Variations in the Metabolome of Unaged and Aged Beef from Black-and-White Cows and Heifers by 1H NMR Spectroscopy
Abstract
:1. Introduction
2. Materials and Methods
2.1. Raw Material
2.2. Beef Aging
2.3. Sample Preparation
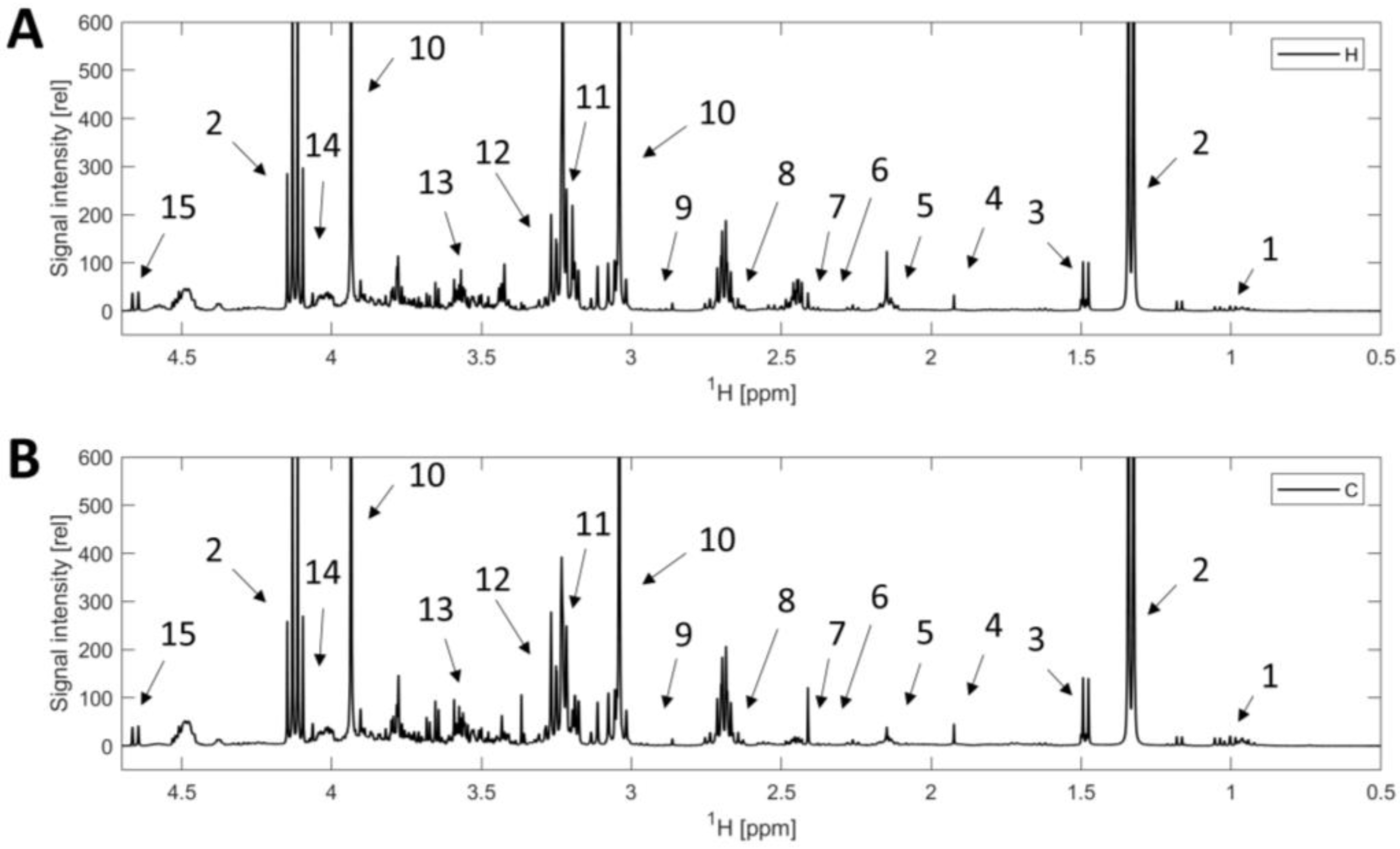
2.4. NMR Spectroscopy
2.5. Data Analysis
2.5.1. Non-Targeted Analysis
2.5.2. Targeted Analysis
3. Results and Discussion
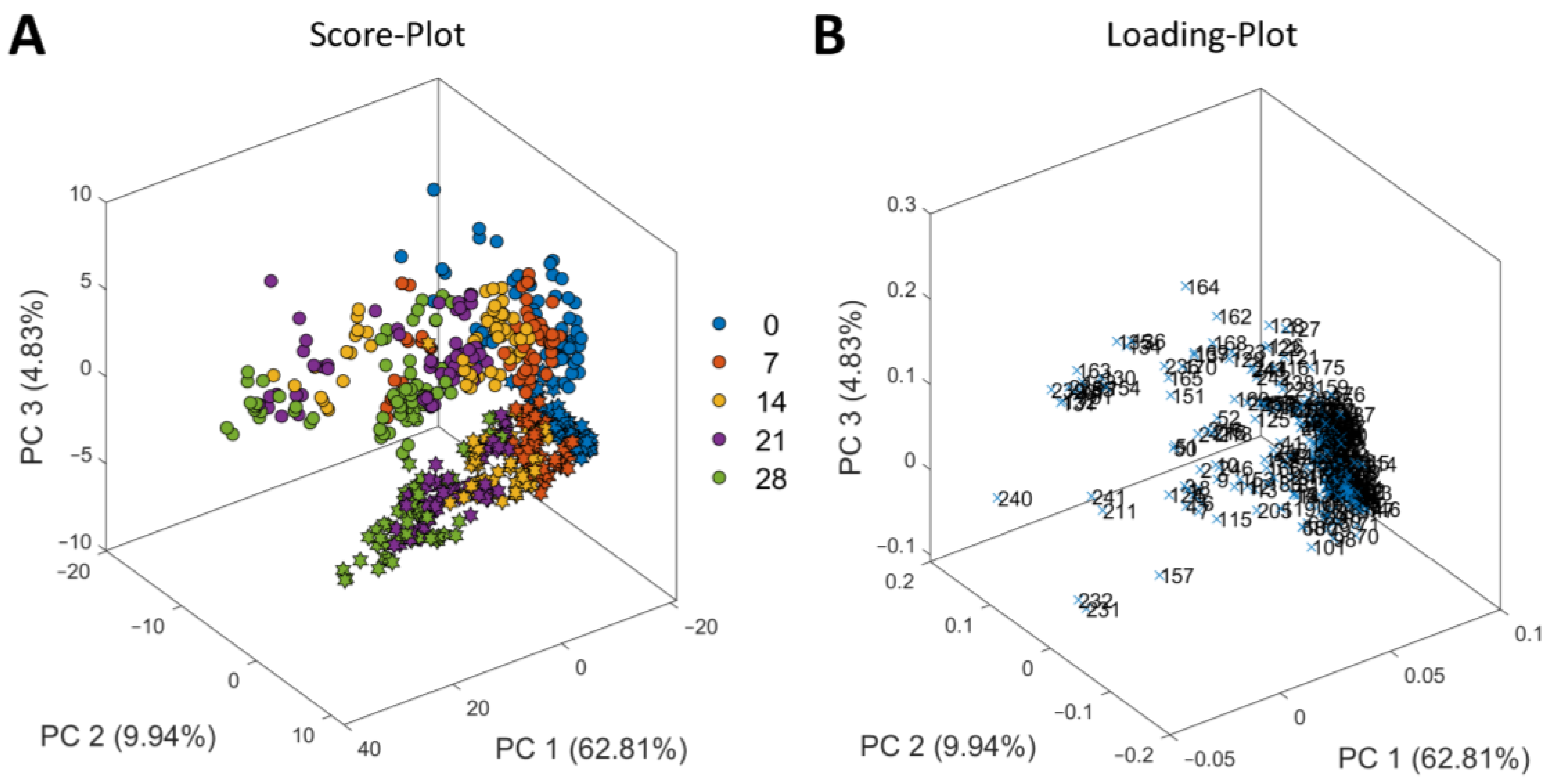
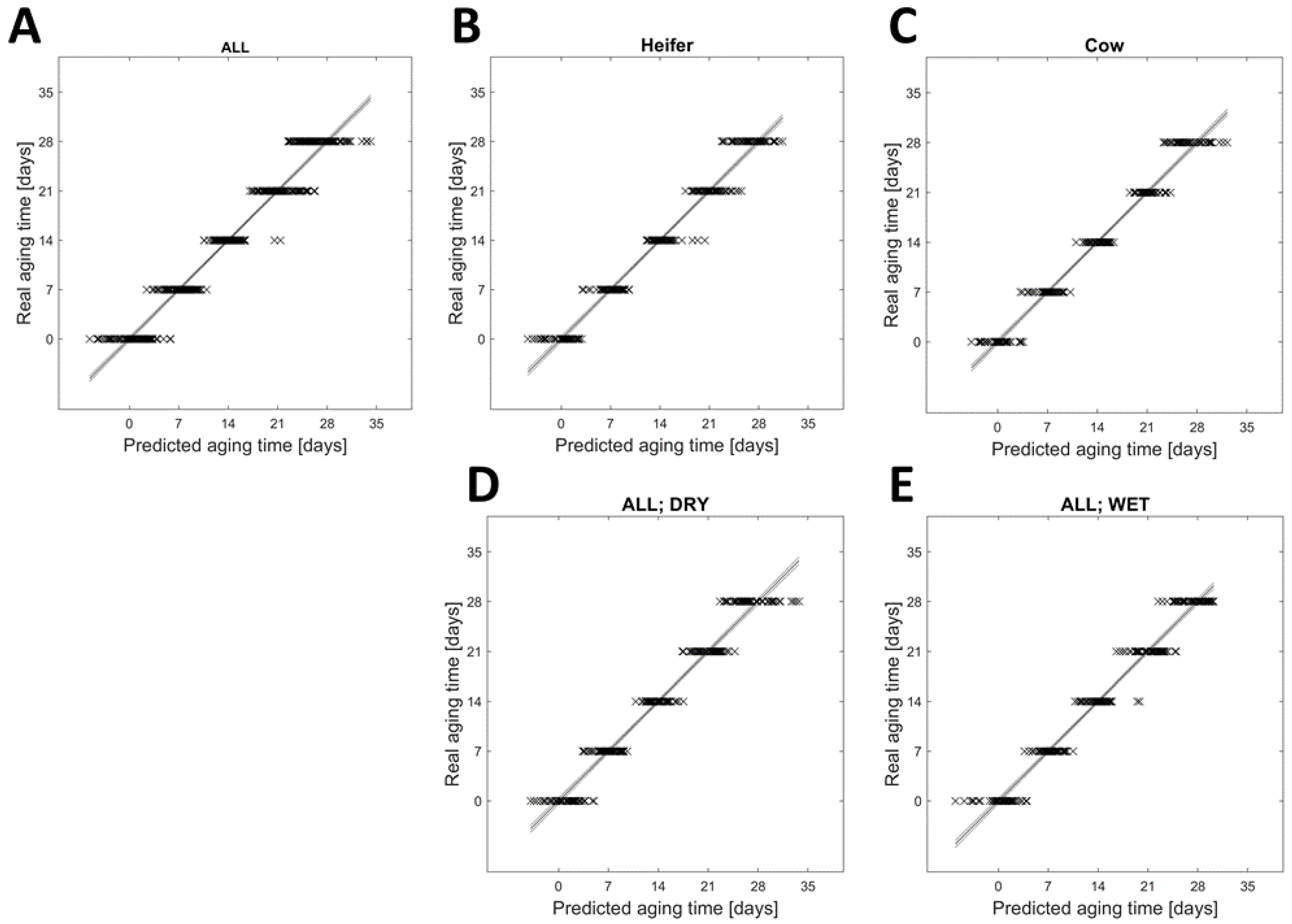
3.1. Influence of Aging Time on Beef Metabolome
