Possible Alternatives: Identifying and Quantifying Adulteration in Buffalo, Goat, and Camel Milk Using Mid-Infrared Spectroscopy Combined with Modern Statistical Machine Learning Methods
Abstract
:1. Introduction
2. Materials and Methods
2.1. Milk Samples
2.2. Data Analysis
2.2.1. Spectral Pretreatment
2.2.2. Machine Learning Algorithms
2.2.3. Performance Evaluation Methods and Metrics
3. Results and Discussion
3.1. Quality Parameter Evaluation and FTIR Spectral Characteristics of the Milks
3.2. Models of Cow Milk or Water Adulteration in Buffalo Milk
3.2.1. Binary Classification Model for Identifying Buffalo–Cow Milk Mixtures or Buffalo Milk–Water Mixtures
3.2.2. Multi-Classification Models for Identifying High or Low Adulterant Level
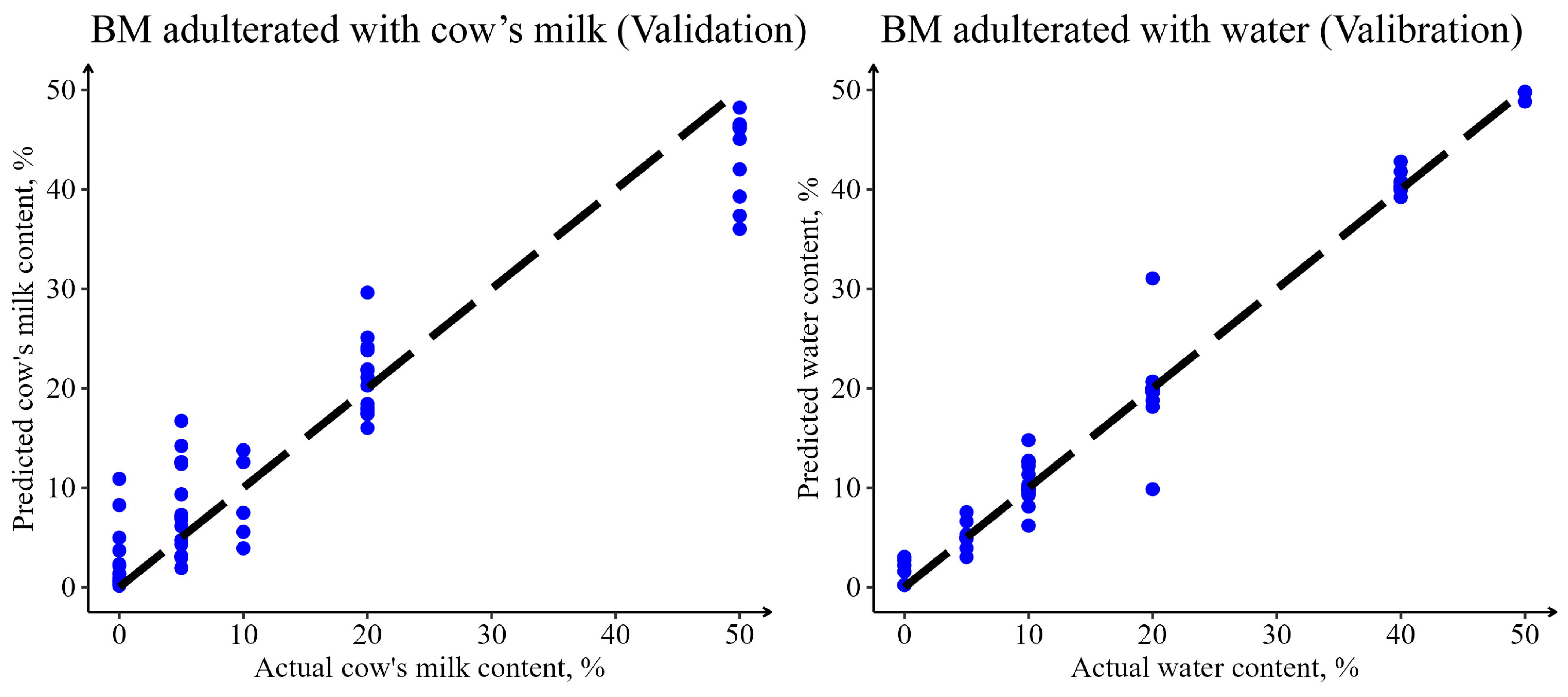
3.2.3. Quantitative Prediction of Adulteration in Buffalo Milk with Cow’s Milk and Water
3.3. Models of Cow Milk or Water Adulteration in Goat and Camel Milk
3.3.1. Classification Model for Identifying Goat Milk Adulterated with Cow Milk or Water and Its Level of Adulteration (High or Low)
3.3.2. Classification Model for Identifying Camel Milk Adulterated with Cow Milk or Water and Its Level of Adulteration (High or Low)
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Spink, J.; Moyer, D.C. Defining the public health threat of food fraud. J. Food Sci. 2011, 76, R157–R163. [Google Scholar] [CrossRef] [PubMed]
- Valand, R.; Tanna, S.; Lawson, G.; Bengtström, L. A review of Fourier Transform Infrared (FTIR) spectroscopy used in food adulteration and authenticity investigations. Food Addit. Contam. Part A 2020, 37, 19–38. [Google Scholar] [CrossRef] [PubMed]
- Nicolaou, N.; Xu, Y.; Goodacre, R. Fourier transform infrared spectroscopy and multivariate analysis for the detection and quantification of different milk species. J. Dairy Sci. 2010, 93, 5651–5660. [Google Scholar] [CrossRef] [PubMed]
- Yazgan, N.N.; Genis, H.E.; Bulat, T.; Topcu, A.; Durna, S.; Yetisemiyen, A.; Boyaci, I.H. Discrimination of milk species using Raman spectroscopy coupled with partial least squares discriminant analysis in raw and pasteurized milk. J. Sci. Food Agric. 2020, 100, 4756–4765. [Google Scholar] [CrossRef]
- Chafen, J.J.; Newberry, S.J.; Riedl, M.A.; Bravata, D.M.; Maglione, M.; Suttorp, M.J.; Sundaram, V.; Paige, N.M.; Towfigh, A.; Hulley, B.J.; et al. Diagnosing and managing common food allergies: A systematic review. J. Am. Med. Assoc. 2010, 303, 1848–1856. [Google Scholar] [CrossRef]
- Trimboli, F.; Costanzo, N.; Lopreiato, V.; Ceniti, C.; Morittu, V.M.; Spina, A.; Britti, D. Detection of buffalo milk adulteration with cow milk by capillary electrophoresis analysis. J. Dairy Sci. 2019, 102, 5962–5970. [Google Scholar] [CrossRef]
