Simultaneous Quantitative Determination of Low-Concentration Preservatives and Heavy Metals in Tricholoma Matsutakes Based on SERS and FLU Spectral Data Fusion
Abstract
:1. Introduction
2. Materials and Methods
2.1. Sample Preparation
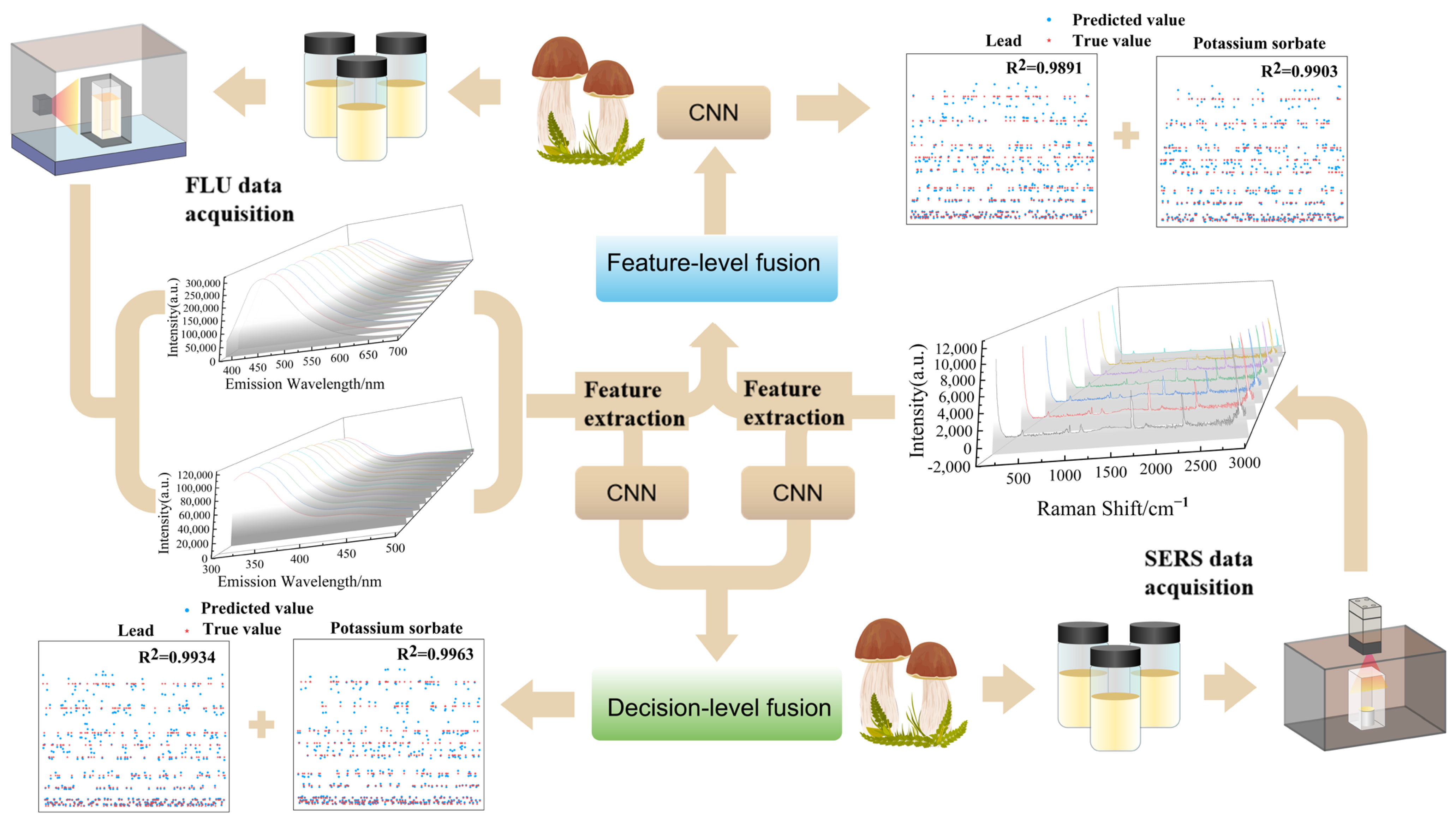
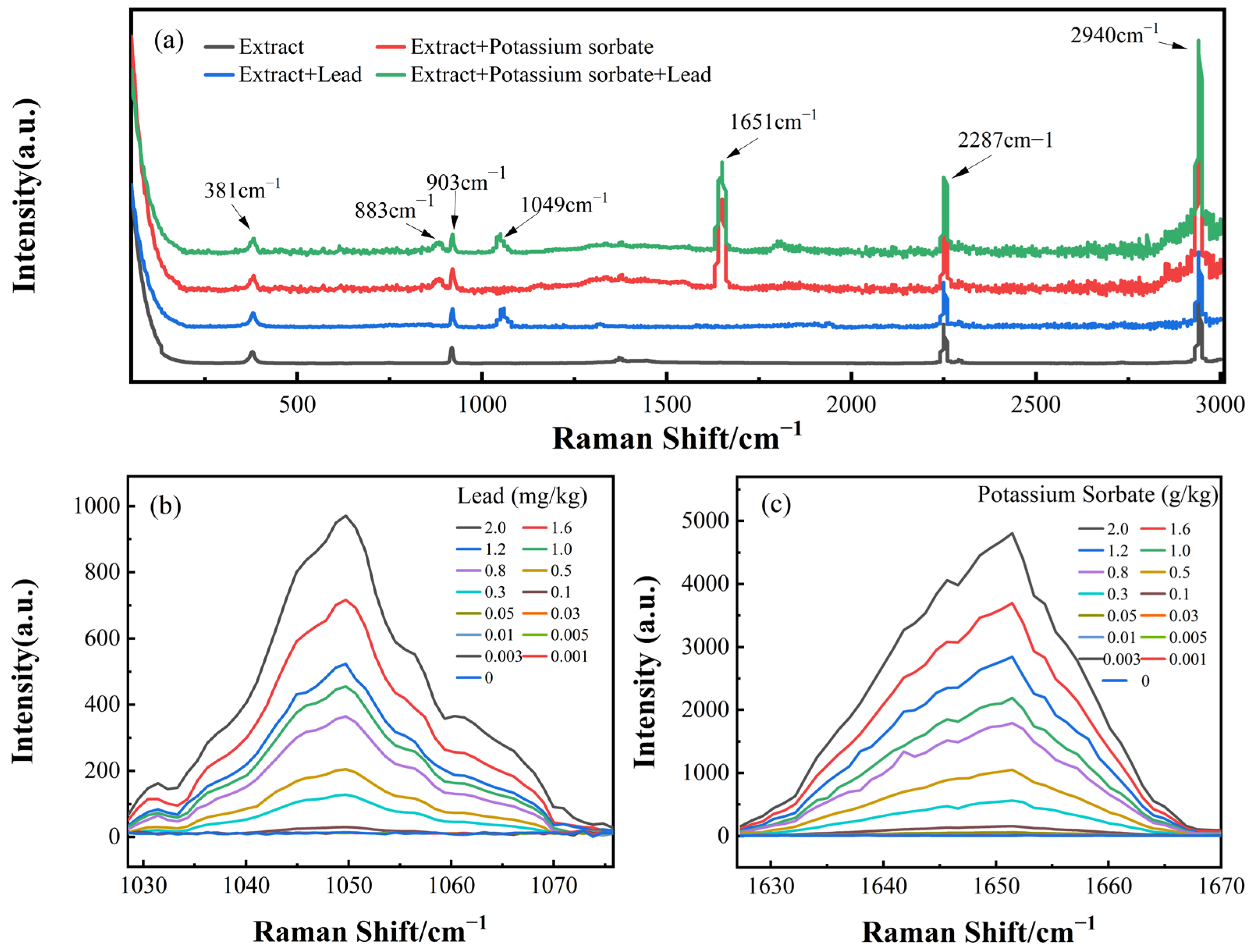
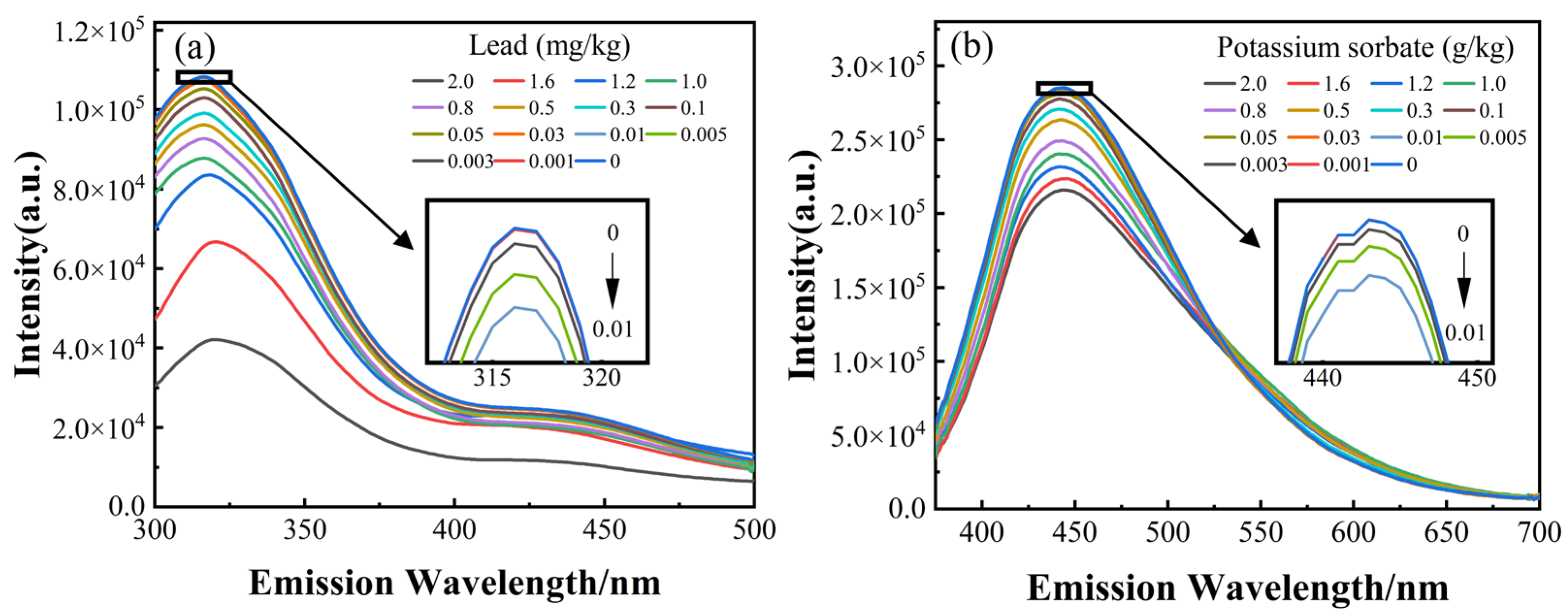
2.2. Spectroscopy Data Acquisition
2.3. Data Analysis Methods
2.3.1. Quantitative Models and Evaluation
2.3.2. Data Processing and Feature Extraction
2.3.3. Data Fusion
3. Results and Discussion
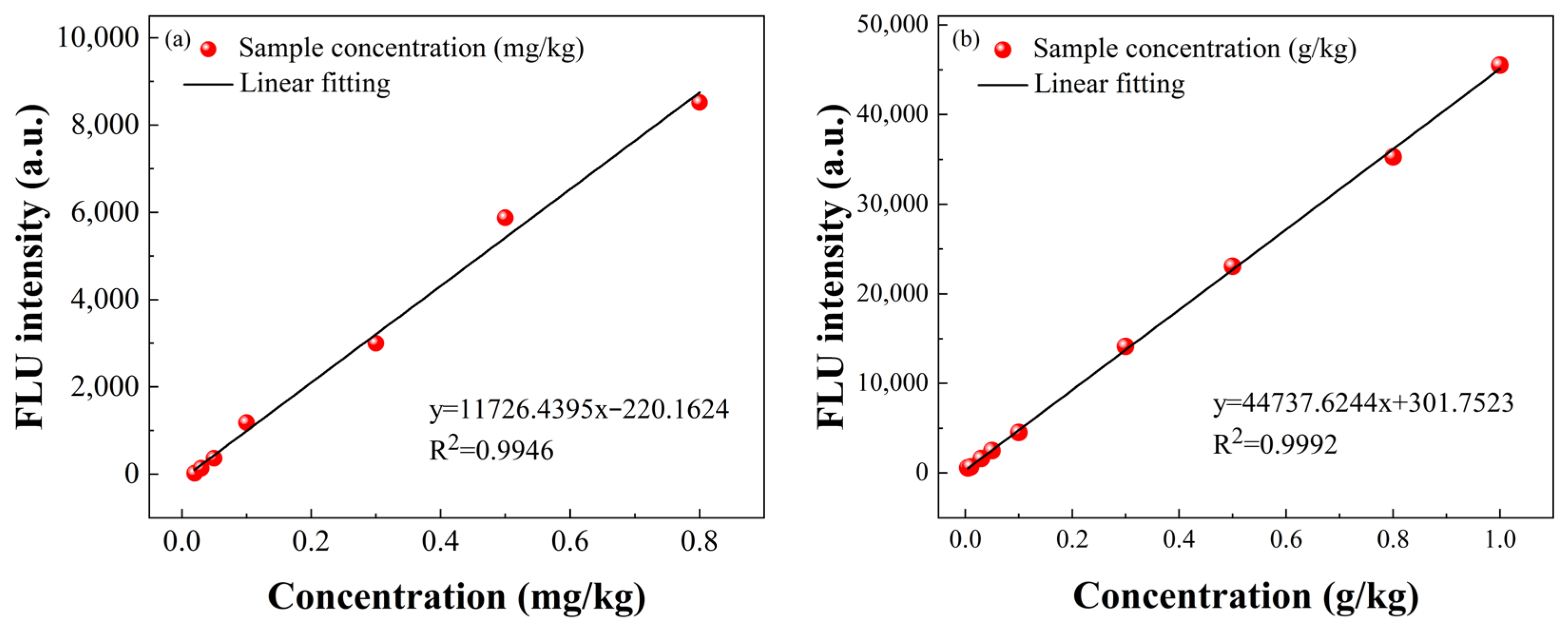
3.1. Spectral Curve
3.2. Modeling and Analysis of the Individual Spectra
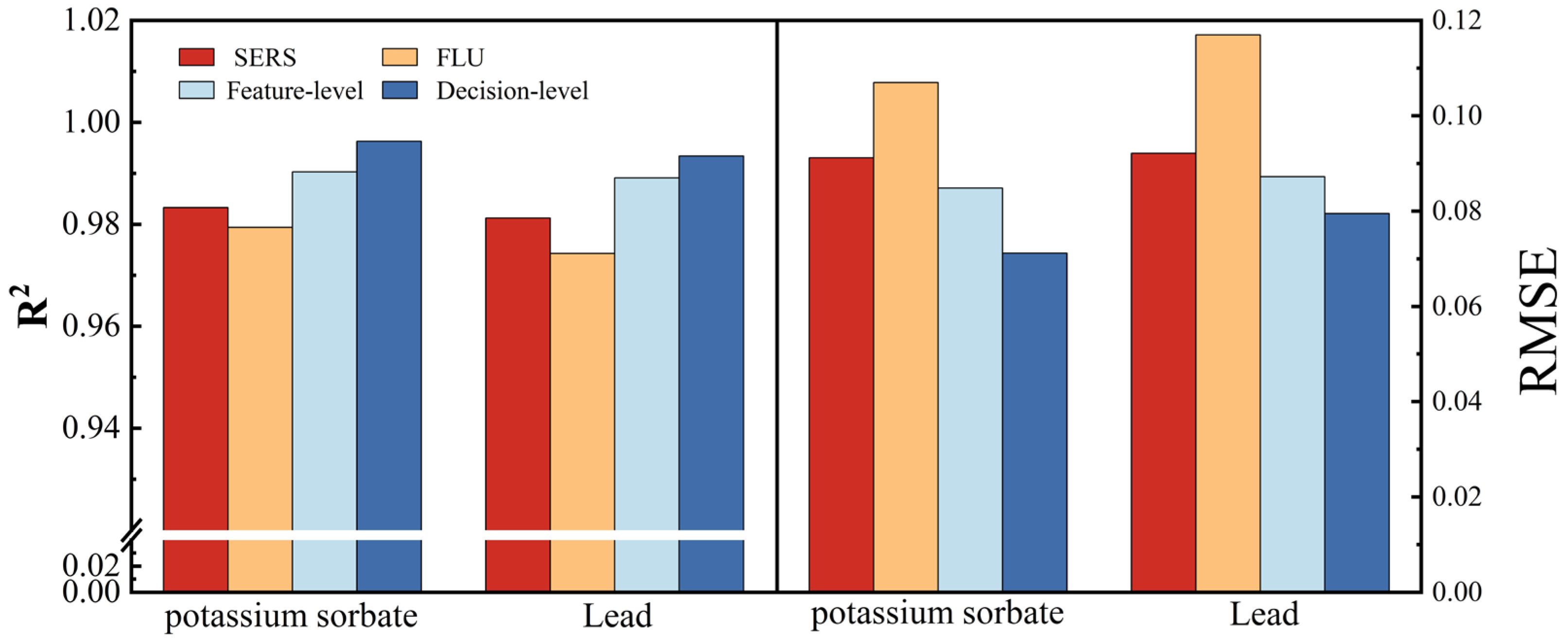
3.3. Data Fusion
3.3.1. Modeling and Analysis of Feature-Level Data Fusion
3.3.2. Modeling and Analysis of Decision-Level Data Fusion
4. Conclusions
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Zhou, Y.F.; El-Seedi, H.R.; Xu, B.J. Insights into health promoting effects and myochemical profiles of pine mushroom Tricholoma matsutake. Crit. Rev. Food Sci. Nutr. 2021, 63, 5698–5723. [Google Scholar] [CrossRef]
- Ronda, O.; Grzadka, E.; Ostolska, I.; Orzel, J.; Cieslik, B.M. Accumulation of radioisotopes and heavy metals in selected species of mushrooms. Food Chem. 2022, 367, 130670. [Google Scholar] [CrossRef]
- Liu, S.; Liu, H.G.; Li, J.Q.; Wang, Y.Z. Research Progress on Elements of Wild Edible Mushrooms. J. Fungi 2022, 8, 964. [Google Scholar] [CrossRef]
- Yang, Z.H.; Xu, J.C.; Yang, L.; Zhang, X.S. Optimized Dynamic Monitoring and Quality Management System for Post-Harvest Matsutake of Different Preservation Packaging in Cold Chain. Foods 2022, 11, 2646. [Google Scholar] [CrossRef]