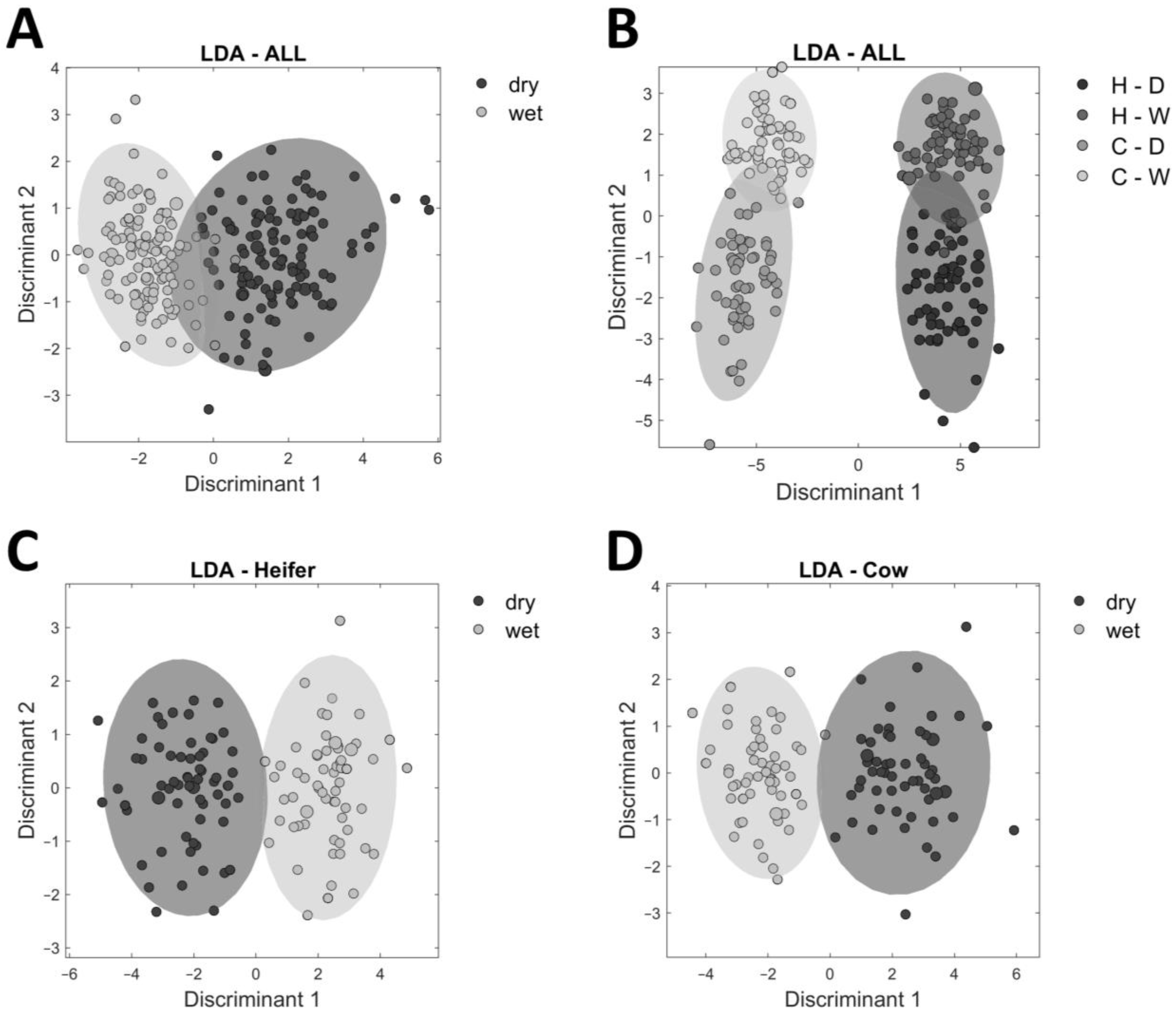
3.2. Differences in the Metabolome of Beef between Cows and Heifers
3.3. Influence of Aging Type on the Metabolome
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Appendix A
| Metabolite | Cattle Type | Aging Type | Concentration in µmol/g Wet Meat over Aging Time | Spearman’s Correlation rsp | ||||
|---|---|---|---|---|---|---|---|---|
| 0 Days | 7 Days | 14 Days | 21 Days | 28 Days | ||||
| Amino acids | ||||||||