- Sen, S.; Dundar, Z.; Uncu, O.; Ozen, B. Potential of Fourier-transform infrared spectroscopy in adulteration detection and quality assessment in buffalo and goat milks. Microchem. J. 2021, 166, 106207. [Google Scholar] [CrossRef]
- Dal Bosco, C.; Panero, S.; Navarra, M.A.; Tomai, P.; Curini, R.; Gentili, A. Screening and assessment of low-molecular-weight biomarkers of milk from cow and water buffalo: An alternative approach for the rapid identification of adulterated water buffalo mozzarellas. J. Agric. Food Chem. 2018, 66, 5410–5417. [Google Scholar] [CrossRef]
- European Union. Commission Regulation (EC) No 273/2008 of 5 March 2008 laying down detailed rules for the application of Council Regulation (EC) No 1255/1999 as regards methods for the analysis and quality evaluation of milk and milk products. Off. J. 2008, 88, 1–115. [Google Scholar]
- Hassoun, A.; Måge, I.; Schmidt, W.F.; Temiz, H.T.; Li, L.; Kim, H.Y.; Nilsen, H.; Biancolillo, A.; Aït-Kaddour, A.; Sikorski, M.; et al. Fraud in Animal Origin Food Products: Advances in Emerging Spectroscopic Detection Methods over the Past Five Years. Foods 2020, 9, 1069. [Google Scholar] [CrossRef]
- Ayvaz, H.; Temizkan, R. Quick vacuum drying of liquid samples prior to ATR-FTIR spectral collection improves the quantitative prediction: A case study of milk adulteration. Int. J. Food Sci. Technol. 2018, 53, 2482–2489. [Google Scholar] [CrossRef]
- Moncayo, S.; Manzoor, S.; Rosales, J.D.; Anzano, J.; Caceres, J.O. Qualitative and quantitative analysis of milk for the detection of adulteration by laser induced breakdown spectroscopy (LIBS). Food Chem. 2017, 232, 322–328. [Google Scholar] [CrossRef]
- Sezer, B.; Durna, S.; Bilge, G.; Berkkan, A.; Yetisemiyen, A.; Boyaci, I.H. Identification of milk fraud using laser-induced breakdown spectroscopy (LIBS). Int. Dairy J. 2018, 81, 1–7. [Google Scholar] [CrossRef]
- Mabood, F.; Jabeen, F.; Ahmed, M.; Hussain, J.; Al Mashaykhi, S.A.A.; Al Rubaiey, Z.M.A.; Farooq, S.; Boqué, R.; Ali, L.; Hussain, Z.; et al. Development of new NIR-spectroscopy method combined with multivariate analysis for detection of adulteration in camel milk with goat milk. Food Chem. 2017, 221, 746–750. [Google Scholar] [CrossRef] [PubMed]
- Nicolaou, N.; Goodacre, R. Rapid and quantitative detection of the microbial spoilage in milk using Fourier transform infrared spectroscopy and chemometrics. Analyst 2008, 133, 1424–1431. [Google Scholar] [CrossRef]
- Ceniti, C.; Spina, A.A.; Piras, C.; Oppedisano, F.; Tilocca, B.; Roncada, P.; Britti, D.; Morittu, V.M. Recent Advances in the Determination of Milk Adulterants and Contaminants by Mid-Infrared Spectroscopy. Foods 2023, 12, 2917. [Google Scholar] [CrossRef] [PubMed]