- Kalantari, R.; Moghimi, A.; Azizinezhad, F. Simultaneous Green Separation/Preconcentration and Determination of Lead Ions in Water Samples Via Graphite Furnace Atomic Absorption Spectrometry. J. Appl. Spectrosc. 2023, 90, 686–695. [Google Scholar] [CrossRef]
- Kim, Y.J.; Lee, S.-Y.; Hur, M. Back to the Basics of Liquid Chromatography-Mass Spectrometry. Ann. Lab. Med. 2022, 42, 119–120. [Google Scholar] [CrossRef] [PubMed]
- Wang, L.Y.; Peng, X.L.; Fu, H.J.; Huang, C.; Li, Y.P.; Liu, Z.M. Recent advances in the development of electrochemical aptasensors for detection of heavy metals in food. Biosens. Bioelectron. 2020, 147, 111777. [Google Scholar] [CrossRef] [PubMed]
- Fang, X.Q.; Peng, Y.K.; Wang, W.X.; Zheng, X.C.; Li, Y.Y.; Bu, X.P. Rapid and Simultaneous Detection of Sodium Benzoate and Potassium Sorbate in Cocktail Based on Surface-Enhanced Raman Spectroscopy. Spectrosc. Spectr. Anal. 2018, 38, 2794–2799. [Google Scholar]
- Wang, S.T.; Liu, S.Y.; Wang, Z.F.; Zhang, J.K.; Kong, D.M.; Wang, Y.T. The Determination of Potassium Sorbate Concentration Based on ICSO-SVM Combining Three-Dimensional Fluorescence Spectra. Spectrosc. Spectr. Anal. 2020, 40, 1614–1619. [Google Scholar]
- An, H.; Zhai, C.; Zhang, F.; Ma, Q.; Sun, J.; Tang, Y.; Wang, W. Quantitative analysis of Chinese steamed bread staling using NIR, MIR, and Raman spectral data fusion. Food Chem. 2023, 405, 134821. [Google Scholar] [CrossRef]
- Qingya, W.; Li, F.; Jiang, X.; Hao, J.; Zhao, Y.; Wu, S.; Cai, Y.; Huang, W. Quantitative analysis of soil cadmium content based on the fusion of XRF and Vis-NIR data. Chemom. Intell. Lab. Syst. 2022, 226, 104578. [Google Scholar] [CrossRef]
