Recent Advances in Personal Glucose Meter-Based Biosensors for Food Safety Hazard Detection
Abstract
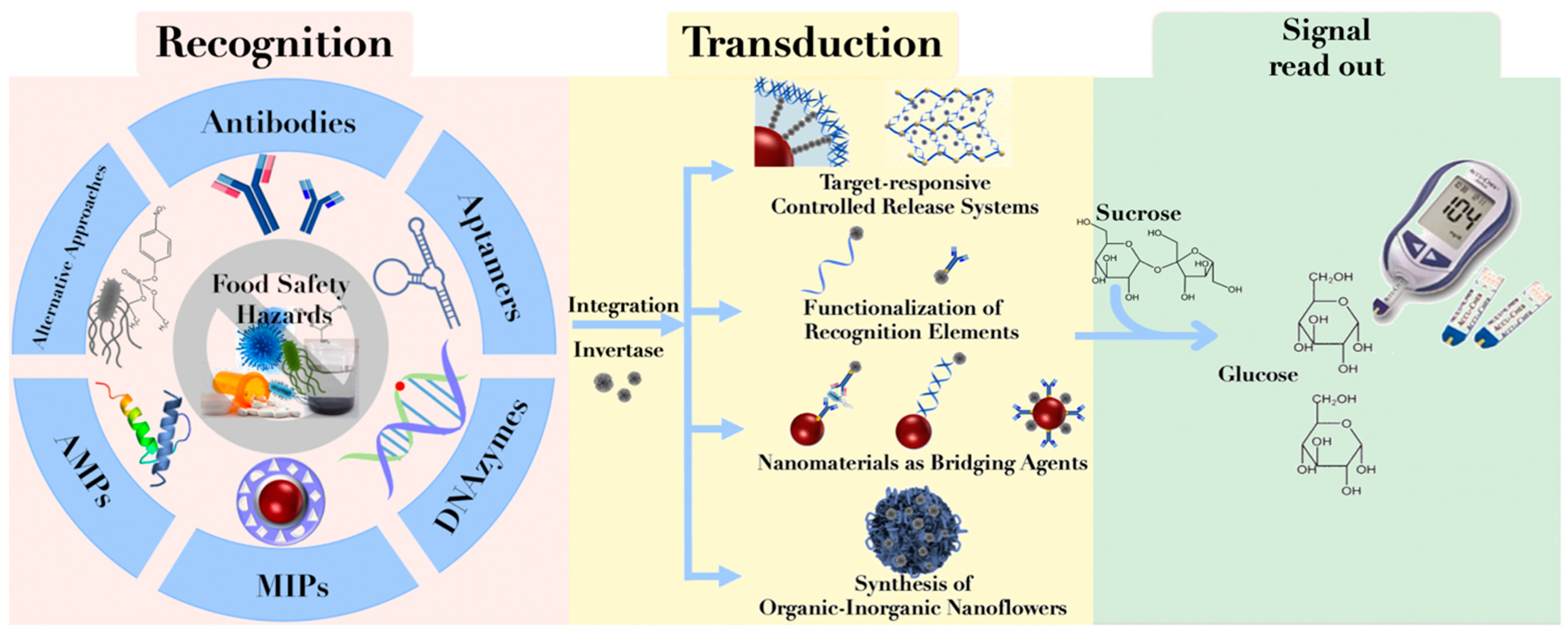
:1. Introduction
2. Construction of PGM-Based Biosensors
2.1. Recognition Elements
2.1.1. Antibodies
2.1.2. Aptamers
2.1.3. DNAzymes
2.1.4. Molecularly Imprinted Polymers
2.1.5. Antimicrobial Peptides
2.1.6. Alternative Approaches
2.2. Signal Transduction Elements
2.3. Integration Strategies of Recognition and Signal Transduction Elements
2.3.1. Target-Responsive Controlled Release Systems
2.3.2. Functionalization of Recognition Elements
2.3.3. Nanomaterials as Bridging Agents
2.3.4. Synthesis of Organic–Inorganic Nanoflowers
3. Application of PGMs in Food Safety Hazard Detection
3.1. Detection of Foodborne Pathogens
3.2. Detection of Mycotoxins
3.3. Detection of Pesticide and Veterinary Drug Residues
3.4. Detection of Heavy Metal Ions
3.5. Detection of Illegal Additives
4. Prospects of PGM-Based Biosensors in Food Safety Hazard Detection
- (1)
- The development of additional recognition mechanisms is necessary. Antibodies, aptamers, and DNAzymes have been widely employed as direct recognition elements in the construction of PGM-based sensors. Given the diverse range of food safety hazards, it is essential to develop alternative, more efficient, and applicable recognition mechanisms to broaden their applicability, such as phages’ highly specific recognition [87] and click chemistry [88].