- De Marchi, M.; Toffanin, V.; Cassandro, M.; Penasa, M. Invited review: Mid-infrared spectroscopy as phenotyping tool for milk traits. J. Dairy Sci. 2014, 97, 1171–1186. [Google Scholar] [CrossRef]
- Spina, A.A.; Ceniti, C.; Piras, C.; Tilocca, B.; Britti, D.; Morittu, V.M. Mid-infrared (MIR) spectroscopy for the detection of cow’s milk in buffalo milk. J. Anim. Sci. Technol. 2022, 64, 531–538. [Google Scholar] [CrossRef]
- Souhassou, S.; Bassbasi, M.; Hirri, A.; Kzaiber, F. Detection of camel milk adulteration using Fourier transformed infrared spectroscopy FT-IR coupled with chemometrics methods. Int. Food Res. J. 2018, 25, 1213–1218. [Google Scholar]
- Boukria, O.; Boudalia, S.; Bhat, Z.F.; Hassoun, A.; Aït-Kaddour, A. Evaluation of the adulteration of camel milk by non-camel milk using multispectral image, fluorescence and infrared spectroscopy. Spectrochim. Acta Part A Mol. Biomol. Spectrosc. 2023, 300, 122932. [Google Scholar] [CrossRef]
- Ferragina, A.; Cipolat-Gotet, C.; Cecchinato, A.; Pazzola, M.; Dettori, M.L.; Vacca, G.M.; Bittante, G. Prediction and repeatability of milk coagulation properties and curd-firming modeling parameters of ovine milk using Fourier-transform infrared spectroscopy and Bayesian models. J. Dairy Sci. 2017, 100, 3526–3538. [Google Scholar] [CrossRef] [PubMed]
- Shadpour, S.; Chud TC, S.; Hailemariam, D.; Plastow, G.; Oliveira, H.R.; Stothard, P.; Lassen, J.; Miglior, F.; Baes, C.F.; Tulpan, D.; et al. Predicting methane emission in Canadian Holstein dairy cattle using milk mid-infrared reflectance spectroscopy and other commonly available predictors via artificial neural networks. J. Dairy Sci. 2022, 105, 8272–8285. [Google Scholar] [CrossRef]
- Frizzarin, M.; Gormley, I.C.; Berry, D.P.; Murphy, T.B.; Casa, A.; Lynch, A.; McParland, S. Predicting cow milk quality traits from routinely available milk spectra using statistical machine learning methods. J. Dairy Sci. 2021, 104, 7438–7447. [Google Scholar] [CrossRef] [PubMed]
- Bonfatti, V.; Tiezzi, F.; Miglior, F.; Carnier, P. Comparison of Bayesian regression models and partial least squares regression for the development of infrared prediction equations. J. Dairy Sci. 2017, 100, 7306–7319. [Google Scholar] [CrossRef] [PubMed]
- Rinnan, A.; Norgaard, L.; van den Berg, F.; Thygesen, J.; Bro, R.; Engelsen, S.B. Data pre-processing. In Infrared Spectroscopy for Food Quality Analysis and Control; Academic Press: Cambridge, MA, USA, 2009; pp. 29–50. [Google Scholar]
- R Core Team. R: A Language and Environment for Statistical Computing; R Foundation for Statistical Computing: Vienna, Austria, 2020; Available online: https://www.r-project.org (accessed on 17 July 2023).
