Comparative Study of Quercetin and Hyperoside: Antimicrobial Potential towards Food Spoilage Bacteria, Mode of Action and Molecular Docking
Abstract
:1. Introduction
2. Materials and Methods
2.1. Materials
2.2. Study on Antibacterial Activities of Quercetin and Hyperoside on Spoilage Bacteria
2.2.1. Bacterial Strains
2.2.2. Minimum Inhibitory Concentration (MIC) and Minimum Bactericidal Concentration (MBC) of Quercetin and Hyperoside
2.3. Study on Time–Kill Kinetics and Cell Leakage of Spoilage Bacteria for Quercetin and Hyperoside
2.3.1. Time–Kill Kinetics
2.3.2. Triphenyl-2H Tetrazolium Chloride (TTC) Dehydrogenase Activity
2.3.3. Potassium (K+) and Magnesium (Mg2+) Ion Leakage
2.3.4. Conductivity
2.3.5. Malondialdehyde (MDA) Content
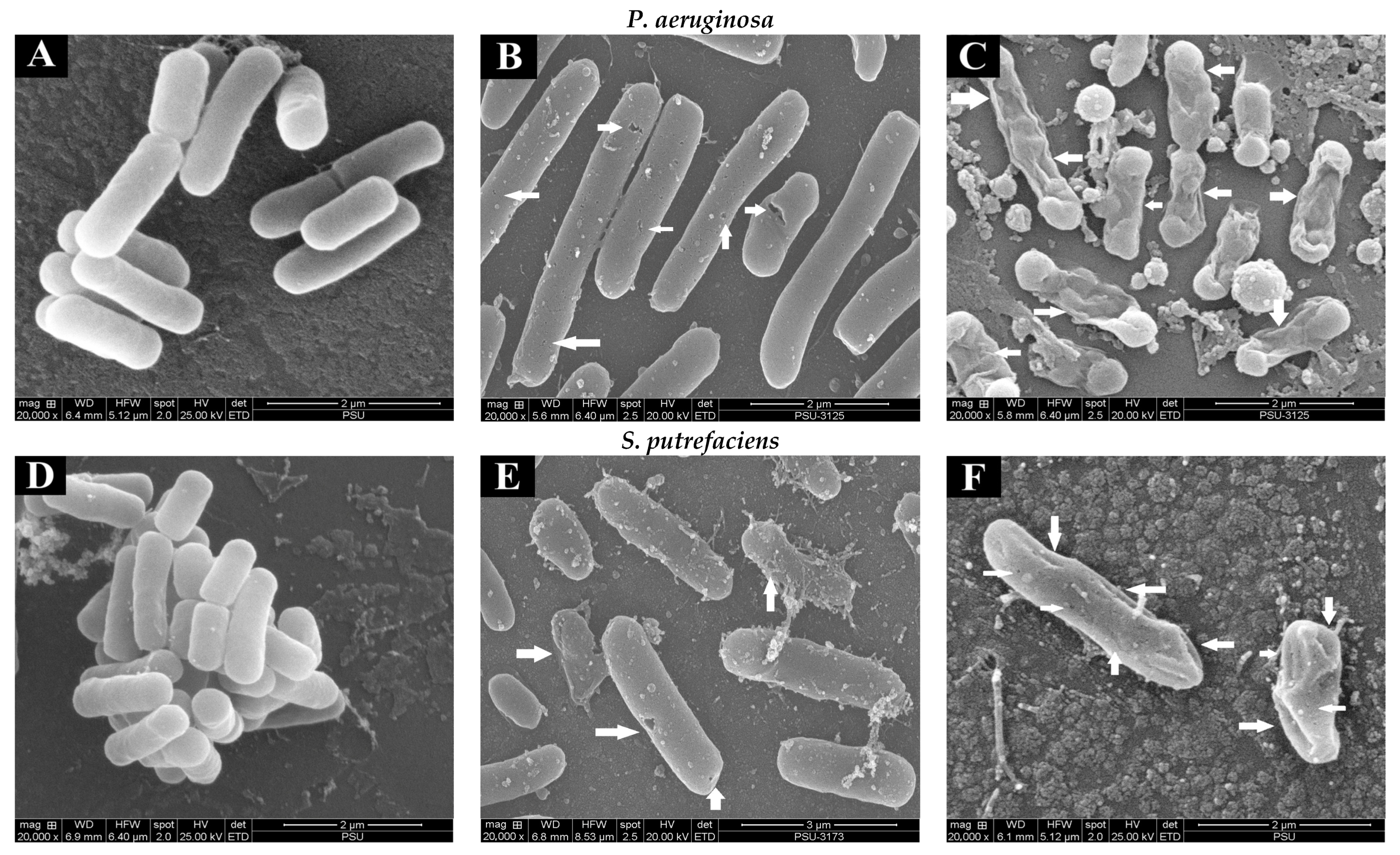
2.3.6. Scanning Electron Microscopic (SEM) Images
2.4. Study on Anti-Swimming and Swarming Activity and Antibiofilm Formation of Quercetin and Hyperoside towards Spoilage Bacteria
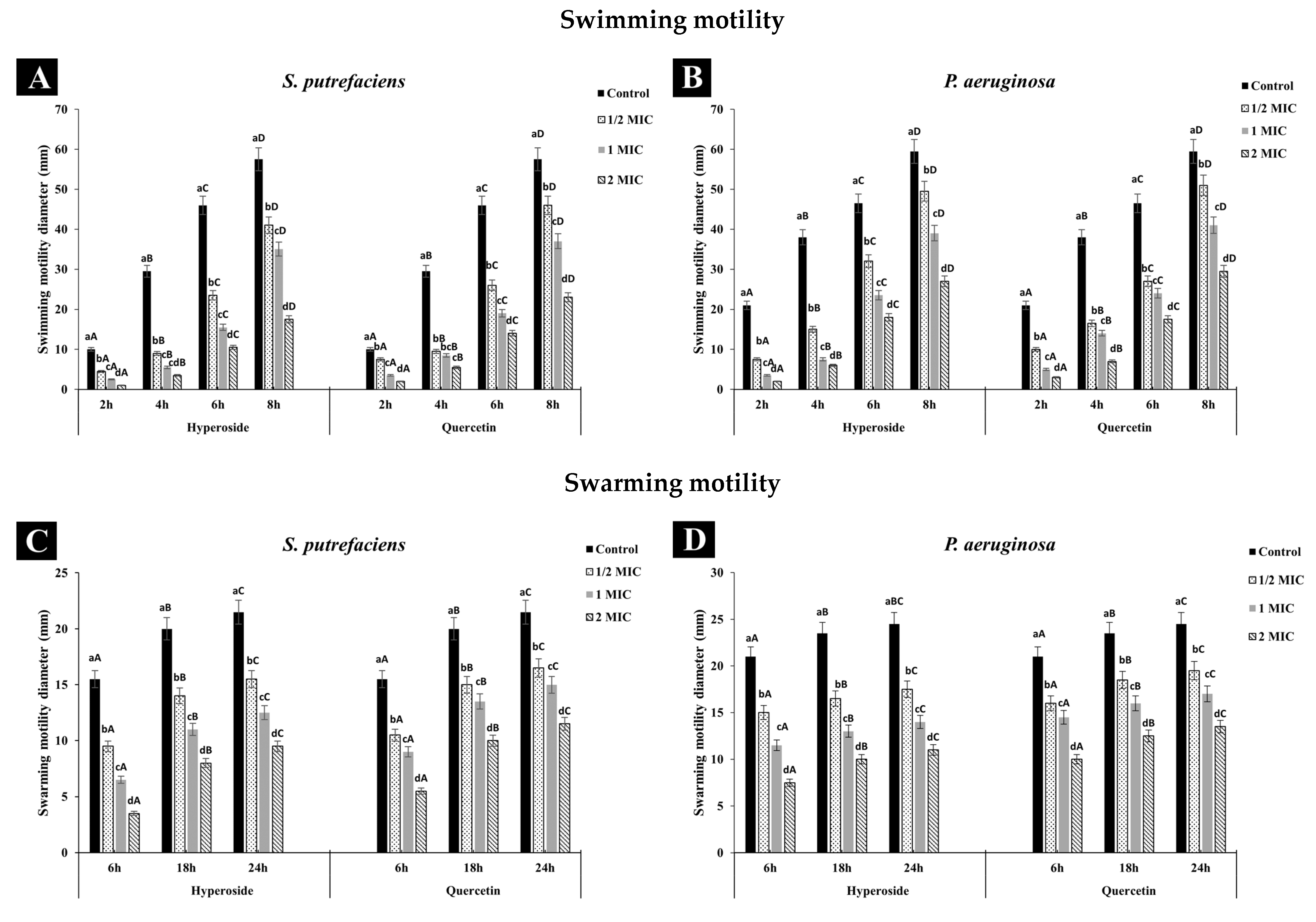
2.4.1. Anti-Swimming and Swarming Motility
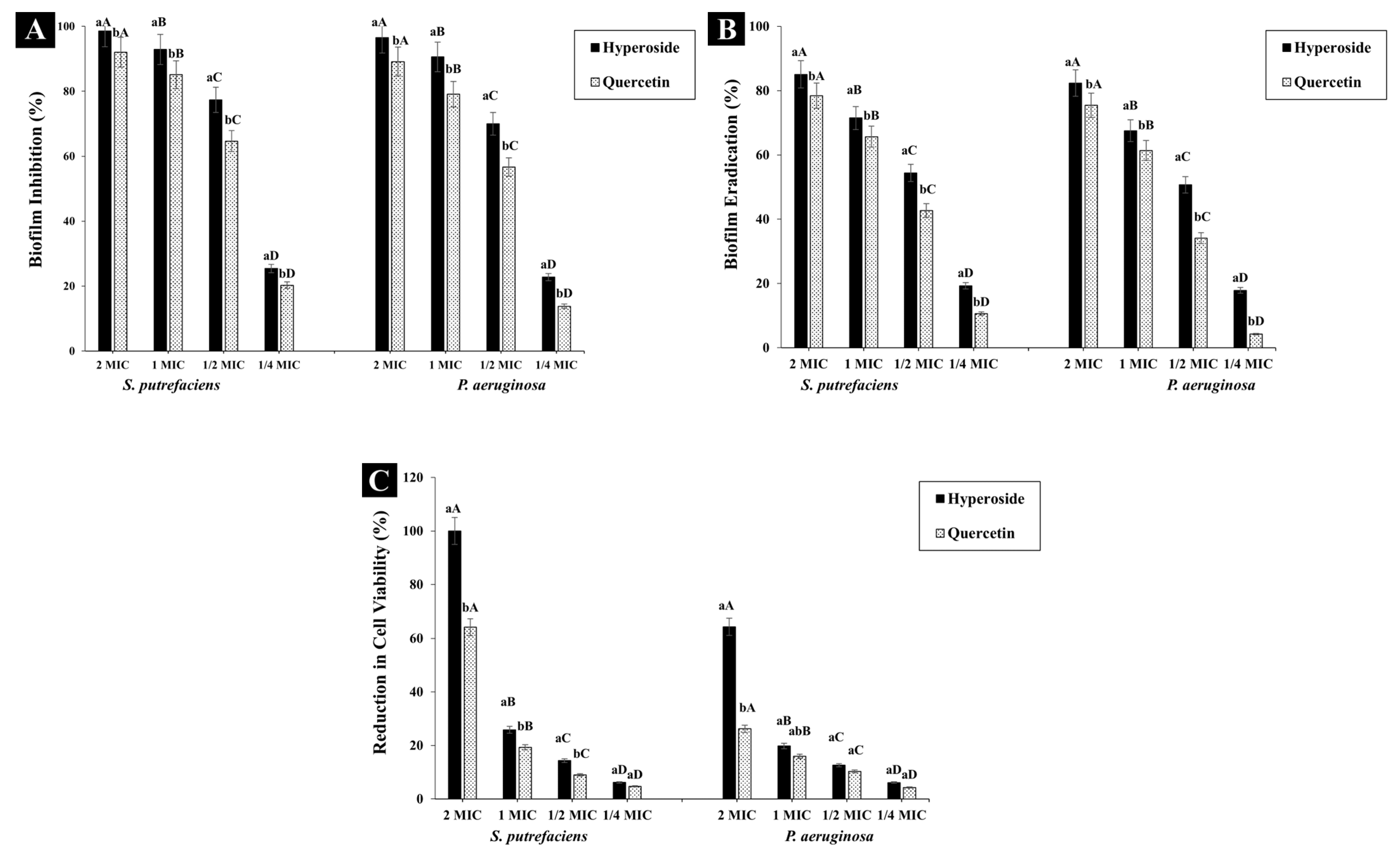
2.4.2. Biofilm Inhibition
2.4.3. Biofilm Eradication
2.4.4. Biofilm Cell Viability
2.4.5. Extracellular Polymeric Substance (EPS) Content
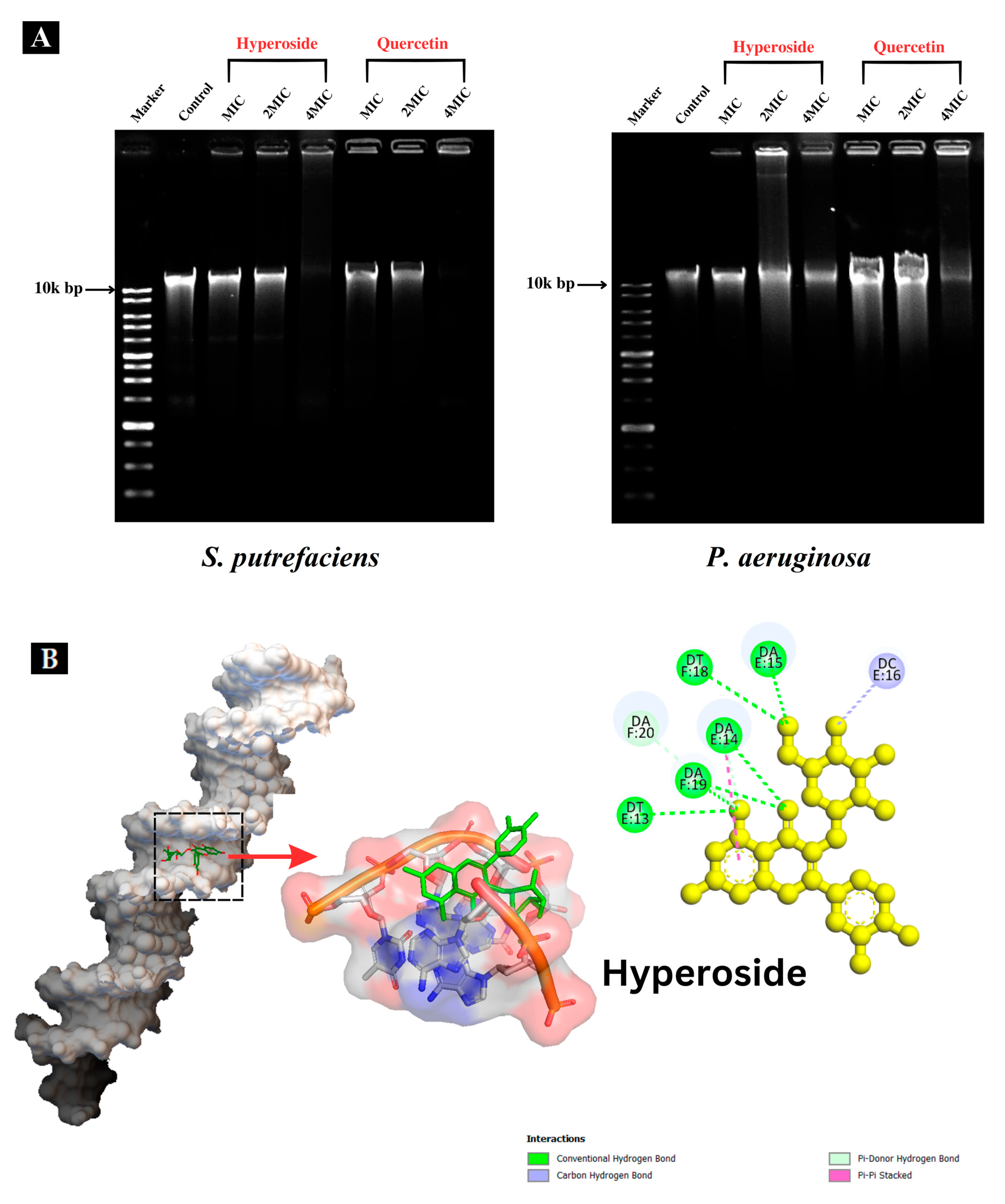
2.5. Study on Microbial DNA of Spoilage Bacteria Treated with Quercetin and Hyperoside
2.5.1. DNA Interaction
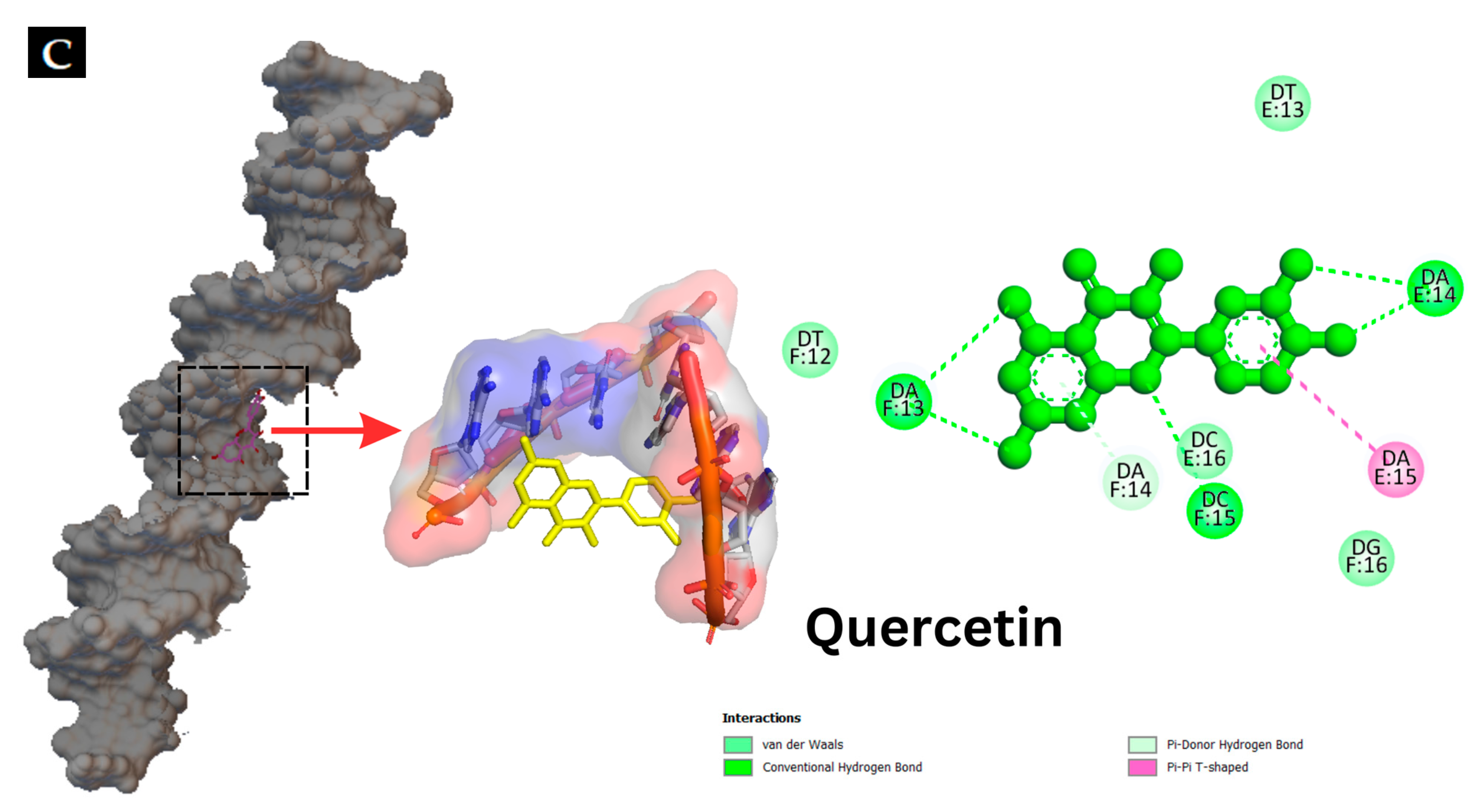
2.5.2. Molecular Docking of Hyperoside–DNA and Quercetin–DNA Interactions
2.6. Statistical Analysis
3. Results and Discussion
3.1. Antibacterial Activity of Quercetin and Hyperoside toward Spoilage Bacteria
3.1.1. MIC and MBC
3.1.2. Time–Kill Kinetics
3.1.3. TTC-Dehydrogenase Activity
3.1.4. Leakage of Potassium (K+) and Magnesium (Mg2+) Ions
3.1.5. Conductivity
3.1.6. Malondialdehyde (MDA) Content
3.1.7. Scanning Electron Microscopic (SEM) Images
3.2. Anti-Swimming, Anti-Swarming and Antibiofilm Activities of Quercetin and Hyperoside
3.2.1. Anti-Swimming and Anti-Swarming Motility
3.2.2. Antibiofilm Activity
3.2.3. Extracellular Polymeric Substance (EPS) Content
3.3. Microbial DNA Damages of Spoilage Bacteria by Quercetin and Hyperoside
3.3.1. Interaction with Genomic DNA
3.3.2. Molecular Docking