- (2)
- The coupling strategy between recognition elements and signal transduction components requires further optimization. The binding of oligonucleotides, antibodies, and other recognition elements to enzymes often leads to reduced enzymatic efficiency and escalated experimental costs. Efforts have been directed toward the design and development of antibody-enzyme fusion proteins to ascertain their potential universality and commercial viability [52].
- (3)
- The fabrication of LFA-PGM sensing platforms faces certain constraints. Firstly, the current setup still necessitates manual intervention in the cutting of LFA detection lines and their integration with PGMs. Seamlessly bridging the gap between LFA and PGMs warrants careful consideration. Secondly, the potential loss of enzymes within the paper-based chromatography of LFA could potentially compromise sensitivity.
- (4)
- Tedious pre-processing steps impact detection time. Although MNPs find extensive application in PGM biosensors, the intricate magnetic separation and washing procedures pose challenges to rapid on-site testing. DNA nanoflowers with recognition and separation capabilities [89] hold promise as alternatives to MNPs.
- (5)
- While PGMs have achieved significant commercialization, research into PGM-based biosensors remains primarily within the laboratory domain. Currently, there is a dearth of matured and enhanced PGM detection equipment suitable for practical commercial applications. Factors such as cost, storage requirements, shelf life, and strategies for mitigating interference from endogenous sugar sources are critical determinants in facilitating the commercialization of PGM-based biosensors [12].
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Sontag, G.; Pinto, M.I.; Noronha, J.P.; Burrows, H.D. Analysis of Food by High Performance Liquid Chromatography Coupled with Coulometric Detection and Related Techniques: A Review. J. Agric. Food Chem. 2019, 67, 4113–4144. [Google Scholar] [CrossRef] [PubMed]
- Robbins, K.S.; Shah, R.; MacMahon, S.; de Jager, L.S. Development of a Liquid Chromatography–Tandem Mass Spectrometry Method for the Determination of Sulfite in Food. J. Agric. Food Chem. 2015, 63, 5126–5132. [Google Scholar] [CrossRef] [PubMed]
- Chu, P.W.S.; Chan, K.M.; Cheung, S.T.C.; Wong, Y.-C. Review of analytical techniques used in proficiency-testing programs for melamine in animal feed and milk. TrAC Trends Anal. Chem. 2010, 29, 1014–1026. [Google Scholar] [CrossRef]
- Shao, B.; Li, H.; Shen, J.; Wu, Y. Nontargeted Detection Methods for Food Safety and Integrity. Annu. Rev. Food Sci. Technol. 2019, 10, 429–455. [Google Scholar] [CrossRef]
- Maduraiveeran, G.; Sasidharan, M.; Ganesan, V. Electrochemical sensor and biosensor platforms based on advanced nanomaterials for biological and biomedical applications. Biosens. Bioelectron. Int. J. Prof. Involv. Res. Technol. Appl. Biosensers Relat. Devices 2018, 103, 113–129. [Google Scholar] [CrossRef]
- Guilbault, G.G.; Pravda, M.; Kreuzer, M.; O’Sullivan, C.K. Biosensors—42 years and counting. Anal. Lett. 2004, 37, 1481–1496. [Google Scholar] [CrossRef]
- Wackerlig, J.; Lieberzeit, P.A. Molecularly imprinted polymer nanoparticles in chemical sensing—Synthesis, characterisation and application. Sens. Actuators B-Chem. 2015, 207, 144–157. [Google Scholar] [CrossRef]
- Mahmood Khan, I.; Niazi, S.; Akhtar, W.; Yue, L.; Pasha, I.; Khan, M.K.I.; Mohsin, A.; Waheed Iqbal, M.; Zhang, Y.; Wang, Z. Surface functionalized AuNCs optical biosensor as an emerging food safety indicator: Fundamental mechanism to future prospects. Coord. Chem. Rev. 2023, 474, 214842. [Google Scholar] [CrossRef]