- Ren, L.; Tian, Y.; Yang, X.; Wang, Q.; Wang, L.; Geng, X.; Wang, K.; Du, Z.; Li, Y.; Lin, H. Rapid identification of fish species by laser-induced breakdown spectroscopy and Raman spectroscopy coupled with machine learning methods. Food Chem. 2023, 400, 134043. [Google Scholar] [CrossRef] [PubMed]
- Zhao, Q.; Yu, Y.; Hao, N.; Miao, P.; Li, X.; Liu, C.; Li, Z. Data fusion of Laser-induced breakdown spectroscopy and Near-infrared spectroscopy to quantitatively detect heavy metals in lily. Microchem. J. 2023, 190, 108670. [Google Scholar] [CrossRef]
- Li, X.; Cai, M.; Li, M.; Wei, X.; Liu, Z.; Wang, J.; Jia, K.; Han, Y. Combining Vis-NIR and NIR hyperspectral imaging techniques with a data fusion strategy for the rapid qualitative evaluation of multiple qualities in chicken. Food Control 2023, 145, 109416. [Google Scholar] [CrossRef]
- Li, Y.; Huang, Y.; Xia, J.; Xiong, Y.; Min, S. Quantitative analysis of honey adulteration by spectrum analysis combined with several high-level data fusion strategies. Vib. Spectrosc. 2020, 108, 103060. [Google Scholar] [CrossRef]
- Xu, Z.; Li, X.; Cheng, W.; Zhao, G.; Tang, L.; Yang, Y.; Wu, Y.; Zhang, P.; Wang, Q. Data fusion strategy based on ultraviolet–visible spectra and near-infrared spectra for simultaneous and accurate determination of key parameters in surface water. Spectrochim. Acta Part A Mol. Biomol. Spectrosc. 2023, 302, 123007. [Google Scholar] [CrossRef]
- Li, D.; Yue, W.; Gao, P.; Gong, T.; Wang, C.; Luo, X. Surface-enhanced Raman spectroscopy (SERS) for the characterization of atmospheric aerosols: Current status and challenges. TrAC Trends Anal. Chem. 2023, 170, 117426. [Google Scholar] [CrossRef]