- El Jabri, M.; Sanchez, M.P.; Trossat, P.; Laithier, C.; Wolf, V.; Grosperrin, P.; Beuvier, E.; Rolet-Répécaud, O.; Gavoye, S.; Gaüzère, Y.; et al. Comparison of Bayesian and partial least squares regression methods for mid-infrared prediction of cheese-making properties in Montbéliarde cows. J. Dairy Sci. 2019, 102, 6943–6958. [Google Scholar] [CrossRef]
- Manley, M. Near-infrared spectroscopy and hyperspectral imaging: Non-destructive analysis of biological materials. Chem. Soc. Rev. 2014, 43, 8200–8214. [Google Scholar] [CrossRef]
- Cirak, O.; Icyer, N.; Durak, M. Rapid detection of adulteration of milks from different species using Fourier Transform Infrared Spectroscopy (FTIR). J. Dairy Sci. 2018, 85, 222–225. [Google Scholar] [CrossRef]
- Kaylegian, K.E.; Lynch, J.M.; Fleming, J.R.; Barbano, D.M. Influence of fatty acid chain length and unsaturation on mid-infrared milk analysis. J. Dairy Sci. 2009, 92, 2485–2501. [Google Scholar] [CrossRef]
- Silva, L.K.R.; Gonçalves, B.R.F.; da Hora, F.F.; Santos, L.S.; Ferrão, S.P.B. Spectroscopic method (FTIR-ATR) and chemometric tools to detect cow’s milk addition to buffalo’s milk. Rev. Mex. De Ing. Química 2020, 19, 11–20. [Google Scholar] [CrossRef]
- Soyeurt, H.; Dehareng, F.; Gengler, N.; McParland, S.; Wall, E.; Berry, D.P.; Coffey, M.; Dardenne, P. Mid-infrared prediction of bovine milk fatty acids across multiple breeds, production systems, and countries. J. Dairy Sci. 2011, 94, 1657–1667. [Google Scholar] [CrossRef]
- Delhez, P.; Ho, P.N.; Gengler, N.; Soyeurt, H.; Pryce, J.E. Diagnosing the pregnancy status of dairy cows: How useful is milk mid-infrared spectroscopy? J. Dairy Sci. 2020, 103, 3264–3274. [Google Scholar] [CrossRef] [PubMed]
- Gonçalves, B.-H.R.F.; Silva, G.J.; de Jesus, J.C.; Conceição, D.G.; Santos, L.S.; Ferrão, S.P.B. Fast verification of buffalo’s milk authenticity by mid-infrared spectroscopy, analytical measurements and multivariate calibration. J. Braz. Chem. Soc. 2020, 31, 1453–1460. [Google Scholar] [CrossRef]
- Dórea, J.R.R.; Rosa, G.J.M.; Weld, K.A.; Armentano, L.E. Mining data from milk infrared spectroscopy to improve feed intake predictions in lactating dairy cows. J. Dairy Sci. 2018, 101, 5878–5889. [Google Scholar] [CrossRef] [PubMed]
- Shadpour, S.; Chud, T.C.S.; Hailemariam, D.; Oliveira, H.R.; Plastow, G.; Stothard, P.; Lassen, J.; Baldwin, R.; Miglior, F.; Baes, C.F.; et al. Predicting dry matter intake in Canadian Holstein dairy cattle using milk mid-infrared reflectance spectroscopy and other commonly available predictors via artificial neural networks. J. Dairy Sci. 2022, 105, 8257–8271. [Google Scholar] [CrossRef] [PubMed]