- Zhang, C.; Lai, Q.; Chen, W.; Zhang, Y.; Mo, L.; Liu, Z. Three-Dimensional Electrochemical Sensors for Food Safety Applications. Biosensors 2023, 13, 529. [Google Scholar] [CrossRef]
- Fritsche, J. Recent Developments and Digital Perspectives in Food Safety and Authenticity. J. Agric. Food Chem. 2018, 66, 7562–7567. [Google Scholar] [CrossRef]
- Lisi, F.; Peterson, J.R.; Gooding, J.J. The application of personal glucose meters as universal point-of-care diagnostic tools. Biosens. Bioelectron. 2020, 148, 111835. [Google Scholar] [CrossRef] [PubMed]
- Xing, X.; Yao, L.; Yan, C.; Xu, Z.; Xu, J.; Liu, G.; Yao, B.; Chen, W. Recent progress of personal glucose meters integrated methods in food safety hazards detection. Crit. Rev. Food Sci. Nutr. 2022, 62, 7413–7426. [Google Scholar] [CrossRef] [PubMed]
- Xiang, Y.; Lu, Y. Using personal glucose meters and functional DNA sensors to quantify a variety of analytical targets. Nat. Chem. 2011, 3, 697–703. [Google Scholar] [CrossRef] [PubMed]
- Kreuzer, M.P.; McCarthy, R.; Pravda, M.; Guilbault, G.G. Development of electrochemical immunosensor for progesterone analysis in milk. Anal. Lett. 2004, 37, 943–956. [Google Scholar] [CrossRef]
- Sharma, S.; Byrne, H.; O’Kennedy, R.J. Antibodies and antibody-derived analytical biosensors. Essays Biochem. 2016, 60, 9–18. [Google Scholar]
- Yalow, R.S.; Berson, S.A. Assay of plasma insulin in human subjects by immunological methods. Nature 1959, 184 (Suppl. 21), 1648–1649. [Google Scholar] [CrossRef]
- Huang, H.; Zhao, G.; Dou, W. Portable and quantitative point-of-care monitoring of Escherichia coli O157:H7 using a personal glucose meter based on immunochromatographic assay. Biosens. Bioelectron. 2018, 107, 266–271. [Google Scholar] [CrossRef]
- Gopinath, S.C.; Lakshmipriya, T.; Chen, Y.; Phang, W.M.; Hashim, U. Aptamer-based ‘point-of-care testing’. Biotechnol. Adv. 2016, 34, 198–208. [Google Scholar] [CrossRef]
- Ellington, A.D.; Szostak, J.W. In vitro selection of RNA molecules that bind specific ligands. Nature 1990, 346, 818–822. [Google Scholar] [CrossRef]
- Ma, X.; Yue, X.; Han, J.; Li, J. Method for Predicting Protein-aptamer Interaction. Med. Inf. 2020, 33, 3. [Google Scholar]
- Conidi, A.; van den Berghe, V.; Huylebroeck, D. Aptamers and their potential to selectively target aspects of EGF, Wnt/β-catenin and TGFβ-smad family signaling. Int. J. Mol. Sci. 2013, 14, 6690–6719. [Google Scholar] [CrossRef] [PubMed]
- Gu, C.; Lan, T.; Shi, H.; Lu, Y. Portable Detection of Melamine in Milk Using a Personal Glucose Meter Based on an in Vitro Selected Structure-Switching Aptamer. Anal. Chem. 2015, 87, 7676–7682. [Google Scholar] [CrossRef]
- Yang, Y.; Wu, T.; Xu, L.P.; Zhang, X. Portable detection of Staphylococcus aureus using personal glucose meter based on hybridization chain reaction strategy. Talanta 2021, 226, 122132. [Google Scholar] [CrossRef] [PubMed]
- Breaker, R.R.; Joyce, G.F. A DNA enzyme that cleaves RNA. Chem. Biol. 1994, 1, 223–229. [Google Scholar] [CrossRef] [PubMed]
- Kyzas, G.Z.; Bikiaris, D.N. Molecular Imprinting for High-Added Value Metals: An Overview of Recent Environmental Applications. Adv. Mater. Sci. Eng. 2014, 2014, 932637. [Google Scholar] [CrossRef]
- Khan, S.; Burciu, B.; Filipe, C.D.M.; Li, Y.; Dellinger, K.; Didar, T.F. DNAzyme-Based Biosensors: Immobilization Strategies, Applications, and Future Prospective. ACS Nano 2021, 15, 13943–13969. [Google Scholar] [CrossRef]