- Zhang, Y.; Guo, S.; Wei, C. Fluorescence Spectroscopy: Part I Principles. In Encyclopedia of Soils in the Environment, 2nd ed.; Goss, M.J., Oliver, M., Eds.; Academic Press: Oxford, UK, 2023; pp. 544–551. [Google Scholar]
- Zeng, S.; Zhang, Z.; Cheng, X.; Cai, X.; Cao, M.; Guo, W. Prediction of soluble solids content using near-infrared spectra and optical properties of intact apple and pulp applying PLSR and CNN. Spectrochim. Acta Part A Mol. Biomol. Spectrosc. 2024, 304, 123402. [Google Scholar] [CrossRef]
- Liu, P.; Wang, X.; Yin, L.; Liu, B. Flat random forest: A new ensemble learning method towards better training efficiency and adaptive model size to deep forest. Int. J. Mach. Learn. Cybern. 2020, 11, 2501–2513. [Google Scholar] [CrossRef]
- Camara, C.; Peris-Lopez, P.; Safkhani, M.; Bagheri, N. ECG Identification Based on the Gramian Angular Field and Tested with Individuals in Resting and Activity States. Sensors 2023, 23, 937. [Google Scholar] [CrossRef]
- Li, R.; Wu, Y.; Wu, Q.; Dey, N.; González Crespo, R.; Shi, F. Emotion stimuli-based surface electromyography signal classification employing Markov transition field and deep neural networks. Measurement 2022, 189, 110470. [Google Scholar] [CrossRef]
- Chen, W.; Shi, K. A deep learning framework for time series classification using Relative Position Matrix and Convolutional Neural Network. Neurocomputing 2019, 359, 384–394. [Google Scholar] [CrossRef]
- Bai, R.; Meng, Z.; Xu, Q.; Fan, F. Fractional Fourier and time domain recurrence plot fusion combining convolutional neural network for bearing fault diagnosis under variable working conditions. Reliab. Eng. Syst. Saf. 2023, 232, 109076. [Google Scholar] [CrossRef]
