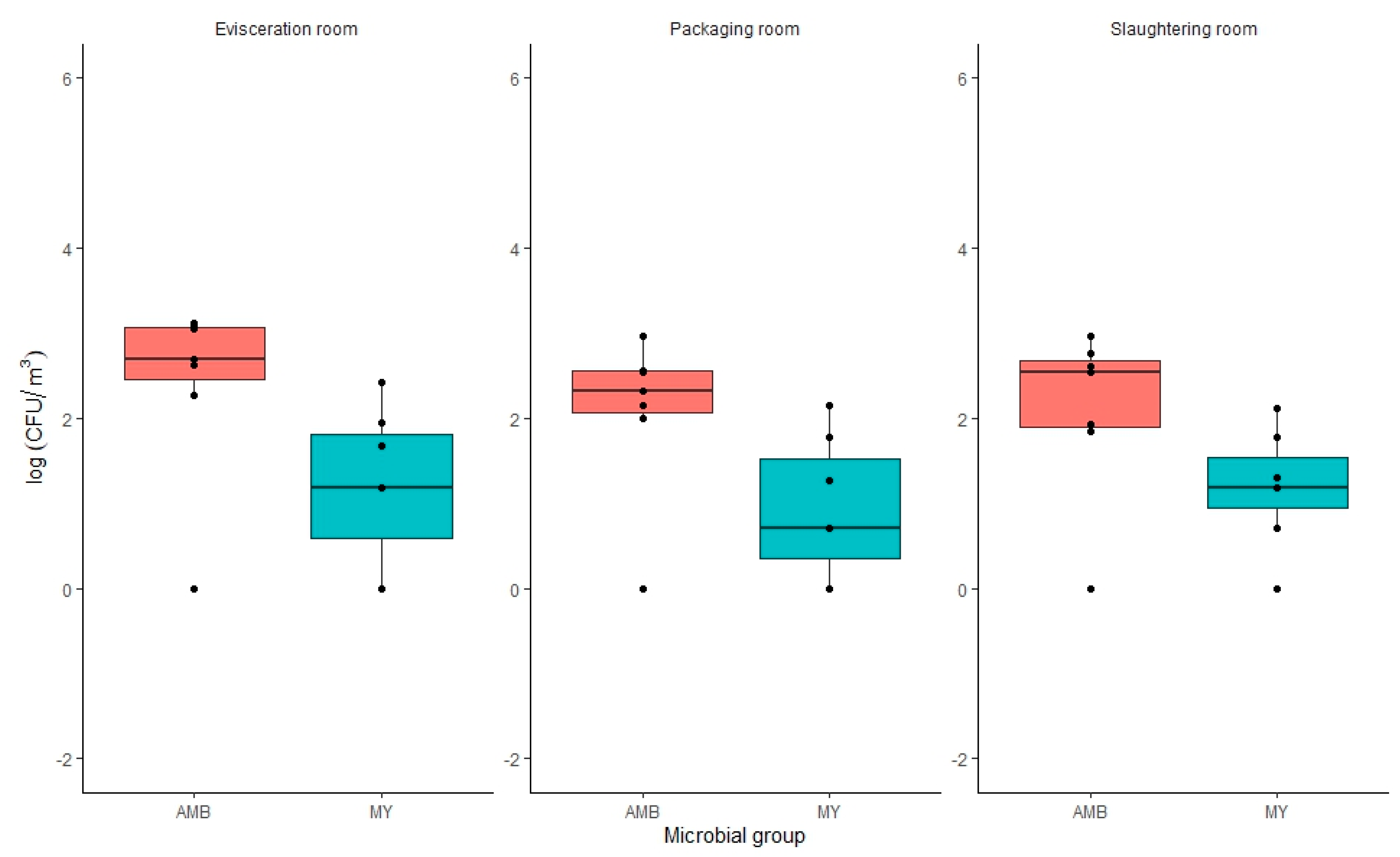
3.2.2. Air Samples
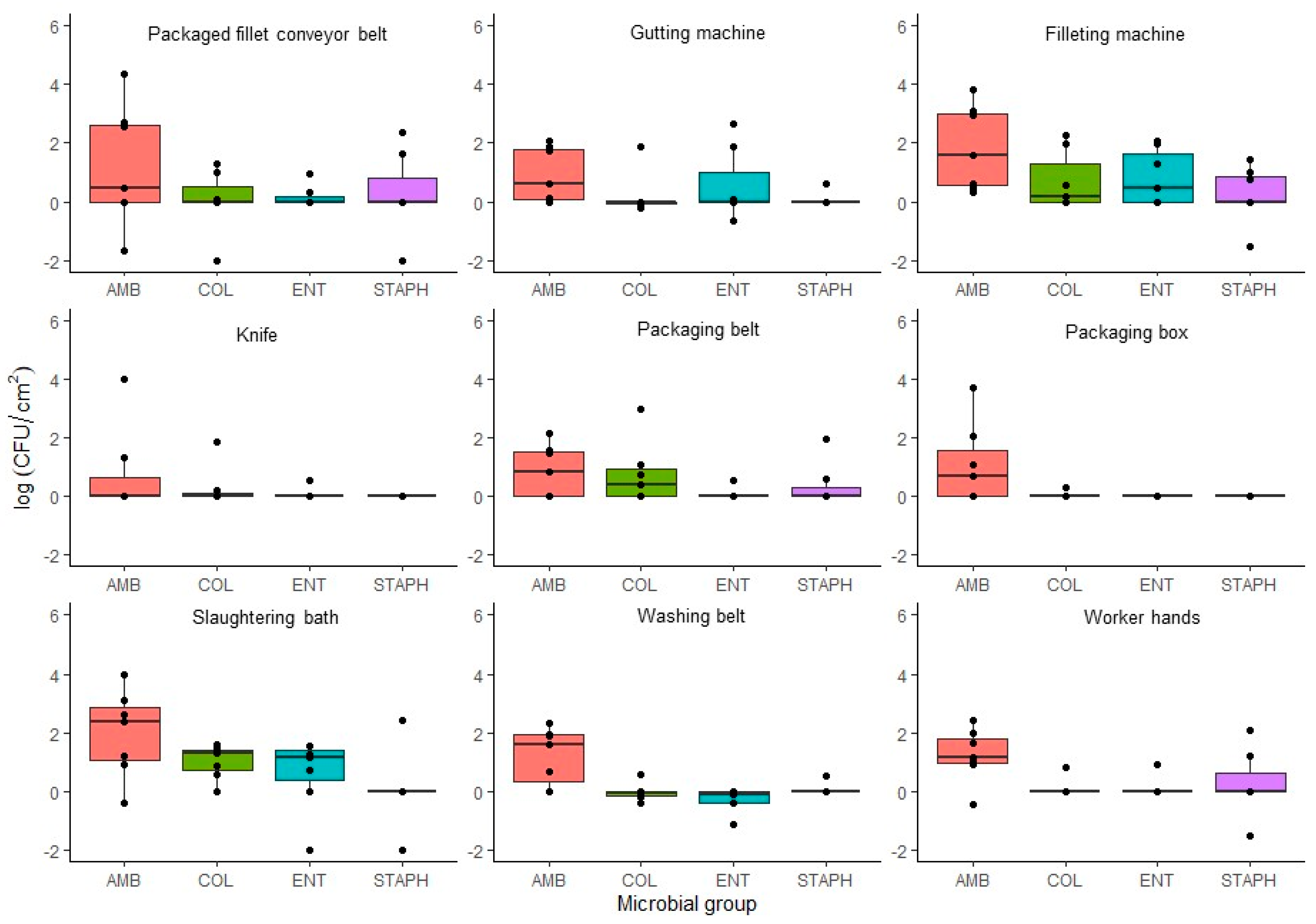
3.2.3. Food-Contact Surfaces (FCS) Samples
Bacterial Genus Presence in Food-Contact Surfaces
3.3. Rainbow Trout Samples
Bacterial Genus Presence in Rainbow Trout Samples
3.4. Distribution of Bacterial Genus among Type of Samples
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Spanish Aquaculture Producers Association (APROMAR-OPP30). Aquaculture in Spain 2021. Available online: https://apromar.es/wp-content/uploads/2021/12/La-Acuicultura-en-Espana-2021.pdf (accessed on 7 September 2023).
- Valero Gaspar, T.; Rodríguez Alonso, P.; Ruíz Moreno, E.; Ávila Torres, J.M.; Valera Moreiras, G. Rainbow trout (Oncorhynchus mykiss). In La Alimentación Española. Características Nutricionales de los Principales Alimentos de Nuestra Dieta, 2nd ed.; Ministerio de Agricultura, Pesca y Alimentación, Secretaría General Técnica, Centro de Publicaciones, Imprenta ROAL, S.L.: Madrid, Spain, 2018; pp. 519–520. ISBN 978-84-491-1506-6. Available online: https://www.fen.org.es/storage/app/media/imgPublicaciones/2018/libro-la-alimentacion-espanola.pdf (accessed on 6 October 2023).
- Spanish Aquaculture Producers Association (APROMAR-OPP30). Aquaculture in Spain 2022. 2022. Available online: https://apromar.es/wp-content/uploads/2022/10/Aquaculture-in-Spain-2022_APROMAR.pdf (accessed on 4 July 2023).
- Food and Agriculture Organization of the United Nations (F.A.O.). Practical Manual for Rainbow Trout Farming. 2014. Available online: http://www.fao.org/3/bc354s/bc354s.pdf (accessed on 7 September 2023).
- Barba Salcedo, R.; Martínez Pérez, J.M. La Acuicultura Continental en Andalucía; Consejería de Medio Ambiente de la Junta de Andalucía: Andalucía, Spain, 2010; ISBN 9788495785435. Available online: https://www.juntadeandalucia.es/medioambiente/consolidado/publicacionesdigitales/CA-220-4_LA_ACUICULTURA_CONTINENTAL_EN_ANDALUCIA/CA-220-4.htm (accessed on 6 October 2023).