- Fu, L.; Zhuang, J.; Lai, W.; Que, X.; Lu, M.; Tang, D. Portable and quantitative monitoring of heavy metal ions using DNAzyme-capped mesoporous silica nanoparticles with a glucometer readout. J. Mater. Chem. B 2013, 1, 6123–6128. [Google Scholar] [CrossRef]
- Wulff, G.; Sarhan, A.; Zabrocki, K. Enzyme-analogue built polymers and their use for the resolution of racemates. Tetrahedron Lett. 1973, 14, 4329–4332. [Google Scholar] [CrossRef]
- Andersson, L.; Sellergren, B.; Mosbach, K. Imprinting of amino acid derivatives in macroporous polymers. Tetrahedron Lett. 1984, 25, 5211–5214. [Google Scholar] [CrossRef]
- Vlatakis, G.; Andersson, L.I.; Müller, R.; Mosbach, K. Drug assay using antibody mimics made by molecular imprinting. Nature 1993, 361, 645–647. [Google Scholar] [CrossRef]
- Zhang, Q.; Shi, J.; Deng, Q.; Jing, Z. Molecular Imprinting Polymer. Mater. Rep. 2003, S1, 194–196. [Google Scholar]
- Chen, L.; Xu, S.; Li, J. Recent advances in molecular imprinting technology: Current status, challenges and highlighted applications. Chem. Soc. Rev. 2011, 40, 2922–2942. [Google Scholar] [CrossRef] [PubMed]
- Bhogal, S.; Kaur, K.; Malik, A.K.; Sonne, C. Core-Shell Structured Molecularly Imprinted Materials for Sensing Applications. TrAC Trends Anal. Chem. 2020, 133, 116043. [Google Scholar] [CrossRef]
- Chen, S.; Gan, N.; Zhang, H.; Hu, F.; Li, T.; Cui, H.; Cao, Y.; Jiang, Q. A portable and antibody-free sandwich assay for determination of chloramphenicol in food based on a personal glucose meter. Anal. Bioanal. Chem. 2015, 407, 2499–2507. [Google Scholar] [CrossRef]
- Zhang, J. Application of Antimicrobial Peptides-Based Biosensor Methods in Detection of Foodborne Pathogens. Chemistry 2021, 84, 1300–1305. [Google Scholar]
- Steiner, H.; Hultmark, D.; Engström, A.; Bennich, H.; Boman, H.G. Sequence and specificity of two antibacterial proteins involved in insect immunity. Nature 1981, 292, 246–248. [Google Scholar] [CrossRef]
- Liu, Q.; Wang, J.; Boyd, B.J. Peptide-based biosensors. Talanta 2015, 136, 114–127. [Google Scholar] [CrossRef]
- Bai, H.; Bu, S.; Wang, C.; Ma, C.; Li, Z.; Hao, Z.; Wan, J.; Han, Y. Sandwich immunoassay based on antimicrobial peptide-mediated nanocomposite pair for determination of Escherichia coli O157:H7 using personal glucose meter as readout. Mikrochim. Acta 2020, 187, 220. [Google Scholar] [CrossRef]
- Chavali, R.; Gunda, N.S.K.; Naicker, S.; Mitra, S.K. Detection of Escherichia coli in potable water using personal glucose meters. Anal. Methods 2014, 6, 6223–6227. [Google Scholar] [CrossRef]
- Tang, W.; Yang, J.; Wang, F.; Wang, J.; Li, Z. Thiocholine-triggered reaction in personal glucose meters for portable quantitative detection of organophosphorus pesticide. Anal. Chim. Acta 2019, 1060, 97–102. [Google Scholar] [CrossRef]
- Montagnana, M.; Caputo, M.; Giavarina, D.; Lippi, G. Overview on self-monitoring of blood glucose. Clin. Chim. Acta 2009, 402, 7–13. [Google Scholar] [CrossRef] [PubMed]
- Xu, X.T.; Liang, K.Y.; Zeng, J.Y. Portable and sensitive quantitative detection of DNA based on personal glucose meters and isothermal circular strand-displacement polymerization reaction. Biosens. Bioelectron. 2015, 64, 671–675. [Google Scholar] [PubMed]
- Fu, X.H.; Xu, K.; Ye, J.; Chen, J.; Feng, X.Y. Glucoamylase-labeled nanogold flowers for in situ enhanced sensitivity of a glucometer-based enzyme immunoassay. Anal. Methods 2015, 7, 507–512. [Google Scholar] [CrossRef]
- Kim, J.J.; Park, K. Modulated insulin delivery from glucose-sensitive hydrogel dosage forms. J. Control. Release 2001, 77, 39–47. [Google Scholar] [CrossRef] [PubMed]