- Tang, R.; Chen, X.; Li, C. Detection of Nitrogen Content in Rubber Leaves Using Near-Infrared (NIR) Spectroscopy with Correlation-Based Successive Projections Algorithm (SPA). Appl. Spectrosc. 2018, 72, 740–749. [Google Scholar] [CrossRef] [PubMed]
- Lakshmanan, M.K.; Boelt, B.; Gislum, R. A chemometric method for the viability analysis of spinach seeds by near infrared spectroscopy with variable selection using successive projections algorithm. J. Near Infrared Spectrosc. 2023, 31, 24–32. [Google Scholar] [CrossRef]
- Wang, Z. Research on Feature Selection Methods based on Random Forest. Tehnicki Vjesn.-Tech. Gaz. 2023, 30, 623–633. [Google Scholar] [CrossRef]
- Li, C.; Ma, X.; Teng, Y.; Li, S.; Jin, Y.; Du, J.; Jiang, L. Quantitative Analysis of Forest Water COD Value Based on UV-vis and FLU Spectral Information Fusion. Forests 2023, 14, 1361. [Google Scholar] [CrossRef]
- Lin, H.; Tang, C. Analysis and Optimization of Urban Public Transport Lines Based on Multiobjective Adaptive Particle Swarm Optimization. IEEE Trans. Intell. Transp. Syst. 2022, 23, 16786–16798. [Google Scholar] [CrossRef]
- Nie, L.; Dai, Z.; Ma, S. Enhanced Accuracy of Near-Infrared Spectroscopy for Traditional Chinese Medicine with Competitive Adaptive Reweighted Sampling. Anal. Lett. 2016, 49, 2259–2267. [Google Scholar] [CrossRef]
- Agjee, N.e.H.; Ismail, R.; Mutanga, O. Identifying relevant hyperspectral bands using Boruta: A temporal analysis of water hyacinth biocontrol. J. Appl. Remote Sens. 2016, 10, 042002. [Google Scholar] [CrossRef]