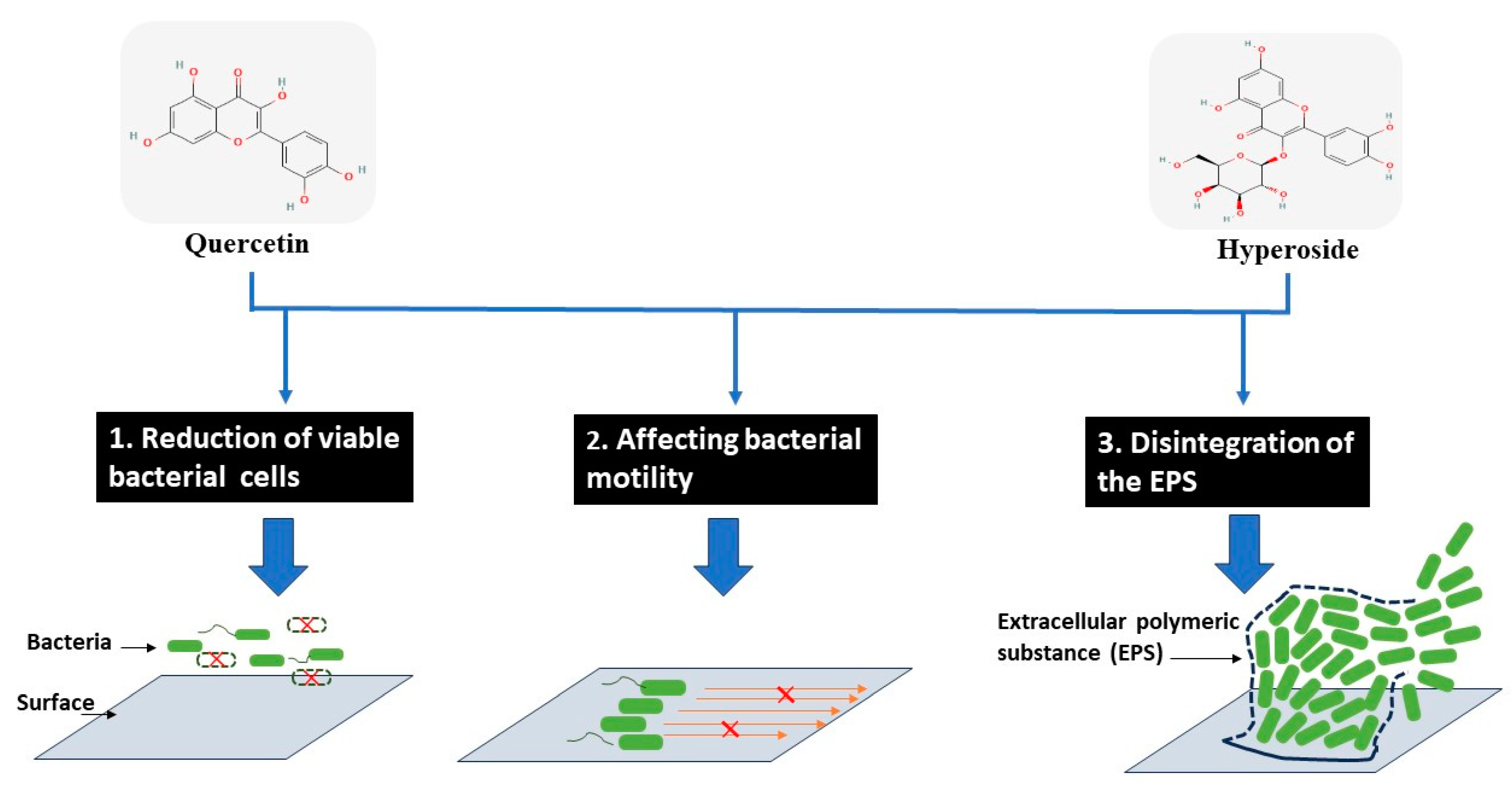
4. Conclusions
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Tagrida, M.; Benjakul, S. Betel (Piper betle L.) leaf ethanolic extracts dechlorophyllized using different methods: Antioxidant and antibacterial activities, and application for shelf-life extension of Nile tilapia (Oreochromis niloticus) fillets. RSC Adv. 2021, 11, 17630–17641. [Google Scholar] [CrossRef] [PubMed]
- He, Y.; Sang, S.; Tang, H.; Ou, C. In vitro mechanism of antibacterial activity of eucalyptus essential oil against specific spoilage organisms in aquatic products. J. Food Process. Preserv. 2022, 46, e16349. [Google Scholar] [CrossRef]
- Liu, X.; Yuan, W.; Liu, Y. Antibacterial effects of brown algae extract against tilapia spoilage bacteria Pseudomonas fluorescens and Shewanella putrefaciens. BioResources 2023, 18, 2897–2912. [Google Scholar] [CrossRef]
- Anand, S.; Sati, N. Artificial preservatives and their harmful effects: Looking toward nature for safer alternatives. Int. J. Pharm. Sci. Res. 2013, 4, 2496. [Google Scholar]
- Olatunde, O.O.; Benjakul, S. Natural preservatives for extending the shelf-life of seafood: A revisit. Compr. Rev. Food Sci. Food Saf. 2018, 17, 1595–1612. [Google Scholar] [CrossRef]
- Carocho, M.; Morales, P.; Ferreira, I.C. Natural food additives: Quo vadis? Trends Food Sci. Technol. 2015, 45, 284–295. [Google Scholar] [CrossRef]
- Leri, M.; Scuto, M.; Ontario, M.L.; Calabrese, V.; Calabrese, E.J.; Bucciantini, M.; Stefani, M. Healthy effects of plant polyphenols: Molecular mechanisms. Int. J. Mol. Sci. 2020, 21, 1250. [Google Scholar] [CrossRef]
- Bouarab Chibane, L.; Degraeve, P.; Ferhout, H.; Bouajila, J.; Oulahal, N. Plant antimicrobial polyphenols as potential natural food preservatives. J. Sci. Food Agric. 2019, 99, 1457–1474. [Google Scholar] [CrossRef]
- Li, W.; Liu, M.; Xu, Y.-F.; Feng, Y.; Che, J.-P.; Wang, G.-C.; Zheng, J.-H. Combination of quercetin and hyperoside has anticancer effects on renal cancer cells through inhibition of oncogenic microRNA-27a. Oncol. Rep. 2014, 31, 117–124. [Google Scholar] [CrossRef]
- Singh, P.; Arif, Y.; Bajguz, A.; Hayat, S. The role of quercetin in plants. Plant Physiol. Biochem. 2021, 166, 10–19. [Google Scholar] [CrossRef]