- Siepmann, F.; Siepmann, J.; Walther, M.; MacRae, R.J.; Bodmeier, R. Polymer blends for controlled release coatings. J. Control. Release 2008, 125, 1–15. [Google Scholar] [CrossRef]
- Hou, L.; Zhu, C.; Wu, X.; Chen, G.; Tang, D. Bioresponsive controlled release from mesoporous silica nanocontainers with glucometer readout. Chem. Commun. 2014, 50, 1441–1443. [Google Scholar] [CrossRef]
- Hernandez, R.; Tseng, H.R.; Wong, J.W.; Stoddart, J.F.; Zink, J.I. An operational supramolecular nanovalve. J. Am. Chem. Soc. 2004, 126, 3370–3371. [Google Scholar] [CrossRef]
- Schlossbauer, A.; Kecht, J.; Bein, T. Biotin-avidin as a protease-responsive cap system for controlled guest release from colloidal mesoporous silica. Angew. Chem. Int. Ed. Engl. 2009, 48, 3092–3095. [Google Scholar] [CrossRef]
- Wang, L.; Zhu, F.; Chen, M.; Xiong, Y.; Zhu, Y.; Xie, S.; Liu, Q.; Yang, H.; Chen, X. Development of a “Dual Gates” Locked, Target-Triggered Nanodevice for Point-of-Care Testing with a Glucometer Readout. ACS Sens. 2019, 4, 968–976. [Google Scholar] [CrossRef]
- Yan, L.; Zhu, Z.; Zou, Y.; Huang, Y.; Liu, D.; Jia, S.; Xu, D.; Wu, M.; Zhou, Y.; Zhou, S.; et al. Target-Responsive “Sweet” Hydrogel with Glucometer Readout for Portable and Quantitative Detection of Non-Glucose Targets. J. Am. Chem. Soc. 2013, 135, 3748–3751. [Google Scholar] [CrossRef]
- Xiang, Y.; Lu, Y. Using commercially available personal glucose meters for portable quantification of DNA. Anal. Chem. 2012, 84, 1975–1980. [Google Scholar] [CrossRef] [PubMed]
- Leonard, E.K.; Pellitero, M.A.; Juelg, B.; Spangler, J.B.; Arroyo-Curras, N. Antibody-Invertase Fusion Protein Enables Quantitative Detection of SARS-CoV-2 Antibodies Using Widely Available Glucometers. J. Am. Chem. Soc. 2022, 144, 11226–11237. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.; Shen, Z.; Xiang, Y.; Lu, Y. Integration of Solution-Based Assays onto Lateral Flow Device for One-Step Quantitative Point-of-Care Diagnostics Using Personal Glucose Meter. ACS Sens. 2016, 1, 1091–1096. [Google Scholar] [CrossRef]
- Zheng, J.; Cheng, G.; Feng, W.; He, P.; Fang, Y. Application of Gold Nanoparticle and Magnetic Nanoparticle in the Biosensor. Chem. World 2010, 51, 4. [Google Scholar]
- Shen, G.; Liang, X. Progress on Biosensors Modified by Gold-based Nanoparticles. Mater. Rep. 2012, 26, 4. [Google Scholar]
- Joo, J.; Kwon, D.; Shin, H.H.; Park, K.-H.; Cha, H.J.; Jeon, S. A facile and sensitive method for detecting pathogenic bacteria using personal glucose meters. Sens. Actuators B-Chem. 2013, 188, 1250–1254. [Google Scholar] [CrossRef]
- Ge, J.; Lei, J.; Zare, R.N. Protein-inorganic hybrid nanoflowers. Nat. Nanotechnol. 2012, 7, 428–432. [Google Scholar] [CrossRef]
- Ye, R.; Zhu, C.; Song, Y.; Song, J.; Fu, S.; Lu, Q.; Yang, X.; Zhu, M.-J.; Du, D.; Li, H.; et al. One-pot bioinspired synthesis of all-inclusive protein-protein nanoflowers for point-of-care bioassay: Detection of E. coli O157:H7 from milk. Nanoscale 2016, 8, 18980–18986. [Google Scholar] [CrossRef]
- Huang, F.; Zhang, H.; Wang, L.; Lai, W.; Lin, J. A sensitive biosensor using double-layer capillary based immunomagnetic separation and invertase-nanocluster based signal amplification for rapid detection of foodborne pathogen. Biosens. Bioelectron. 2018, 100, 583–590. [Google Scholar] [CrossRef]
- Ye, L.; Zhao, G.; Dou, W. An ultrasensitive sandwich immunoassay with a glucometer readout for portable and quantitative detection of Cronobacter sakazakii. Anal. Methods 2017, 9, 6286–6292. [Google Scholar] [CrossRef]