- Liu, L.; Wan, X.; Li, J.; Wang, W.; Gao, Z. An Improved Entropy-Weighted Topsis Method for Decision-Level Fusion Evaluation System of Multi-Source Data. Sensors 2022, 22, 6391. [Google Scholar] [CrossRef]
- Fu, C.; Li, M. Data Fusion-Based Structural Damage Identification Approach Integrating Fractal and RCPN. Appl. Sci. 2023, 13, 5289. [Google Scholar] [CrossRef]
- Jing, Z.; Pan, H.; Qin, Y. Current progress of information fusion in China. Chin. Sci. Bull. 2013, 58, 4533–4540. [Google Scholar] [CrossRef]
- Meng, Q.; Zhang, C.; Song, T.; Li, N. The Application of the Improved TOPSIS Method in Bid Evaluation of Highway Construction. In Proceedings of the 2nd International Conference on Civil Engineering, Architecture and Building Materials (CEABM 2012), Yantai, China, 25–27 May 2012. [Google Scholar]
- Speiser, J.L.; Miller, M.E.; Tooze, J.; Ip, E. A comparison of random forest variable selection methods for classification prediction modeling. Expert Syst. Appl. 2019, 134, 93–101. [Google Scholar] [CrossRef]
- Chen, X.; Ishwaran, H. Random forests for genomic data analysis. Genomics 2012, 99, 323–329. [Google Scholar] [CrossRef]
- Jiang, H.; Chen, J.; Deng, J.; Zhao, X.; Xu, L. Quantitative determination of heavy metal Pb content in soybean oil based on microwave detection technique combined with multivariate analysis. Sens. Actuators A Phys. 2023, 363, 114771. [Google Scholar] [CrossRef]
- Wang, S.T.; Zhang, C.; Zhang, Q.; Wang, Z.; Zhu, C.; Yang, X. The determination of potassium sorbate based on improved least squares support vector machine combining fluorescence spectra. Opt. Tech. 2018, 44, 188–193. [Google Scholar] [CrossRef]
| Serial Number | Potassium Sorbate (g·kg−1) | Lead Element (mg·kg−1) |
|---|---|---|