- Roberts, T.A.; Cordier, J.L.; Gram, L.; Tompkin, R.B.; Pitt, J.I.; Gorris, L.G.M.; Swanson, K.M.J. Microorganisms in Foods 6: Microbial Ecology of Food Commodities, 2nd ed.; Springer: London, UK, 2005. [Google Scholar]
- Austin, B. The bacterial microflora of fish, revised. Sci. World J. 2006, 6, 931–945. [Google Scholar] [CrossRef] [PubMed]
- Ward, D.R. Microbiology of aquaculture products. Food Technol. 1989, 43, 82–85. [Google Scholar]
- Nedoluha, P.C.; Westhoff, D. Microbiological analysis of striped bass (Morone saxatilis) grown in flow-through tanks. J. Food Prot. 1995, 58, 1363–1368. [Google Scholar] [CrossRef] [PubMed]
- Gram, L.; Huss, H.H. Microbiological spoilage of fish and fish products. Int. J. Food Microbiol. 1996, 33, 121–137. [Google Scholar] [CrossRef] [PubMed]
- Wittwer, G. Caracterización Bacteriana de Intestino de Salmón del Atlántico Adulto. Bachelor’s Thesis, Universidad Austral de Chile, Valdivia, Chile, 2012; 94p. [Google Scholar]
- Painter, J.A.; Hoekstra, R.M.; Ayers, T.; Tauxe, R.V.; Braden, C.R.; Angulo, F.J.; Griffin, P.M. Attribution of foodborne illnesses, hospitalizations, and deaths to food commodities by using outbreak data, United States, 1998–2008. Emerg. Infect. Dis. 2013, 19, 407–415. [Google Scholar] [CrossRef]
- Svanevik, C.S.; Roiha, I.S.; Levsen, A.; Lunestad, B.T. Microbiological assessment along the fish production chain of the Norwegian pelagic fisheries sector—Results from a spot sampling programme. Food Microbiol. 2015, 51, 144–153. [Google Scholar] [CrossRef]
- Maji, U.; Mohanty, S.; Mahapatra, A.; Maiti, N. Diversity and probiotic potentials of putative lactic acid bacteria for application in freshwater aquaculture. Turk. J. Fish. Aquat. Sci. 2016, 16, 805–818. [Google Scholar] [CrossRef]
- Ghafir, Y.; China, B.; Dierick, K.; De Zutter, L.; Daube, G. Hygiene indicator microorganisms for selected pathogens on beef, pork, and poultry meats in Belgium. J. Food Prot. 2008, 71, 35–45. [Google Scholar] [CrossRef]
- Tortorello, M.L. Indicator organisms for safety and quality-uses and methods for detection: Minireview. J. AOAC Int. 2003, 86, 1208–1217. Available online: https://www.scopus.com/record/display.uri?eid=2-s2.0-1342329968&origin=inward&txGid=1a8a73e7af604fc94f2b6ebc5991be77 (accessed on 6 October 2023). [CrossRef]
- Koutsoumanis, K.; Nychas, G.J.E. Application of a systematic experimental procedure to develop a microbial model for rapid fish shelf life predictions. Int. J. Food Microbiol. 2000, 60, 171–185. [Google Scholar] [CrossRef]
- Anihouvi, D.G.H.; Kpoclou, Y.E.; Abdel Massih, M.; Iko Afé, O.H.; Assogba, M.F.; Covo, M.; Scippo, M.L.; Hounhouigan, D.J.; Anihouvi, V.; Mahillon, J. Microbiological characteristics of smoked and smoked–dried fish processed in Benin. Food Sci. Nutr. 2019, 7, 1821–1827. [Google Scholar] [CrossRef] [PubMed]
- Eizenberga, I.; Terentjeva, M.; Valcina, O.; Novoslavskij, A.; Strazdina, V.; Ošmjana, J.; Bērziņš, A. Microbiological quality of raw fish at retail market in Latvia. Nordic View to Sustainable Rural Development. In Proceedings of the 25th NJF Congress, Riga, Latvia, 16–18 June 2015; Volume 1, pp. 324–328. [Google Scholar]
- Papadopoulou, C.; Economou, E.; Zakas, G.; Salamoura, C.; Dontorou, C.; Apostolou, J. Microbiological and pathogenic contaminants of seafood in Greece. J. Food Qual. 2007, 30, 28–42. [Google Scholar] [CrossRef]
- Smoot, L.M.; Pierson, M.D. Indicator microorganisms and microbiological criteria. In Food Microbiology: Fundamentals and Frontiers; ASM Press: Washington, DC, USA, 1997; pp. 66–80. [Google Scholar]
- Popovic, N.T.; Skukan, A.B.; Dzidara, P.; Coz-Rakovac, R.; Strunjak-Perovic, I.; Kozacinski, L.; Jadan, M.; Brlek-Gorski, D. Microbiological quality of marketed fresh and frozen seafood caught off the Adriatic coast of Croatia. Vet. Med. 2010, 55, 233–241. [Google Scholar] [CrossRef]
- Araújo, C.; Muñoz, E.; Nahuelquína, Y.; Poeta, P.; Igrejas, G.; Hernández, P.; Herranz, C.; Cintas, L. Inhibition of fish pathogens by the microbiota from rainbow trout (Oncorhynchus mykiss, Walbaum) and rearing environment. Anaerobe 2015, 32, 7–14. [Google Scholar] [CrossRef]
- Muthukumar, P.; Kandeepan, C. Isolation, identification and characterization of probiotic organisms from intestine of fresh water fishes. Int. J. Curr. Microbiol. Appl. Sci. 2015, 4, 607–616. [Google Scholar]
- Quispe, W.; Mantilla, B.; Ccama, A.; Ortega, Y.; Sandoval, N. Aislamiento de bacterias nativas de Oncorhynchus mykiss con potencial probiótico frente a Yersinia ruckeri. Rev. Investig. Vet. Perú 2020, 31, e19024. [Google Scholar] [CrossRef]
- Henríquez, C.P. Caracterización de Propiedades Probióticas de Microorganismos del Tracto Digestivo de Salmónidos. Master’s Thesis, Universidad de Chile, Santiago de Chile, Chile, 2013; 43p. [Google Scholar]
- Balcázar, J.L.; Blas, I.; Ruiz-Zarzuela, I.; Vendrell, D.; Gironés, O.; Múzquiz, L. Sequencing of variable regions of the 16S rRNA gene for identification of lactic acid bacteria isolated from the intestinal microbiota of healthy salmonids. Comp. Immunol. Microbiol. Infect. Dis. 2007, 30, 111–118. [Google Scholar] [CrossRef] [PubMed]