- Sharma, R.; Verma, A.; Shinde, N.; Mann, B.; Gandhi, K.; Wichers, J.H.; van Amerongen, A. Adulteration of cow’s milk with buffalo’s milk detected by an on-site carbon nanoparticles-based lateral flow immunoassay. Food Chem. 2021, 351, 129311. [Google Scholar] [CrossRef] [PubMed]
- Ullah, R.; Khan, S.; Ali, H.; Bilal, M. Potentiality of using front face fluorescence spectroscopy for quantitative analysis of cow milk adulteration in buffalo milk. Spectrochim. Acta Part A Mol. Biomol. Spectrosc. 2020, 225, 117518. [Google Scholar] [CrossRef]


| Type | Proportion of Adulteration of Cow Milk (vol/vol) 1 | n | Proportion of Adulteration of Water (vol/vol) 1 | n |
|---|---|---|---|---|
| Buffalo milk | 0% | 198 | 0% | 187 |
| 5% | 59 | 5% | 68 | |
| 10% | 34 | 10% | 67 | |
| 20% | 82 | 20% | 119 | |
| 50% | 49 | 40% | 52 | |
| 50% | 111 | |||
| Goat milk | 0% | 40 | 0% | 33 |
| 20% | 40 | 20% | 19 | |
| 50% | 40 | 50% | 33 | |
| Camel milk | 0% | 97 | 0% | 97 |
| 20% | 95 | 20% | 84 | |
| 50% | 70 | 50% | 97 |
| Model Type | Pure Milk–Adulterants | Calibration Sets | Validation Sets |
|---|---|---|---|
| Binary classification models | Buffalo milk–cow milk | 338 (158 pure and 180 mixtures) | 84 (40 pure and 44 mixtures) |
| Buffalo milk–water | 484 (150 pure and 334 mixtures) | 120 (37 pure and 83 mixtures) | |
| Goat milk–cow milk | 96 (33 pure and 63 mixtures) | 24 (7 pure and 17 mixtures) | |
| Goat milk–water | 68 (29 pure and 39 mixtures) | 17 (4 pure and 13 mixtures) | |
| Camel milk–cow milk | 210 (78 pure and 132 mixtures) | 52 (19 pure and 33 mixtures) | |
| Camel milk–water | 223 (77 pure and 146 mixtures) | 55 (20 pure and 35 mixtures) | |
| Multi-classification models | Buffalo milk–cow milk | 338 (158 pure, 140 low, and 40 high) | 84 (40 pure, 35 low, and 9 high) |
| Buffalo milk–water | 484 (141 pure, 212 low, and 131 high) | 120 (46 pure, 42 low, and 32 high) | |
| Goat milk–cow milk | 96 (32 pure, 32 low, and 32 high) | 24 (8 pure, 8 low, and 8 high) | |
| Goat milk–water | 69 (25 pure, 17 low, and 27 high) | 16 (8 pure, 2 low, and 6 high) | |
| Camel milk–cow milk | 210 (78 pure, 76 low, and 56 high) | 52 (19 pure, 19 low, and 14 high) | |
| Camel milk–water | 223 (84 pure, 61 low, and 78 high) | 55 (13 pure, 23 low, and 19 high) | |
| Quantitative regression models | Buffalo milk–cow milk | 339 | 83 |
| Buffalo milk–water | 484 | 120 |
| Traits | Cow Milk | Buffalo Milk | Goat Milk | Camel Milk |