- Fang, Y.; Lu, Y.; Zang, X.; Wu, T.; Qi, X.; Pan, S.; Xu, X. 3D-QSAR and docking studies of flavonoids as potent Escherichia coli inhibitors. Sci. Rep. 2016, 6, 23634. [Google Scholar] [CrossRef] [PubMed]
- Lavado, G.; Ladero, L.; Cava, R. Cork oak (Quercus suber L.) leaf extracts potential use as natural antioxidants in cooked meat. Indust. Crop. Prod. 2021, 160, 113086. [Google Scholar] [CrossRef]
- Sun, K.; Luo, J.; Jing, X.; Xiang, W.; Guo, J.; Yao, X.; Liang, S.; Guo, F.; Xu, T. Hyperoside ameliorates the progression of osteoarthritis: An in vitro and in vivo study. Phytomedicine 2021, 80, 153387. [Google Scholar] [CrossRef] [PubMed]
- Hwang, H.V.; Tran, D.T.; Rebuffatti, M.N.; Li, C.-S.; Knowlton, A.A. Investigation of quercetin and hyperoside as senolytics in adult human endothelial cells. PLoS ONE 2018, 13, e0190374. [Google Scholar] [CrossRef] [PubMed]
- Wang, Q.; Wei, H.C.; Zhou, S.J.; Li, Y.; Zheng, T.T.; Zhou, C.Z.; Wan, X.H. Hyperoside: A review on its sources, biological activities, and molecular mechanisms. Phytother. Res. 2022, 36, 2779–2802. [Google Scholar] [CrossRef] [PubMed]
- Singh, A.; Mehta, S.; Patil, U.; Nuthong, P.; Benjakul, S. Hyperoside improves the textural and rheological properties of threadfin bream surimi gel. Int. J. Food Sci. Tech. 2023, 58, 4829–4840. [Google Scholar] [CrossRef]
- Buamard, N.; Singh, A.; Zhang, B.; Hong, H.; Singh, P.; Benjakul, S. Ethanolic extract of Duea Ching fruit: Extraction, characterization and its effect on the properties and storage stability of sardine surimi gel. Foods 2023, 12, 1635. [Google Scholar] [CrossRef]
- Olatunde, O.O.; Benjakul, S.; Vongkamjan, K. Coconut husk extract: Antibacterial properties and its application for shelf-life extension of Asian sea bass slices. Int. J. Food Sci. Tech. 2019, 54, 810–822. [Google Scholar] [CrossRef]
- Mehring, A.; Erdmann, N.; Walther, J.; Stiefelmaier, J.; Strieth, D.; Ulber, R. A simple and low-cost resazurin assay for vitality assessment across species. J. Biotechnol. 2021, 333, 63–66. [Google Scholar] [CrossRef]
- Tagrida, M.; Benjakul, S. Liposomes loaded with betel leaf (Piper betle L.) extract: Antibacterial activity and preservative effect in combination with hurdle technologies on tilapia slices. Food Control 2022, 138, 108999. [Google Scholar] [CrossRef]