- Luo, Y.; Dou, W.; Zhao, G. Rapid electrochemical quantification of Salmonella Pullorum and Salmonella Gallinarum based on glucose oxidase and antibody-modified silica nanoparticles. Anal. Bioanal. Chem. 2017, 409, 4139–4147. [Google Scholar] [CrossRef] [PubMed]
- Yang, X.; Shi, D.; Zhu, S.; Wang, B.; Zhang, X.; Wang, G. Portable Aptasensor of Aflatoxin B1 in Bread Based on a Personal Glucose Meter and DNA Walking Machine. Acs Sens. 2018, 3, 1368–1375. [Google Scholar] [CrossRef] [PubMed]
- Tang, J.; Huang, Y.; Liu, H.; Zhang, C.; Tang, D. Novel glucometer-based immunosensing strategy suitable for complex systems with signal amplification using surfactant-responsive cargo release from glucose-encapsulated liposome nanocarriers. Biosens. Bioelectron. 2016, 79, 508–514. [Google Scholar] [CrossRef] [PubMed]
- Gu, C.; Long, F.; Zhou, X.; Shi, H. Portable detection of ochratoxin A in red wine based on a structure-switching aptamer using a personal glucometer. RSC Adv. 2016, 6, 29563–29569. [Google Scholar] [CrossRef]
- Qiu, S.; Yuan, L.; Wei, Y.; Zhang, D.; Chen, Q.; Lin, Z.; Luo, L. DNA template-mediated click chemistry-based portable signal-on sensor for ochratoxin A detection. Food Chem. 2019, 297, 124929. [Google Scholar] [CrossRef]
- Zhang, S.; Luan, Y.; Xiong, M.; Zhang, J.; Lake, R.; Lu, Y. DNAzyme Amplified Aptasensing Platform for Ochratoxin A Detection Using a Personal Glucose Meter. ACS Appl. Mater. Interfaces 2021, 13, 9472–9481. [Google Scholar] [CrossRef]
- Nie, D.; Zhang, Z.; Guo, D.; Tang, Y.; Hu, X.; Huang, Q.; Zhao, Z.; Han, Z. A flexible assay strategy for non-glucose targets based on sulfhydryl-terminated liposomes combined with personal glucometer. Biosens. Bioelectron. 2021, 175, 112884. [Google Scholar] [CrossRef]
- Kwon, D.; Lee, H.; Yoo, H.; Hwang, J.; Lee, D.; Jeon, S. Facile method for enrofloxacin detection in milk using a personal glucose meter. Sens. Actuators B-Chem. 2018, 254, 935–939. [Google Scholar] [CrossRef]
- Li, F.; Li, X.; Zhu, N.; Li, R.; Kang, H.; Zhang, Q. An aptasensor for the detection of ampicillin in milk using a personal glucose meter. Anal. Methods 2020, 12, 3376–3381. [Google Scholar] [CrossRef]
- Gao, S.; Hao, J.; Su, D.; Wu, T.; Gao, J.; Hu, G. Facile and sensitive detection of norfloxacin in animal-derived foods using immuno-personal glucose meter. Eur. Food Res. Technol. 2021, 247, 2635–2644. [Google Scholar] [CrossRef]
- Zhang, J.; Tang, Y.; Teng, L.; Lu, M.; Tang, D. Low-cost and highly efficient DNA biosensor for heavy metal ion using specific DNAzyme-modified microplate and portable glucometer-based detection mode. Biosens. Bioelectron. 2015, 68, 232–238. [Google Scholar] [CrossRef]
- Zeng, L.; Gong, J.; Rong, P.; Liu, C.; Chen, J. A portable and quantitative biosensor for cadmium detection using glucometer as the point-of-use device. Talanta 2019, 198, 412–416. [Google Scholar] [CrossRef]
- Li, F.; Zhang, R.; Kang, H.; Hu, Y.; Liu, Y.; Zhu, J. Facile and sensitive detection of clenbuterol in pork using a personal glucose meter. Anal. Methods 2017, 9, 6507–6512. [Google Scholar] [CrossRef]
- Li, G.Z.; Tang, D.P. Bioresponsive controlled glucose release from TiO2 nanotube arrays: A simple and portable biosensing system for cocaine with a glucometer readout. J. Mater. Chem. B 2017, 5, 5573–5579. [Google Scholar] [CrossRef]
- Fuzawa, M.; Smith, R.L.; Ku, K.M.; Shisler, J.L.; Feng, H.; Juvik, J.A.; Nguyen, T.H. Roles of Vegetable Surface Properties and Sanitizer Type on Annual Disease Burden of Rotavirus Illness by Consumption of Rotavirus-Contaminated Fresh Vegetables: A Quantitative Microbial Risk Assessment. Risk Anal. 2020, 40, 741–757. [Google Scholar] [CrossRef] [PubMed]