|---|---|---|---|---|
| Fat, % | 3.54 ± 1.00 c | 7.76 ± 1.79 a | 5.39 ± 4.10 b | 5.52 ± 0.48 b |
| Protein, % | 3.59 ± 0.31 b | 4.83 ± 1.21 a | 3.29 ± 0.42 c | 3.71 ± 0.26 b |
| Lactose, % | 4.89 ± 0.31 bc | 4.97 ± 0.53 b | 4.77 ± 0.45 c | 5.11 ± 0.18 a |
| SNF, % | 9.10 ± 0.46 c | 10.72 ± 1.16 a | 8.58 ± 0.81 d | 9.59 ± 0.34 b |
| TS, % | 12.67 ± 1.22 d | 18.21 ± 2.38 a | 14.27 ± 4.61 c | 15.29 ± 0.72 b |
| MUN, mg/100 g | 10.52 ± 1.80 c | 12.11 ± 9.14 c | 31.95 ± 0.45 b | 39.43 ± 2.42 a |
| Milk Product 2 | Preprocessing 3 | Best Method 4 | RSD, % 5 | Calibration 5 | Validation 5 | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| AUC | Acc | Sen | Spe | PPV | NPV | AUC | Acc | Sen | Spe | PPV | NPV | ||||
| adulterated with cow milk | |||||||||||||||
| BM | 1D | PLSDA | 0.49 | 1.00 | 1.00 | 1.00 | 0.99 | 0.99 | 1.00 | 1.00 | 1.00 | 1.00 | 1.00 | 1.00 | 1.00 |
| 1D | LSVM | 1.94 | 1.00 | 1.00 | 1.00 | 0.99 | 0.99 | 1.00 | 1.00 | 1.00 | 1.00 | 1.00 | 1.00 | 1.00 | |
| 2D | RSVM | 3.86 | 1.00 | 0.99 | 0.99 | 0.99 | 0.99 | 0.99 | 0.98 | 0.92 | 0.95 | 0.88 | 0.89 | 0.95 | |
| GM | 2D | PLSDA | 2.47 | 1.00 | 0.99 | 0.98 | 1.00 | 1.00 | 0.97 | 0.99 | 0.96 | 0.94 | 1.00 | 1.00 | 0.86 |
| 1D | LSVM | 3.54 | 1.00 | 1.00 | 1.00 | 1.00 | 1.00 | 1.00 | 1.00 | 1.00 | 1.00 | 1.00 | 1.00 | 1.00 | |
| 2D | RSVM | 10.61 | 1.00 | 0.99 | 0.98 | 1.00 | 1.00 | 0.97 | 0.98 | 0.92 | 0.94 | 0.86 | 0.94 | 0.86 | |
| CM | 2D | PLSDA | 0.24 | 1.00 | 0.99 | 0.98 | 1.00 | 1.00 | 0.96 | 1.00 | 1.00 | 1.00 | 1.00 | 1.00 | 1.00 |
| 1D/2D | LSVM | 0.00 | 1.00 | 1.00 | 1.00 | 1.00 | 1.00 | 1.00 | 1.00 | 1.00 | 1.00 | 1.00 | 1.00 | 1.00 | |
| 2D | RSVM | 0.98 | 1.00 | 1.00 | 1.00 | 1.00 | 1.00 | 1.00 | 0.99 | 0.95 | 0.97 | 0.90 | 0.95 | 0.95 | |
| adulterated with water | |||||||||||||||
| BM | 2D | PLSDA | 0.55 | 1.00 | 1.00 | 1.00 | 0.99 | 1.00 | 0.99 | 1.00 | 1.00 | 1.00 | 1.00 | 1.00 | 1.00 |
| 1D | LSVM | 1.03 | 1.00 | 1.00 | 1.00 | 0.99 | 0.99 | 1.00 | 1.00 | 0.99 | 0.99 | 1.00 | 1.00 | 0.97 | |
| 1D | RSVM | 0.74 | 1.00 | 1.00 | 1.00 | 1.00 | 1.00 | 1.00 | 1.00 | 0.98 | 0.98 | 0.97 | 0.99 | 0.95 | |
| GM | SNV | PLSDA | 6.84 | 1.00 | 0.97 | 0.95 | 1.00 | 1.00 | 0.94 | 1.00 | 0.94 | 0.92 | 1.00 | 1.00 | 0.80 |
| 1D | LSVM | 9.58 | 0.97 | 0.97 | 0.95 | 1.00 | 1.00 | 0.94 | 1.00 | 1.00 | 1.00 | 1.00 | 1.00 | 1.00 | |
| 1D | RSVM | 10.95 | 1.00 | 0.97 | 0.95 | 1.00 | 1.00 | 0.94 | 1.00 | 0.94 | 0.92 | 1.00 | 1.00 | 0.80 | |
| CM | 1D | PLSDA | 0.00 | 1.00 | 1.00 | 1.00 | 1.00 | 1.00 | 1.00 | 1.00 | 1.00 | 1.00 | 1.00 | 1.00 | 1.00 |