- Jayeoye, T.J.; Nwabor, O.F.; Rujiralai, T. Synthesis of highly stable and dispersed silver nanoparticles/poly (vinyl alcohol-co-ethylene glycol)/poly (3-aminophenyl boronic acid) nanocomposite: Characterization and antibacterial, hemolytic and cytotoxicity studies. J. Ind. Eng. Chem. 2020, 89, 288–300. [Google Scholar] [CrossRef]
- Palamae, S.; Mittal, A.; Buatong, J.; Zhang, B.; Hong, H.; Benjakul, S. Chitooligosaccharide-catechin conjugate: Antimicrobial mechanisms toward Vibrio parahaemolyticus and its use in shucked Asian green mussel. Food Control 2023, 151, 109794. [Google Scholar] [CrossRef]
- Tang, Q.; Su, Y.-W.; Xian, C.J. Determining oxidative damage by lipid peroxidation assay in rat serum. Bio-Protocol 2019, 9, e3263. [Google Scholar] [CrossRef] [PubMed]
- Mohamed, S.; Mohamed, M.; Khalil, M.; Azmy, M.; Mabrouk, M. Combination of essential oil and ciprofloxacin to inhibit/eradicate biofilms in multidrug-resistant Klebsiella pneumoniae. J. Appl. Microbiol. 2018, 125, 84–95. [Google Scholar] [CrossRef] [PubMed]
- Mah, T.-F. Establishing the minimal bactericidal concentration of an antimicrobial agent for planktonic cells (MBC-P) and biofilm cells (MBC-B). J. Vis. Exp. 2014, 83, e50854. [Google Scholar]
- Liu, H.; Wang, Y.; Cao, J.; Jiang, H.; Yao, J.; Gong, G.; Chen, X.; Xu, W.; He, X. Antimicrobial activity and virulence attenuation of citral against the fish pathogen Vibrio alginolyticus. Aquaculture 2020, 515, 734578. [Google Scholar] [CrossRef]
- Fang, M.; Wang, R.; Agyekumwaa, A.K.; Yu, Y.; Xiao, X. Antibacterial effect of phenyllactic acid against Vibrio parahaemolyticus and its application on raw salmon fillets. LWT Food Sci. Technol. 2022, 154, 112586. [Google Scholar] [CrossRef]
- Niu, D.; Wang, Q.-Y.; Ren, E.-F.; Zeng, X.-A.; Wang, L.-H.; He, T.-F.; Wen, Q.-H.; Brennan, C.S. Multi-target antibacterial mechanism of eugenol and its combined inactivation with pulsed electric fields in a hurdle strategy on Escherichia coli. Food Control 2019, 106, 106742. [Google Scholar]
- Xie, Y.; Chen, J.; Xiao, A.; Liu, L. Antibacterial activity of polyphenols: Structure-activity relationship and influence of hyperglycemic condition. Molecules 2017, 22, 1913. [Google Scholar] [CrossRef]