| 1 | 0 | 0 |
| 2 | 0.001 | 0.001 |
| 3 | 0.003 | 0.003 |
| 4 | 0.005 | 0.005 |
| 5 | 0.01 | 0.01 |
| 6 | 0.03 | 0.03 |
| 7 | 0.05 | 0.05 |
| 8 | 0.1 | 0.1 |
| 9 | 0.3 | 0.3 |
| 10 | 0.5 | 0.5 |
| 11 | 0.8 | 0.8 |
| 12 | 1.0 | 1.0 |
| 13 | 1.2 | 1.2 |
| 14 | 1.6 | 1.6 |
| 15 | 2.0 | 2.0 |
| Methods | Model | Lead Element | Potassium Sorbate | ||
|---|---|---|---|---|---|
| R2 | RMSE (mg·kg−1) | R2 | RMSE (g·kg−1) | ||
| none | PLSR | 0.9604 | 0.1227 | 0.9668 | 0.1202 |
| SPA | 0.9681 | 0.1172 | 0.9724 | 0.1095 | |
| Boruta | 0.9652 | 0.1191 | 0.9688 | 0.1143 | |
| CARS | 0.9702 | 0.1125 | 0.9725 | 0.1090 | |
| none | DF | 0.9677 | 0.1147 | 0.9714 | 0.1109 |
| SPA | 0.9714 | 0.1085 | 0.9783 | 0.1026 | |
| Boruta | 0.9685 | 0.1097 | 0.9735 | 0.1078 | |
| CARS | 0.9725 | 0.1066 | 0.9803 | 0.0997 | |
| SPA-GAF | CNN | 0.9801 | 0.0894 | 0.9833 | 0.0841 |
| Boruta-GAF | 0.9782 | 0.0972 | 0.9781 | 0.0931 | |
| CARS-GAF | 0.9812 | 0.0875 | 0.9829 | 0.0852 | |
| SPA-MTF | 0.9741 | 0.1012 | 0.9779 | 0.0967 | |
| Boruta-MTF | 0.9688 | 0.1097 | 0.9751 | 0.1002 | |
| CARS-MTF | 0.9748 | 0.0962 | 0.9775 | 0.0972 | |
| SPA-RP | 0.9785 | 0.0923 | 0.9812 | 0.0895 | |
| Boruta-RP | 0.9698 | 0.1067 | 0.9766 | 0.1021 | |
| CARS-RP | 0.9792 | 0.0901 | 0.9810 | 0.0899 | |
| SPA-RPB | 0.9765 | 0.0992 | 0.9804 | 0.0907 | |
| Boruta-RPB | 0.9724 | 0.1075 | 0.9789 | 0.0931 | |
| CARS-RPB | 0.9766 | 0.0990 | 0.9799 | 0.0918 | |
| Analyte | Spectra | Models | R2 | RMSE |
|---|---|---|---|---|
| Lead element | SERS | CARS-GAF-CNN | 0.9812 | 0.0875 |
| Potassium sorbate | SPA-GAF-CNN | 0.9833 | 0.0841 | |
| Lead element | FLU | CARS-GAF-CNN | 0.9743 | 0.1117 |