| 2D | LSVM | 0.00 | 1.00 | 1.00 | 1.00 | 1.00 | 1.00 | 1.00 | 1.00 | 1.00 | 1.00 | 1.00 | 1.00 | 1.00 | |
| 2D | RSVM | 0.35 | 1.00 | 1.00 | 1.00 | 1.00 | 1.00 | 1.00 | 1.00 | 1.00 | 1.00 | 1.00 | 1.00 | 1.00 | |
| Type 1 | Model 2 | Adulterated with Cow’s Milk | Adulterated with Water | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Preprocessing 3 | RSD, % 4 | Accc | Kappac | Accv | Kappav | Preprocessing 3 | RSD, % 4 | Accv | Kappav | Accv | Kappav | ||
| BM | PLSDA | 1D | 3.39 | 0.97 | 0.96 | 0.91 | 0.84 | 1D | 2.60 | 0.98 | 0.97 | 0.96 | 0.94 |
| LSVM | 1D | 3.49 | 0.99 | 0.98 | 0.94 | 0.90 | 1D | 1.02 | 1.00 | 1.00 | 0.99 | 0.99 | |
| RSVM | 2D | 6.63 | 0.96 | 0.94 | 0.83 | 0.71 | 1D | 1.97 | 1.00 | 1.00 | 0.99 | 0.99 | |
| GM | PLSDA | None | 12.64 | 0.99 | 0.98 | 0.96 | 0.94 | 1D | 18.17 | 1.00 | 1.00 | 0.94 | 0.90 |
| LSVM | 2D | 7.06 | 1.00 | 1.00 | 1.00 | 1.00 | None | 9.54 | 0.97 | 0.96 | 1.00 | 1.00 | |
| RSVM | 2D | 17.97 | 0.99 | 0.98 | 0.92 | 0.88 | 2D | 19.81 | 0.96 | 0.93 | 1.00 | 1.00 | |
| CM | PLSDA | 1D | 5.39 | 0.99 | 0.98 | 1.00 | 1.00 | 1D | 1.51 | 1.00 | 1.00 | 1.00 | 1.00 |
| LSVM | 1D | 0.00 | 1.00 | 1.00 | 1.00 | 1.00 | 2D | 1.77 | 1.00 | 1.00 | 1.00 | 1.00 | |
| RSVM | 1D | 5.12 | 1.00 | 1.00 | 0.98 | 0.97 | 2D | 2.19 | 1.00 | 1.00 | 1.00 | 1.00 | |
| Modeling 2 | Pre 3 | RSD 1 | Calibration Set 1 | Validation Set 1 | ||||||
|---|---|---|---|---|---|---|---|---|---|---|
| RMSEC | MAEC | RC2 | RPDC | RMSEV | MAEV | RV2 | RPDV | |||
| adulterated with cow’s milk | ||||||||||
| PLSR | SNV | 10.18 | 6.53 | 4.89 | 0.83 | 2.46 | 7.56 | 5.83 | 0.76 | 2.06 |
| LSVM | SNV | 9.41 | 6.68 | 4.66 | 0.84 | 2.40 | 7.88 | 5.87 | 0.74 | 1.98 |
| RSVM | 2D | 9.36 | 4.60 | 2.51 | 0.92 | 3.49 | 7.21 | 5.12 | 0.79 | 2.16 |
| SSR | SNV | 12.32 | 8.33 | 6.27 | 0.74 | 1.93 | 8.22 | 6.38 | 0.72 | 1.89 |
| PPR | None | 10.15 | 2.75 | 1.23 | 0.97 | 5.84 | 7.37 | 3.73 | 0.77 | 2.11 |
| CART | SG | 40.40 | 8.21 | 4.90 | 0.74 | 1.95 | 11.51 | 6.80 | 0.49 | 1.35 |
| BRNN | SG | 5.56 | 4.94 | 3.27 | 0.91 | 3.25 | 6.02 | 4.09 | 0.85 | 2.59 |
| RR | SNV | 11.06 | 8.24 | 6.23 | 0.75 | 1.95 | 8.28 | 6.46 | 0.71 | 1.88 |
| EN | SNV | 8.47 | 6.41 | 4.90 | 0.84 | 2.50 | 7.45 | 5.69 | 0.77 | 2.09 |
| LASSO | SNV | 7.84 | 6.32 | 4.83 | 0.84 | 2.54 | 7.36 | 5.63 | 0.77 | 2.12 |
| RF | 2D | 12.69 | 3.77 | 2.35 | 0.96 | 4.25 | 8.25 | 5.70 | 0.72 | 1.89 |
| GBM | 1D | 14.19 | 4.15 | 2.82 | 0.93 | 3.87 | 8.81 | 6.01 | 0.70 | 1.77 |
| PCA+BRNN | 1D | 6.71 | 5.74 | 3.88 | 0.87 | 2.80 | 5.42 | 3.65 | 0.88 | 2.87 |
| adulterated with water | ||||||||||