- Bouarab-Chibane, L.; Forquet, V.; Lantéri, P.; Clément, Y.; Léonard-Akkari, L.; Oulahal, N.; Degraeve, P.; Bordes, C. Antibacterial properties of polyphenols: Characterization and QSAR (Quantitative structure–activity relationship) models. Front. Microbiol. 2019, 10, 829. [Google Scholar] [CrossRef]
- Mueller, E.A.; Levin, P.A. Bacterial cell wall quality control during environmental stress. mBio 2020, 11, e02456-20. [Google Scholar] [CrossRef] [PubMed]
- Cushnie, T.T.; O’Driscoll, N.H.; Lamb, A.J. Morphological and ultrastructural changes in bacterial cells as an indicator of antibacterial mechanism of action. Cell. Mol. Life Sci. 2016, 73, 4471–4492. [Google Scholar] [CrossRef] [PubMed]
- Karadeniz, F.; Oh, J.H.; Seo, Y.; Yang, J.; Lee, H.; Kong, C.-S. Quercetin 3-o-galactoside isolated from Limonium tetragonum inhibits melanogenesis by regulating PKA/MITF signaling and ERK activation. Int. J. Mol. Sci. 2023, 24, 3064. [Google Scholar] [CrossRef] [PubMed]
- Reygaert, W.C. An overview of the antimicrobial resistance mechanisms of bacteria. AIMS Microbiol. 2018, 4, 482. [Google Scholar] [CrossRef] [PubMed]
- Heikal, A.; Nakatani, Y.; Dunn, E.; Weimar, M.R.; Day, C.L.; Baker, E.N.; Lott, J.S.; Sazanov, L.A.; Cook, G.M. Structure of the bacterial type II NADH dehydrogenase: A monotopic membrane protein with an essential role in energy generation. Mol. Microbiol. 2014, 91, 950–964. [Google Scholar] [CrossRef] [PubMed]
- Song, L.; Hu, X.; Ren, X.; Liu, J.; Liu, X. Antibacterial modes of herbal flavonoids combat resistant bacteria. Front. Pharmacol. 2022, 13, 873374. [Google Scholar] [CrossRef] [PubMed]
- Nierhaus, K.H. Mg2+, K+, and the ribosome. J. Bacteriol. 2014, 196, 3817–3819. [Google Scholar] [CrossRef] [PubMed]
- Gründling, A. Potassium uptake systems in Staphylococcus aureus: New stories about ancient systems. mBio 2013, 4, e00784-13. [Google Scholar] [CrossRef]
- Thomas, K.J.; Rice, C.V. Revised model of calcium and magnesium binding to the bacterial cell wall. BioMetals 2014, 27, 1361–1370. [Google Scholar] [CrossRef]