- Pullela, S.; Fernandes, C.F.; Flick, G.J.; Libey, G.S.; Smith, S.A.; Coale, C.W. Indicative and pathogenic microbiological quality of aquaculture finfish grown in different production systems. J. Food Prot. 1998, 61, 205–210. [Google Scholar] [CrossRef] [PubMed]
- Pond, M.J.; Stone, D.M.; Alderman, D.J. Comparison of conventional and molecular techniques to investigate the intestinal microflora of rainbow trout (Oncorhynchus mykiss). Aquaculture 2006, 261, 194–203. [Google Scholar] [CrossRef]
- Reyes-Rodríguez, N.E.; Salgado-Miranda, C.; Flores-Valle, I.T.; González-Gómez, M.; Soriano-Vargas, E.; Peláez-Acero, A.; Vega-Sánchez, V. Molecular identification and virulence potential of the genus Aeromonas isolated from wild rainbow trout (Oncorhynchus mykiss) in Mexico. J. Food Prot. 2019, 82, 1706–1713. [Google Scholar] [CrossRef]
- Flores Durán, K. Determinación de la Diversidad Fenotípica de Yersinia ruckeri en Aislados de Truchas Arcoíris (Oncorhynchus mykiss) de Cultivo de las Regiones de Junín, Ancash y Huancavelica. Ph.D. Thesis, Universidad Nacional Mayor de San Marcos, Lima, Perú, 2013. [Google Scholar]
- Ghaly, A.E.; Dave, D.; Budge, S.; Brooks, M.S. Fish spoilage mechanisms and preservation techniques. Am. J. Appl. Sci. 2010, 7, 859–877. [Google Scholar] [CrossRef]
- Koutsoumanis, K. Predictive modeling of the shelf life of fish under nonisothermal conditions. Appl. Environ. Microbiol. 2001, 67, 1821–1829. [Google Scholar] [CrossRef] [PubMed]
- Tryfinopoulou, P.; Tsakalidou, E.; Nychas, G.J. Characterization of Pseudomonas spp. associated with spoilage of gilt-head sea bream stored under various conditions. Appl. Environ. Microbiol. 2002, 68, 65–72. [Google Scholar] [CrossRef]
- Antunes, P.; Campos, J.; Mourão, J.; Pereira, J.; Novais, C.; Peixe, L. Inflow water is a major source of trout farming contamination with Salmonella and multidrug resistant bacteria. Sci. Total Environ. 2018, 642, 1163–1171. [Google Scholar] [CrossRef]
- Miettinen, H.; Wirtanen, G. Prevalence and location of Listeria monocytogenes in farmed rainbow trout. Int. J. Food Microbiol. 2005, 104, 135–143. [Google Scholar] [CrossRef]
- ISO 4833-1:2013; Microbiology of the Food Chain—Horizontal Method for the Enumeration of Microorganisms—Part 1: Colony Count at 30 °C by the Pour Plate Technique. Food Microbiology, 1st ed. International Organization for Standardization: Geneva, Switzerland, 2013; pp. 1–9.
- ISO 6888-1:2021; Microbiology of the Food Chain—Horizontal Method for the Enumeration of Coagulase-Positive Staphylococci (Staphylococcus aureus and Other Species)—Part 1: Method Using Baird-Parker Agar Medium. Food Microbiology, 2nd ed. International Organization for Standardization: Geneva, Switzerland, 2021; pp. 1–20.
- ISO 15214:1998; Microbiology of Food and Animal Feeding Stuffs—Horizontal Method for the Enumeration of Mesophilic Lactic Acid Bacteria—Colony-Count Technique at 30 °C. Food Microbiology, 1st ed. International Organization for Standardization: Geneva, Switzerland, 1998; pp. 1–7.
- ISO 21527-1:2008; Microbiology of Food and Animal Feeding Stuffs—Horizontal Method for the Enumeration of Yeasts and Moulds—Part 1: Colony Count Technique in Products with Water Activity Greater than 0.95. Food Microbiology, 1st ed. International Organization for Standardization: Geneva, Switzerland, 2008; pp. 1–8.
- ISO 4832:2006; Microbiology of Food and Animal Feeding Stuffs—Horizontal Method for the Enumeration of Coliforms—Colony-Count Technique. Food Microbiology, 3rd ed. International Organization for Standardization: Geneva, Switzerland, 2006; pp. 1–6.
- ISO 21528-2:2017; Microbiology of the Food Chain—Horizontal Method for the Detection and Enumeration of Enterobacteriaceae—Part 2: Colony-Count Technique. Food Microbiology, 2nd ed. International Organization for Standardization: Geneva, Switzerland, 2018; pp. 1–15.
- ISO 15213-1:2023; Microbiology of the Food Chain—Horizontal Method for the Detection and Enumeration of Clostridium spp.—Part 1: Enumeration of Sulfite-Reducing Clostridium spp. by Colony-Count Technique. Food Microbiology, 1st ed. International Organization for Standardization: Geneva, Switzerland, 2023; pp. 1–21.
- ISO 17410:2019; Microbiology of the Food Chain—Horizontal Method for the Enumeration of Psychrotrophic Microorganisms. Food Microbiology, 2nd ed. International Organization for Standardization: Geneva, Switzerland, 2019; pp. 1–10.
- ISO 6579-1:2017; Microbiology of the Food Chain—Horizontal Method for the Detection, Enumeration and Serotyping of Salmonella—Part 1: Detection of Salmonella spp. Food Microbiology, 1st ed. International Organization for Standardization: Geneva, Switzerland, 2017; pp. 1–50.
- ISO 11290-1:2017; Microbiology of the Food Chain—Horizontal Method for the Detection and Enumeration of Listeria monocytogenes and of Listeria spp.—Part 1: Detection Method. Food Microbiology, 2nd ed. International Organization for Standardization: Geneva, Switzerland, 2017; pp. 1–36.