| Potassium sorbate | CARS-GAF-CNN | 0.9794 | 0.1070 |
| Analyte | Potassium Sorbate | Lead Element | ||
|---|---|---|---|---|
| Methods | SPA-CNN | CARS-CNN | SPA-CNN | CARS-CNN |
| R2 | 0.9881 | 0.9903 | 0.9852 | 0.9891 |
| RMSE | 0.0902 g·kg−1 | 0.0848 g·kg−1 | 0.0908 mg·kg−1 | 0.0872 mg·kg−1 |
| Analyte | Potassium Sorbate | Lead Element | ||
|---|---|---|---|---|
| Methods | TOPSIS-CNN | RF-CNN | TOPSIS-CNN | RF-CNN |
| R2 | 0.9963 | 0.9952 | 0.9932 | 0.9934 |
| RMSE | 0.0712 g·kg−1 | 0.0741 g·kg−1 | 0.0803 mg·kg−1 | 0.0795 mg·kg−1 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Jin, Y.; Li, C.; Huang, Z.; Jiang, L. Simultaneous Quantitative Determination of Low-Concentration Preservatives and Heavy Metals in Tricholoma Matsutakes Based on SERS and FLU Spectral Data Fusion. Foods 2023, 12, 4267. https://doi.org/10.3390/foods12234267
Jin Y, Li C, Huang Z, Jiang L. Simultaneous Quantitative Determination of Low-Concentration Preservatives and Heavy Metals in Tricholoma Matsutakes Based on SERS and FLU Spectral Data Fusion. Foods. 2023; 12(23):4267. https://doi.org/10.3390/foods12234267
Chicago/Turabian StyleJin, Yuanyin, Chun Li, Zhengwei Huang, and Ling Jiang. 2023. "Simultaneous Quantitative Determination of Low-Concentration Preservatives and Heavy Metals in Tricholoma Matsutakes Based on SERS and FLU Spectral Data Fusion" Foods 12, no. 23: 4267. https://doi.org/10.3390/foods12234267
APA StyleJin, Y., Li, C., Huang, Z., & Jiang, L. (2023). Simultaneous Quantitative Determination of Low-Concentration Preservatives and Heavy Metals in Tricholoma Matsutakes Based on SERS and FLU Spectral Data Fusion. Foods, 12(23), 4267. https://doi.org/10.3390/foods12234267