- Alizadeh, M.; Kheirouri, S. Curcumin reduces malondialdehyde and improves antioxidants in humans with diseased conditions: A comprehensive meta-analysis of randomized controlled trials. BioMedicine 2019, 9, 23. [Google Scholar] [CrossRef]
- Chen, M.; Zhao, Z.; Meng, H.; Yu, S. The antibiotic activity and mechanisms of sugar beet (Beta vulgaris) molasses polyphenols against selected food-borne pathogens. LWT Food Sci. Technol. 2017, 82, 354–360. [Google Scholar] [CrossRef]
- Ignasimuthu, K.; Prakash, R.; Murthy, P.S.; Subban, N. Enhanced bioaccessibility of green tea polyphenols and lipophilic activity of EGCG octaacetate on gram-negative bacteria. LWT Food Sci. Technol. 2019, 105, 103–109. [Google Scholar] [CrossRef]
- Qi, L.; Christopher, G.F. Role of flagella, type IV pili, biosurfactants, and extracellular polymeric substance polysaccharides on the formation of pellicles by Pseudomonas aeruginosa. Langmuir 2019, 35, 5294–5304. [Google Scholar] [CrossRef] [PubMed]
- Yi, Z.; Xie, J. Genomic analysis of two representative strains of Shewanella putrefaciens isolated from bigeye tuna: Biofilm and spoilage-associated behavior. Foods 2022, 11, 1261. [Google Scholar] [CrossRef] [PubMed]
- Colunga Biancatelli, R.M.L.; Berrill, M.; Catravas, J.D.; Marik, P.E. Quercetin and vitamin C: An experimental, synergistic therapy for the prevention and treatment of SARS-CoV-2 related disease (COVID-19). Front. Immunol. 2020, 11, 1451. [Google Scholar] [CrossRef] [PubMed]
- Biharee, A.; Sharma, A.; Kumar, A.; Jaitak, V. Antimicrobial flavonoids as a potential substitute for overcoming antimicrobial resistance. Fitoterapia 2020, 146, 104720. [Google Scholar] [CrossRef] [PubMed]
- Banerjee, A.; Chowdhury, P.; Bauri, K.; Saha, B.; De, P. Inhibition and eradication of bacterial biofilm using polymeric materials. Biomater Sci. 2023, 11, 11–36. [Google Scholar] [CrossRef] [PubMed]
- Su, Y.; Yrastorza, J.T.; Matis, M.; Cusick, J.; Zhao, S.; Wang, G.; Xie, J. Biofilms: Formation, research models, potential targets, and methods for prevention and treatment. Adv. Sci. 2022, 9, 2203291. [Google Scholar] [CrossRef]
- Hamzah, H.; Hertiani, T.; Pratiwi, S.U.T.; Nuryastuti, T.; Murti, Y.B. The biofilm inhibition and eradication activity of curcumin againts polymicrobial biofilm. BIO. Web. Conf. 2020, 28, 04001. [Google Scholar] [CrossRef]