- Al Dagal, M.; Mo, O.; Fung, D.Y.C.; Kastner, C.A. Case study of the influence of microbial quality of air on product shelf life in a meat processing plant. Dairy Food Environ. Sanitat. 1992, 12, 69–70. [Google Scholar]
- Bonilla-Luque, O.M.; Possas, A.; López Cabo, M.; Rodríguez-López, P.; Valero, A. Tracking microbial quality, safety and environmental contamination sources in artisanal goat cheesemaking factories. Food Microbiol. 2023, 114, 104301. [Google Scholar] [CrossRef]
- Royal Decree 3/2023, of January 10, Which Establishes the Technical-Sanitary Criteria for the Quality of Drinking Water, Its Control and Supply. Available online: https://www.boe.es/eli/es/rd/2023/01/10/3/con (accessed on 6 October 2023).
- Grigoryan, K.; Badalyan, G.; Sargsyan, M.; Harutyunyan, A. Assessment of microbiological safety of ground water used in rainbow trout farms. LWT—Food Sci. Technol. 2014, 58, 360–363. [Google Scholar] [CrossRef]
- Zapata, J.E.; Moya, M.; Figueroa, O.A. Hidrólisis enzimática de la proteína de vísceras de trucha arcoíris (Oncorhynchus mykiss). Efecto del tipo de enzima, temperatura, pH y velocidad de agitación. Inf. Tecnol. 2019, 30, 63–72. [Google Scholar] [CrossRef]
- Díaz-Villanueva, J.; Robotham, H. Comparación de dos métodos de sacrificio en trucha arcoíris (Oncorhynchus mykiss). Lat. Am. J. Aquat. Res. 2015, 43, 287–294. [Google Scholar] [CrossRef]
- Del Torre, M.; Carraro, L.; Cardazzo, B.; Fasolato, L.; Betancourt-Barszcz, G.K.; Polese, P.; Stecchini, M.L. Changes of bacterial communities of rainbow trout (Oncorhynchus mykiss) at different processing steps in the production of burgers and their storage life enhancement by an acetate-based preservative. Food Control 2023, 154, 109949. [Google Scholar] [CrossRef]
- Cohen, J.; Shuval, H.I. Coliforms, fecal coliforms, and fecal streptococci as indicators of water pollution. Water Air Soil Pollut. 1973, 2, 85–95. [Google Scholar] [CrossRef]
- Duman, M.; Saticioglu, I.B.; Buyukekiz, A.G.; Balta, F.; Altun, S. Molecular characterization and antimicrobial resistance profile of atypical Citrobacter gillenii and Citrobacter sp. isolated from diseased rainbow trout (Oncorhynchus mykiss). J. Glob. Antimicrob. Resist. 2017, 10, 136–142. [Google Scholar] [CrossRef] [PubMed]
- Sun, H.Y.; Cao, X.H.; Jiang, Y.F.; Ni, L.Y.; Mo, Z.Q.; Qin, Q.W.; Li, Y.W. Outbreak of a novel disease associated with Citrobacter freundii infection in freshwater cultured stingray, Potamotrygon motoro. Aquaculture 2018, 492, 35–39. [Google Scholar] [CrossRef]
- Gerba, C.P. Indicator microorganisms. Environ. Microbiol. 2009, C23, 485–499. [Google Scholar] [CrossRef]
- Gelev, I.; Gelev, E.; Steigerwalt, A.G.; Carter, G.P.; Brenner, D.J. Identification of the bacterium associated with haemorrhagic septicaemia in rainbow trout as Hafnia alvei. Res. Microbiol. 1990, 141, 573–576. [Google Scholar] [CrossRef]
- Ramos Vivas, J.; Tapia, O.; Elexpuru-Zabaleta, M.; Tutusaus Pifarre, K.; Armas Diaz, Y.; Battino, M.; Giampieri, F. The molecular weaponry produced by the bacterium Hafnia alvei in foods. Molecules 2022, 27, 5585. [Google Scholar] [CrossRef]
- Mena, K.D.; Gerba, C.P. Risk assessment of Pseudomonas aeruginosa in water. Rev. Environ. Contam. Toxicol. 2009, 201, 71–115. [Google Scholar] [CrossRef]
- Viswanathan, P.; Kaur, R. Prevalence and growth of pathogens on salad vegetables, fruits and sprouts. Int. J. Hyg. Environ. Health 2001, 203, 205–213. [Google Scholar] [CrossRef]
- Aman, M.; Aneeqha, N.; Bristi, K.; Deeksha, J.; Afza, N.; Sindhuja, V.; Shastry, R.P. Lactic acid bacteria inhibits quorum sensig and biofilm formation of Pseudomonas aeruginosa strain JUPG01 isolated from rancid butter. Biocatal. Agric. Biotechnol. 2021, 36, 102115. [Google Scholar] [CrossRef]
- Hashemi, S.; Anvar, S.; Rahimi, E.; Ahari, H.; Ataee, M. Molecular investigation of prevalence, phenotypic and genotypic diversity, antibiotic resistance, frequency of virulence genes and genome sequencing in Pseudomonas aeruginosa strains isolated from lobster. Int. J. Food Microbiol. 2022, 382, 109901. [Google Scholar] [CrossRef]
- Wu, X.; Yang, L.; Wu, Y.; Li, H.; Shao, B. Spread of multidrug-resistant Pseudomonas aeruginosa in animal-derived foods in Beijing, China. Int. J. Food Microbiol. 2023, 403, 110296. [Google Scholar] [CrossRef] [PubMed]