- Xiong, S.; Zhou, Y.; Huang, X.; Yu, R.; Lai, W.; Xiong, Y. Ultrasensitive direct competitive FLISA using highly luminescent quantum dot beads for tuning affinity of competing antigens to antibodies. Anal. Chim. Acta 2017, 972, 94–101. [Google Scholar] [CrossRef] [PubMed]
- Alshannaq, A.; Yu, J.-H. Occurrence, Toxicity, and Analysis of Major Mycotoxins in Food. Int. J. Environ. Res. Public Health 2017, 14, 632. [Google Scholar] [CrossRef] [PubMed]
- He, F.; Wang, H.; Du, P.; Li, T.; Wang, W.; Tan, T.; Liu, Y.; Ma, Y.; Wang, Y.; El-Aty, A.M.A. Personal glucose meters coupled with signal amplification technologies for quantitative detection of non-glucose targets: Recent progress and challenges in food safety hazards analysis. J. Pharm. Anal. 2023, 13, 223–238. [Google Scholar] [CrossRef] [PubMed]
- Gui, G.; Fan, G.K.; Sang, L.; Lai, N.; Wang, Z.; Chen, Q.; Yang, Z. Research progress on veterinary drug residues and detection methods in natural casing. J. Food Saf. Qual. 2019, 10, 1268–1272. [Google Scholar]
- GB 31650-2019; Maximum Residue Limits for Veterinary Drugs in Foods. Standardization Administration of China: Beijing, China, 2019.
- Zhai, H.; Jin, X.; Yue, J.; Zhao, J. Advances in the Methods of Rapid Determination for Heavy Metals. Hubei Agric. Sci. 2010, 49, 1995–1998. [Google Scholar]
- Järup, L. Hazards of heavy metal contamination. Br. Med. Bull. 2003, 68, 167–182. [Google Scholar] [CrossRef] [PubMed]
- Flora, G.; Gupta, D.; Tiwari, A. Toxicity of lead: A review with recent updates. Interdiscip. Toxicol. 2012, 5, 47–58. [Google Scholar] [CrossRef]
- Guo, X.; Kuang, F.; Liu, Q.; Song, J.; Cao, Q. Safety of Food Additives and Food Safety Detection Technology. Food Sci. Technol. 2021, 000, 137–139. [Google Scholar]
- Shetty, S.; Malik, A.h.; Ali, A.; Yang, Y.C.; Briasoulis, A.; Alvarez, P. Abstract 16794: Characteristics, Trends, Outcomes, and Costs of Stimulant-Related Acute Heart Failure Hospitalizations in the United States. Circulation 2020, 142, A16794. [Google Scholar] [CrossRef]
- Yan, H.; Xu, D.; Meng, H.; Shi, L.; Li, L. Food poisoning by clenbuterol in China. Qual. Assur. Saf. Crops Foods 2015, 7, 27–35. [Google Scholar] [CrossRef]
- Leon-Velarde, C.G.; Jun, J.W.; Skurnik, M. Yersinia Phages and Food Safety. Viruses 2019, 11, 1105. [Google Scholar] [CrossRef] [PubMed]
- Worrell, B.T.; Malik, J.A.; Fokin, V.V. Direct Evidence of a Dinuclear Copper Intermediate in Cu(I)-Catalyzed Azide-Alkyne Cycloadditions. Science 2013, 340, 457–460. [Google Scholar] [CrossRef] [PubMed]
- Liu, M.; Zhang, Q.; Kannan, B.; Botton, G.A.; Yang, J.; Soleymani, L.; Brennan, J.D.; Li, Y. Self-Assembled Functional DNA Superstructures as High-Density and Versatile Recognition Elements for Printed Paper Sensors. Angew. Chem. Int. Ed. Engl. 2018, 57, 12440–12443. [Google Scholar] [CrossRef]





| Target Category | Target | Bioreceptor | Signal Transduction Elements | Signal Transduction Strategies | Detection Samples | LOD | References |
|---|---|---|---|---|---|---|---|
| Foodborne pathogens | E. coli O157:H7 | Antibody | Invertase | Nanomaterials as Bridging Agents | Physiological saline, milk | 6.2 × 104 CFU/mL | [17] |
| Antibody | Invertase | Organic–Inorganic Nanoflowers | Milk | 79 CFU/mL | [59] | ||
| Con A | Invertase | Organic–Inorganic Nanoflowers | Milk | 101 CFU/mL | [58] | ||
| AMPs | Invertase | Organic–Inorganic Nanoflowers | Milk | 10 CFU/mL | [38] | ||