- da Silva, R.A.; Afonina, I.; Kline, K.A. Eradicating biofilm infections: An update on current and prospective approaches. Curr. Opin. Microbiol. 2021, 63, 117–125. [Google Scholar] [CrossRef]
- Tan, Y.; Ma, S.; Leonhard, M.; Moser, D.; Haselmann, G.M.; Wang, J.; Eder, D.; Schneider-Stickler, B. Enhancing antibiofilm activity with functional chitosan nanoparticles targeting biofilm cells and biofilm matrix. Carbohydr. Polym. 2018, 200, 35–42. [Google Scholar] [CrossRef] [PubMed]
- Choudhary, P.; Singh, S.; Agarwal, V. Microbial biofilms. In Bacterial Biofilms; IntechOpen: London, UK, 2020. [Google Scholar]
- Lahiri, D.; Nag, M.; Dutta, B.; Dey, A.; Ray, R.R. Bacterial extracellular polysaccharides in biofilm formation and function. In Application of Biofilms in Applied Microbiology; Elsevier: Amsterdam, The Netherlands, 2022; pp. 1–23. [Google Scholar]
- Wilking, J.N.; Zaburdaev, V.; De Volder, M.; Losick, R.; Brenner, M.P.; Weitz, D.A. Liquid transport facilitated by channels in Bacillus subtilis biofilms. Proc. Natl. Acad. Sci. USA 2013, 110, 848–852. [Google Scholar] [CrossRef] [PubMed]
- Das, S.; Hazarika, Z.; Sarmah, S.; Baruah, K.; Rohman, M.A.; Paul, D.; Jha, A.N.; Roy, A.S. Exploring the interaction of bioactive kaempferol with serum albumin, lysozyme and hemoglobin: A biophysical investigation using multi-spectroscopic, docking and molecular dynamics simulation studies. J. Photochem. Photobiol. B Biol. 2020, 205, 111825. [Google Scholar] [CrossRef] [PubMed]
- Yilmaz, H.; Gultekin Subasi, B.; Celebioglu, H.U.; Ozdal, T.; Capanoglu, E. Chemistry of protein-phenolic interactions toward the microbiota and microbial infections. Front. Nutr. 2022, 9, 914118. [Google Scholar] [CrossRef] [PubMed]
- Guedes, I.A.; de Magalhães, C.S.; Dardenne, L.E. Receptor–ligand molecular docking. Biophys. Rev. 2014, 6, 75–87. [Google Scholar] [CrossRef] [PubMed]
- Stockert, J.C. Predictive Binding Geometry of Ligands to DNA Minor Groove: Isohelicity and hydrogen-bonding pattern. In Methods in Molecular Biology; Humana Press: Totowa, NJ, USA, 2014; pp. 1–12. [Google Scholar]
- Sirajuddin, M.; Ali, S.; Badshah, A. Drug–DNA interactions and their study by UV–Visible, fluorescence spectroscopies and cyclic voltametry. J. Photochem. Photobiol. B Biol. 2013, 124, 1–19. [Google Scholar] [CrossRef] [PubMed]