- Aoi, Y.; Nakata, H.; Kida, H. Isolation of Pseudomonas aeruginosa from Ushubetsu River water in Hokkaido, Japan. Jpn. J. Vet. Res. 2000, 48, 29–34. [Google Scholar] [CrossRef]
- Suzuki, Y.; Kajii, S.; Nishiyama, M.; Iguchi, A. Susceptibility of Pseudomonas aeruginosa isolates collected from river water in Japan to antipseudomonal agents. Sci. Total Environ. 2013, 450–451, 148–154. [Google Scholar] [CrossRef] [PubMed]
- Nasreen, M.; Sarker, A.; Malek, M.A.; Ansaruzzaman, M.D.; Rahman, M. Prevalence and resistance pattern of Pseudomonas aeruginosa isolated from surface water. Adv. Microbiol. 2015, 5, 74–81. [Google Scholar] [CrossRef]
- Toh, S.C.; Lihan, S.; Soh, K.M.; Uyub, N.; Chai, L.C.; Müller, M. Screening and characterization of two strains of Pseudomonas aeruginosa from aquaculture and water environment. Malays. J. Microbiol. 2018, 14, 329–334. [Google Scholar] [CrossRef]
- Feito, J.; Araújo, C.; Gómez-Sala, B.; Contente, D.; Campanero, C.; Arbulu, S.; Saralegui, C.; Peña, N.; Muñoz-Atienza, E.; Borrero, J.; et al. Antimicrobial activity, molecular typing and in vitro safety assessment of Lactococcus garvieae isolates from healthy cultured rainbow trout (Oncorhynchus mykiss; Walbaum) and rearing environment. LWT 2022, 162, 113496. [Google Scholar] [CrossRef]
- Delgado, S.; Rachid, C.; Fernández, E.; Rychlik, T.; Alegría, A.; Peixoto, R.S.; Mayo, B. Diversity of thermophilic bacteria in raw, pasteurized and selectively-cultured milk, as assessed by culturing, PCR-DGGE and pyrosequencing. Food Microbiol. 2013, 36, 103–111. [Google Scholar] [CrossRef]
- Kopermsub, P.; Yunchalard, S. Identification of lactic acid bacteria associated with the production of plaa-som, a traditional fermented fish product of Thailand. Int. J. Food Microbiol. 2010, 138, 200–204. [Google Scholar] [CrossRef]
- Gibello, A.; Galán-Sánchez, F.; Blanco, M.M.; Rodríguez-Iglesias, M.; Domínguez, L.; Fernández-Garayzábal, J.F. The zoonotic potential of Lactococcus garvieae: An overview on microbiology, epidemiology, virulence factors and relationship with its presence in foods. Res. Vet. Sci. 2016, 109, 59–70. [Google Scholar] [CrossRef]
- Chan, J.F.W.; Woo, P.C.Y.; Teng, J.L.L.; Lau, S.K.P.; Leung, S.S.M.; Tam, F.C.C.; Yuen, K.Y. Primary infective spondylodiscitis caused by Lactococcus garvieae and a review of human L. garvieae infections. J. Infect. 2011, 39, 259–264. [Google Scholar] [CrossRef] [PubMed]
- Masotti, F.; Cattaneo, S.; Stuknytė, M.; De Noni, I. Airborne contamination in the food industry: An update on monitoring and disinfection techniques of air. Trends Food Sci. 2019, 90, 147–156. [Google Scholar] [CrossRef]
- Hatakka, M.; Björkroth, K.J.; Asplund, K.N.; Korkeala, H.J. Genotypes and enterotoxicity of Staphylococcus aureus isolated from the hands and nasal cavities of flight-catering employees. J. Food Prot. 2000, 63, 1487–1491. [Google Scholar] [CrossRef] [PubMed]
- Alonso, P.P.V.; Queiroz, M.M.; Gualberto, L.M.; Nascimento, S.M. Klebsiella pneumonia carbapenemase (KPC), methicillin-resistant Staphylococcus aureus (MRSA), and vancomycin-resistant Enterococcus spp. (VRE) in the food production chain and biofilm formation on abiotic surfaces. Curr. Opin. Food Sci. 2019, 26, 79–86. [Google Scholar] [CrossRef]
- Cha, Y.; Son, B.; Ryu, S. Effective removal of staphylococcal biofilms on various food contact surfaces by Staphylococcus aureus phage endolysin LysCSA13. Food Microbiol. 2019, 84, 103245. [Google Scholar] [CrossRef]
- Boulares, M.; Mejri, L.; Hassouna, M. Research note study of the microbial ecology of wild and aquacultured tunisian fresh fish. J. Food Prot. 2011, 74, 1762–1768. [Google Scholar] [CrossRef]
- Broekaert, K.; Heyndrickx, M.; Herman, L.; Devlieghere, F.; Vlaemynck, G. Seafood quality analysis: Molecular identification of dominant microbiota after ice storage on several general growth media. Food Microbiol. 2011, 28, 1162–1169. [Google Scholar] [CrossRef]
- ICMSF. International Commission on Microbiological Specifications for Foods. Preventing abuse of foods after processing. In Micro-Organisms in Foods 6; Springer: Boston, MA, USA, 1998; pp. 577–597. [Google Scholar]