| C. sakazakii | Antibody | Invertase | Nanomaterials as Bridging Agents | Milk powder | 4.2 × 101 CFU/mL | [60] | |
| E. coli | Bacterial glycolysis | Glucose | — | Tap water | 2 × 106 CFU/100 µL | [39] | |
| Salmonella | Antibody | Invertase | Nanomaterials as Bridging Agents | milk | 10 CFU/mL | [56] | |
| Antibody | Glucose oxidase | Nanomaterials as Bridging Agents | Meat broth medium | 7.2 × 101 CFU/mL | [61] | ||
| Staphylococcus aureus | Aptamer | Invertase | Nanomaterials as Bridging Agents | Peach juice, milk, water | 2 CFU/mL | [23] | |
| Mycotoxins | AFB1 | Aptamer | Glucose | Controlled Release Systems | Buffer | 0.02 ng/mL | [49] |
| Aptamer | Invertase | Functionalization of Recognition Elements | Moldy bread | 10 pm | [62] | ||
| Antibody | Glucose | Controlled Release Systems | Buffer | 0.6 pg/mL | [63] | ||
| OTA | Aptamer | Invertase | Functionalization of Recognition Elements | Buffer, red wine | 3.31 μg/L, 3.66 μg/L | [64] | |
| Aptamer | Invertase | Nanomaterials as Bridging Agents | Feed | 72 pg/mL | [65] | ||
| Aptamer | Invertase | Nanomaterials as Bridging Agents | Wine | 0.88 pg/mL | [66] | ||
| patulin | -SH | Glucose | Controlled Release Systems | Grape juice | 0.05 ng/mL | [67] | |
| Agricultural and veterinary drug residues | CAP | MIPs | Invertase | Nanomaterials as Bridging Agents | Fish and pork | 0.16 ng/mL | [34] |
| enrofloxacin | E. coli | Glucose | — | Water and milk | 5 ng/mL | [68] | |
| paraoxon | Acetylcholinesterase | [Fe(CN)6]3− | Thiocholine byproduct | Apple and cucumber | 5 µg/mL | [40] | |
| ampicillin | Aptamer | Invertase | Nanomaterials as Bridging Agents | milk | 2.5 × 10−10 mol/L | [69] | |
| norfloxacin | Antibody | Invertase | Nanomaterials as Bridging Agents | Milk, chicken, pork, shrimp | 0.5 ng/mL | [70] | |
| Heavy metal ions | Pb2+ | DNAzymes | Glucose | Controlled Release Systems | Drinking water | 1 pM | [27] |
| DNAzymes | Invertase | Functionalization of Recognition Elements | Wastewater, drinking water | 1.0 pM | [71] | ||
| Cd2+ | Aptamer | Invertase | Functionalization of Recognition Elements | Lake water, pond water | 5 pM | [72] | |
| Illegal additives | CLB | Antibody | Invertase | Nanomaterials as Bridging Agents | Pork, liver | 0.1 ng/mL | [73] |
| melamine | Aptamer | Invertase | Nanomaterials as Bridging Agents | Buffer, 80% full-fat milk | 0.33 µM, 0.53 µM | [22] | |
| cocaine | Aptamer | Glucoamylase | Controlled Release Systems | — | 3.8 μM | [50] | |
| Aptamer | Glucose | Controlled Release Systems | — | 5.2 nM | [74] |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wang, S.; Huang, H.; Wang, X.; Zhou, Z.; Luo, Y.; Huang, K.; Cheng, N. Recent Advances in Personal Glucose Meter-Based Biosensors for Food Safety Hazard Detection. Foods 2023, 12, 3947. https://doi.org/10.3390/foods12213947
Wang S, Huang H, Wang X, Zhou Z, Luo Y, Huang K, Cheng N. Recent Advances in Personal Glucose Meter-Based Biosensors for Food Safety Hazard Detection. Foods. 2023; 12(21):3947. https://doi.org/10.3390/foods12213947
Chicago/Turabian StyleWang, Su, Huixian Huang, Xin Wang, Ziqi Zhou, Yunbo Luo, Kunlun Huang, and Nan Cheng. 2023. "Recent Advances in Personal Glucose Meter-Based Biosensors for Food Safety Hazard Detection" Foods 12, no. 21: 3947. https://doi.org/10.3390/foods12213947
APA StyleWang, S., Huang, H., Wang, X., Zhou, Z., Luo, Y., Huang, K., & Cheng, N. (2023). Recent Advances in Personal Glucose Meter-Based Biosensors for Food Safety Hazard Detection. Foods, 12(21), 3947. https://doi.org/10.3390/foods12213947