- Bodapati, A.T.S.; Sahoo, B.K.; Ragaiahgari, S.R.; Kandikonda, L.; Madku, S.R. Deciphering the nature of binding of dexlansoprazole with DNA: Biophysical and docking approaches. Int. J. Biol. Macromol. 2022, 217, 1027–1036. [Google Scholar] [CrossRef]

| Activity | Shewanella putrefaciens | Pseudomonas aeruginosa | ||||
|---|---|---|---|---|---|---|
| Control | Quercetin (2 MIC) | Hyperoside (2 MIC) | Control | Quercetin (2 MIC) | Hyperoside (2 MIC) | |
| TTC- dehydrogenase activity (%) | 88.22 ± 0.75 a | 37.31 ± 0.23 b | 30.64 ± 0.44 c | 90.36 ± 0.8 a | 52.74 ± 1.05 b | 41.43 ± 1.14 c |
| K+ ion leakage (mg/L) | 685.65 ± 0.63 c | 1182.4 ± 1.41 b | 1562.35 ± 1.48 a | 777.9 ± 0.56 c | 1208.65 ± 0.49 b | 1676.35 ± 1.34 a |
| Mg+2 ion leakage (mg/L) | 2.307 ± 0.01 c | 6.286 ± 0.01 b | 9.442 ± 0.07 a | 2.992 ± 0.01 c | 6.958 ± 0.06 b | 10.53 ± 0.01 a |
| Conductivity (ms/cm) | 4.75 ± 0.03 c | 8.11 ± 0.08 b | 12.55 ± 0.06 a | 4.97 ± 0.04 c | 9.02 ± 0.02 b | 13.7 ± 0.01 a |
| MDA content (nmol/mL) | 0.88 ± 0.01 c | 4.38 ± 0.02 b | 5.27 ± 0.01 a | 0.94 ± 0.01 c | 4.83 ± 0.01 b | 6.05 ± 0.01 a |
| EPS content reduction (%) | - | 73.04 ± 0.01 b | 78.07 ± 0.01 a | - | 71.55 ± 0.02 b | 75.18 ± 0.00 a |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Tagrida, M.; Palamae, S.; Saetang, J.; Ma, L.; Hong, H.; Benjakul, S. Comparative Study of Quercetin and Hyperoside: Antimicrobial Potential towards Food Spoilage Bacteria, Mode of Action and Molecular Docking. Foods 2023, 12, 4051. https://doi.org/10.3390/foods12224051
Tagrida M, Palamae S, Saetang J, Ma L, Hong H, Benjakul S. Comparative Study of Quercetin and Hyperoside: Antimicrobial Potential towards Food Spoilage Bacteria, Mode of Action and Molecular Docking. Foods. 2023; 12(22):4051. https://doi.org/10.3390/foods12224051
Chicago/Turabian StyleTagrida, Mohamed, Suriya Palamae, Jirakrit Saetang, Lukai Ma, Hui Hong, and Soottawat Benjakul. 2023. "Comparative Study of Quercetin and Hyperoside: Antimicrobial Potential towards Food Spoilage Bacteria, Mode of Action and Molecular Docking" Foods 12, no. 22: 4051. https://doi.org/10.3390/foods12224051