- Merrifield, D.L.; Balcázar, J.L.; Daniels, C.; Zhou, Z.; Carnevali, O.; Sun, Y.-Z.; Hoseinifar, S.H.; Ringø, E. Indigenous lactic acid bacteria in fish and crustaceans. In Aquaculture Nutrition; Merrifield, D., Ringø, E., Eds.; John Wiley & Sons, Ltd.: Hoboken, NJ, USA, 2014. [Google Scholar] [CrossRef]
- Hagi, T.; Tanaka, D.; Iwamura, Y.; Hoshino, T. Diversity and seasonal changes in lactic acid bacteria in the intestinal tract of cultured freshwater fish. Aquaculture 2004, 234, 335–346. [Google Scholar] [CrossRef]
- Possas, A.; Valero, A.; Pérez-Rodríguez, F. New software solutions for microbiological food safety assessment and management. Curr. Opin. Food Sci. 2022, 44, 100814. [Google Scholar] [CrossRef]
- Lalitha, K.V.; Gopakumar, K. Distribution and ecology of Clostridium botulinum in fish and aquatic environments of a tropical region. Food Microbiol. 2000, 17, 535–541. [Google Scholar] [CrossRef]
- Naviner, M.; Giraud, E.; Le Bris, H.; Armand, F.; Mangion, C.; Ganière, J.P. Seasonal variability of intestinal microbiota in rainbow trout (Oncorhynchus mykiss), with a particular attention to Aeromonas spp. as candidate indicator of antimicrobial resistance. Revue Méd. Vét. 2006, 157, 599–604. [Google Scholar]
- Diao, J.; Li, L.; Fan, Y.; Wang, S.; Gai, C.; Wang, Y.; Yu, X.; Wang, X.; Xu, L.; Liu, H.; et al. Recombinant outer membrane protein C of Aeromonas salmonicida subsp. masoucida, a potential vaccine candidate for rainbow trout (Oncorhynchus mykiss). Microb. Pathog. 2020, 145, 104211. [Google Scholar] [CrossRef] [PubMed]
- Iorizzo, M.; Albanese, G.; Testa, B.; Ianiro, M.; Letizia, F.; Succi, M.; Tremonte, P.; D’Andrea, M.; Iaffaldano, N.; Coppola, R. Presence of lactic acid bacteria in the intestinal tract of the mediterranean trout (Salmo macrostigma) in its natural environment. Life 2021, 11, 667. [Google Scholar] [CrossRef]
- Soltani, M.; Naeiji, N.; Zargar, A.; Shohreh, P.; Taherimirghaed, A. Biotyping and serotyping of Lactococcus garvieae isolates in affected farmed rainbow trout (Oncorhynchus mykiss) in north Iran. Iran. J. Fish. Sci. 2021, 20, 1542–1559. [Google Scholar] [CrossRef]
- Musharrafieh, R.; Tacchi, L.; Trujeque, J.; LaPatra, S.; Salinas, I. Staphylococcus warneri, a resident skin commensal of rainbow trout (Oncorhynchus mykiss) with pathobiont characteristics. Vet. Microbiol. 2014, 169, 80–88. [Google Scholar] [CrossRef] [PubMed]





| Microorganism | Media/Enrichment | Media Supplement | ISO Standard | Analysed Samples | Incubation | |
|---|---|---|---|---|---|---|
| T (°C) | Time (h) | |||||
| Aerobic Mesophilic Bacteria | Plate Count Agar | - | 4833-1 [37] | W, P, S, E | 30 | 48 |
| Coagulase-positive Staphylococcus (CPS) | Baird-Parker Agar (BP) | Egg Yolk Tellurite Emulsion | 6888-1 [38] | P, S | 37 | 24–48 |
| Lactic Acid Bacteria (LAB) | De Man, Rogosa, Sharpe Agar (MRS) | - | 15214 [39] | W, P | 30 * | 72 |
| Yeast and molds | Rose-Bengal Chloramphenicol Agar (RBCA) | Chloramphenicol Supplement | 21527-1 [40] | E, P | 30 | 120 |
| Total coliforms | Violet Red Bile Lactose Agar (VRBL) | - | 4832 [41] | W, S | 30 | 24 |
| Enterobacteriaceae | Violet Red Bile Glucose Agar, (VRBG) | - | 21528-2 [42] | W, P, S | 37 | 24 |
| Sulphite-reducing Clostridium | Perfringens Agar Base (TSC & SFP) | Egg Yolk Emulsion and Perfringens (TSC) selective supplement | 15213-1 [43] | W, P | 40 * | 48 |
| Aeromonadaceae | Aeromonas medium base (RYAN) | Ampicillin supplement | - | P | 30–35 | 24 |
| Psychrotrophic bacteria | Plate Count Agar | - | 17410 [44] | P | 7 | 24 |
| Enterococcaceae | MacConkey agar No. 2 | - | - | P | 37 | 24 |
| Salmonella | Xylose-Lysine-Deoxycholate (XLD) Agar | - | 6579 [45] | P | 37 | 24 |
| Rappaport-Vassiliadis Enrichment Broth | 6579 [45] | 41.5 | 24 | |||
| Listeria monocytogenes | Chromogenic Listeria Agar, Ottaviani and Agosti (ALOA) | OCLA Selective Supplement/OCLA Differential | 11290-1/2 [46] | W, P, S | 37 | 24–48 |
| Listeria Selective Agar, Oxford formulation | Listeria selective Supplement | 11290-1/2 [46] | 37 | 24–48 | ||
| Fraser broth/Half-Fraser broth (FF/HF) | Half Fraser Supplement, Fraser Supplement | 11290-1/2 [46] | 37 | 24–48 | ||