| PLSR | None | 1.29 | 2.06 | 1.31 | 0.99 | 9.19 | 2.25 | 1.50 | 0.99 | 8.39 |
| LSVM | 1D | 1.28 | 2.09 | 1.32 | 0.99 | 9.08 | 2.20 | 1.48 | 0.99 | 8.58 |
| RSVM | 1D | 1.25 | 1.66 | 1.29 | 0.99 | 11.40 | 2.76 | 1.86 | 0.98 | 6.85 |
| SSR | 1D | 1.37 | 2.39 | 1.50 | 0.98 | 7.91 | 2.47 | 1.73 | 0.98 | 7.64 |
| PPR | SG | 1.32 | 1.03 | 0.30 | 1.00 | 18.30 | 1.67 | 0.59 | 0.99 | 11.30 |
| CART | 2D | 3.16 | 1.91 | 0.57 | 0.99 | 9.90 | 3.23 | 1.09 | 0.97 | 5.86 |
| BRNN | SG | 1.40 | 1.80 | 1.15 | 0.99 | 10.52 | 2.12 | 1.32 | 0.99 | 8.92 |
| RR | 1D | 1.43 | 2.61 | 1.71 | 0.98 | 7.25 | 2.58 | 1.86 | 0.98 | 7.33 |
| EN | 1D | 1.36 | 2.24 | 1.43 | 0.99 | 8.46 | 2.37 | 1.66 | 0.98 | 7.97 |
| LASSO | 1D | 1.36 | 2.43 | 1.54 | 0.98 | 7.79 | 2.50 | 1.76 | 0.98 | 7.57 |
| RF | 2D | 1.33 | 1.10 | 0.52 | 1.00 | 17.26 | 2.64 | 1.41 | 0.98 | 7.17 |
| GBM | 1D | 0.93 | 0.18 | 0.07 | 1.00 | 104.93 | 2.51 | 1.35 | 0.98 | 7.53 |
| PCA+PPR | 1D | 1.27 | 1.23 | 0.44 | 1.00 | 15.40 | 1.70 | 0.68 | 0.99 | 11.10 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Chu, C.; Wang, H.; Luo, X.; Wen, P.; Nan, L.; Du, C.; Fan, Y.; Gao, D.; Wang, D.; Yang, Z.; et al. Possible Alternatives: Identifying and Quantifying Adulteration in Buffalo, Goat, and Camel Milk Using Mid-Infrared Spectroscopy Combined with Modern Statistical Machine Learning Methods. Foods 2023, 12, 3856. https://doi.org/10.3390/foods12203856
Chu C, Wang H, Luo X, Wen P, Nan L, Du C, Fan Y, Gao D, Wang D, Yang Z, et al. Possible Alternatives: Identifying and Quantifying Adulteration in Buffalo, Goat, and Camel Milk Using Mid-Infrared Spectroscopy Combined with Modern Statistical Machine Learning Methods. Foods. 2023; 12(20):3856. https://doi.org/10.3390/foods12203856
Chicago/Turabian StyleChu, Chu, Haitong Wang, Xuelu Luo, Peipei Wen, Liangkang Nan, Chao Du, Yikai Fan, Dengying Gao, Dongwei Wang, Zhuo Yang, and et al. 2023. "Possible Alternatives: Identifying and Quantifying Adulteration in Buffalo, Goat, and Camel Milk Using Mid-Infrared Spectroscopy Combined with Modern Statistical Machine Learning Methods" Foods 12, no. 20: 3856. https://doi.org/10.3390/foods12203856
APA StyleChu, C., Wang, H., Luo, X., Wen, P., Nan, L., Du, C., Fan, Y., Gao, D., Wang, D., Yang, Z., Yang, G., Liu, L., Li, Y., Hu, B., Abula, Z., & Zhang, S. (2023). Possible Alternatives: Identifying and Quantifying Adulteration in Buffalo, Goat, and Camel Milk Using Mid-Infrared Spectroscopy Combined with Modern Statistical Machine Learning Methods. Foods, 12(20), 3856. https://doi.org/10.3390/foods12203856