| Samples | pH | Conductivity (µS/cm) | Salinity (ppm) | Total Dissolved Solids (TDS) (ppm) | Free Chlorine (ppm) |
|---|---|---|---|---|---|
| Input water | 6.87 ± 0.81 | 783.40 ± 110.60 | 377.40 ± 53.24 | 530.60 ± 80.96 | 0.06 ± 0.04 |
| Processing water | 7.43 ± 0.25 | 870.50 ± 227.94 | 429.75 ± 120.08 | 583.00 ± 153. 22 | 0.35 ± 0.26 |
| Output water | 6.75 ± 0.54 | 853.00 ± 170.73 | 415.20 ± 85.59 | 570.80 ± 113.60 | 0.08 ± 0.12 |
| Ice | 7.05 ± 0.01 | 91.00 ± 0.01 | 50.00 ± 0.01 | 69.00 ± 0.01 | ND * |
| Microorganism | Surface 1 | Surface 2 | r | p |
|---|---|---|---|---|
| Aerobic Mesophilic Bacteria (AMB) | Filleting machine | Packaging box | 0.87 | 0.01 |
| Evisceration machine | Packaging belt | 0.85 | 0.01 | |
| Packaging box | Packaging belt | 0.76 | 0.05 | |
| Coliforms | Knife | Packaging box | 0.99 | 0.00 |
| Washing belt | Filleting machine | −0.95 | 0.00 | |
| Packaging belt | Packaging box | 0.92 | 0.00 | |
| Knife | Packaging belt | 0.90 | 0.01 | |
| Enterobacteriaceae | Knife | Packaging belt | 0.95 | 0.00 |
| Knife | Filleting machine | 0.76 | 0.05 | |
| Coagulase-positive Staphylococcus | Slaughtering bath | Worker hands | 0.92 | 0.00 |
| Evisceration machine | Packaging belt | 0.89 | 0.01 | |
| Slaughtering bath | Filleting machine | 0.81 | 0.03 | |
| Washing belt | Evisceration machine | −0.81 | 0.03 |
| Sample | Microorganism | Mean | S.D. | 95% C.I. |
|---|---|---|---|---|
| Flesh | Aerobic mesophilic bacteria | 3.22 | 1.67 | [2.52–3.92] |
| Psychrotrophic bacteria | 4.28 | 1.71 | [3.56–5.00] | |
| Molds and yeasts | 0.94 | 1.57 | [0.28–1.60] | |
| Lactic Acid Bacteria | 2.70 | 1.77 | [1.94–3.46] | |
| Enterococcus | 2.10 | 1.85 | [1.32–2.88] | |
| Coagulase-positive Staphylococcus | 0.21 | 0.66 | [−0.28–0.48] | |
| Clostridium | 0.03 | 0.15 | [−0.08–0.09] | |
| Enterobacteriaceae | 0.62 | 1.28 | [0.07–1.17] | |
| Aeromonadaceae | 4.57 | 0.52 | [3.82–5.31] | |
| Viscera | Aerobic mesophilic bacteria | 3.97 | 1.77 | [3.21–4.73] |
| Psychrotrophic bacteria | 4.43 | 1.40 | [3.83–5.03] | |
| Molds and yeasts | 1.04 | 1.40 | [0.46–1.63] | |
| Lactic Acid Bacteria | 2.98 | 1.43 | [2.38–3.58] | |
| Enterococcus spp. | 2.61 | 2.00 | [1.75–3.47] | |
| Coagulase-positive Staphylococcus | 0.08 | 0.37 | [−0.19–0.24] | |
| Clostridium | 0.51 | 1.12 | [0.04–0.98] | |
| Enterobacteriaceae | 1.39 | 1.73 | [0.65–2.13] | |
| Aeromonadaceae | 5.16 | 0.35 | [4.75–5.56] |
| Sample | Genus |
|---|---|
| Water | Escherichia, Serratia, Enterococcus |
| Surfaces | Macrococcus, Brevibacterium, Shewanella, Erwinia, Klebsiella, Lysinibacillus, Morganella, Pantoea |
| Product | Aeromonas |
| Water and Product | Enterobacter, Lactococcus |
| Surfaces and Product | Staphylococcus, Providencia, Kluyvera |
| Water and Surfaces and Product | Citrobacter, Hafnia, Pseudomonas |
| Water and Surfaces | Exiguobacterium |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Serrano Heredia, S.M.; Sánchez-Martín, J.; Romero Gil, V.; Arroyo-López, F.N.; Benítez-Cabello, A.; Carrasco Jiménez, E.; Valero Díaz, A. Tracking Microbial Diversity and Hygienic-Sanitary Status during Processing of Farmed Rainbow Trout (Oncorhynchus mykiss). Foods 2023, 12, 3718. https://doi.org/10.3390/foods12203718
Serrano Heredia SM, Sánchez-Martín J, Romero Gil V, Arroyo-López FN, Benítez-Cabello A, Carrasco Jiménez E, Valero Díaz A. Tracking Microbial Diversity and Hygienic-Sanitary Status during Processing of Farmed Rainbow Trout (Oncorhynchus mykiss). Foods. 2023; 12(20):3718. https://doi.org/10.3390/foods12203718
Chicago/Turabian StyleSerrano Heredia, Salud María, Javier Sánchez-Martín, Verónica Romero Gil, Francisco Noé Arroyo-López, Antonio Benítez-Cabello, Elena Carrasco Jiménez, and Antonio Valero Díaz. 2023. "Tracking Microbial Diversity and Hygienic-Sanitary Status during Processing of Farmed Rainbow Trout (Oncorhynchus mykiss)" Foods 12, no. 20: 3718. https://doi.org/10.3390/foods12203718
APA StyleSerrano Heredia, S. M., Sánchez-Martín, J., Romero Gil, V., Arroyo-López, F. N., Benítez-Cabello, A., Carrasco Jiménez, E., & Valero Díaz, A. (2023). Tracking Microbial Diversity and Hygienic-Sanitary Status during Processing of Farmed Rainbow Trout (Oncorhynchus mykiss). Foods, 12(20), 3718. https://doi.org/10.3390/foods12203718