| Alanine (1.47–1.51 ppm) | H | D W | 3.64 ± 0.51 a,A 3.58 ± 0.56 a,A | 3.92 ± 0.75 ab,A 3.85 ± 0.64 ab,A | 4.66 ± 0.83 bc,A 4.46 ± 0.85 bc,A | 5.61 ± 1.39 cd,A 5.21 ± 0.79 cd,A | 6.24 ± 1.11 d,A 5.85 ± 0.72 d,A | 0.78 0.77 |
| C | D W | 3.79 ± 0.32 a,A 4.02 ± 0.37 a,A | 4.16 ± 0.47 a,A 4.20 ± 0.38 a,A | 4.47 ± 0.55 a,A 4.69 ± 0.51 ab,B | 5.45 ± 0.62 b,A 5.33 ± 0.59 bc,A | 6.08 ± 0.73 b,A 5.79 ± 0.54 c,A | 0.80 0.85 | |
| All | 3.75 ± 0.48 a | 4.02 ± 0.60 a | 4.57 ± 0.71 b | 5.40 ± 0.93 c | 6.00 ± 0.82 d | |||
| Aspartate (2.79–2.80 ppm) | H | D W | 0.36 ± 0.21 a,A 0.37 ± 0.17 a,A | 0.47 ± 0.23 a,A 0.55 ± 0.29 ab,B | 0.64 ± 0.26 b,A 0.61 ± 0.20 b,A | 0.69 ± 0.30 b,A 0.70 ± 0.18 bc,A | 0.77 ± 0.23 b,A 0.85 ± 0.20 c,A | 0.58 0.65 |
| C | D W | 0.30 ± 0.08 a,A 0.31 ± 0.09 a,A | 0.35 ± 0.10 ab,A 0.37 ± 0.10 ab,A | 0.46 ± 0.12 bc,A 0.48 ± 0.12 bc,A | 0.54 ± 0.12 cd,A 0.59 ± 0.14 cd,B | 0.73 ± 0.17 d,A 0.77 ± 0.17 d,A | 0.76 0.79 | |
| All | 0.33 ± 0.15 a | 0.44 ± 0.21 b | 0.55 ± 0.20 c | 0.64 ± 0.21 c | 0.78 ± 0.20 d | |||
| Glutamate (2.32–2.39 ppm) | H | D W | 1.21 ± 0.33 a,A 1.24 ± 0.36 a,A | 1.46 ± 0.50 ab,A 1.57 ± 0.52 ab,B | 1.94 ± 0.65 bc,A 2.09 ± 0.75 bc,B | 2.43 ± 0.74 cd,A 2.62 ± 0.69 cd,B | 2.89 ± 0.66 d,A 3.18 ± 0.64 d,B | 0.76 0.81 |
| C | D W | 1.17 ± 0.24 a,A 1.12 ± 0.17 a,A | 1.34 ± 0.26 a,A 1.42 ± 0.22 ab,A | 1.78 ± 0.38 ab,A 1.92 ± 0.38 bc,B | 2.25 ± 0.47 bc,A 2.46 ± 0.54 cd,B | 2.72 ± 0.50 c,A 2.98 ± 0.50 d,B | 0.84 0.87 | |
| All | 1.19 ± 0.29 a | 1.45 ± 0.41 b | 1.94 ± 0.58 c | 2.44 ± 0.63 d | 2.95 ± 0.60 e | |||
| Glutamine (2.11–2.12 ppm) | H | D W | 4.78 ± 1.99 a,A 5.03 ± 2.33 a,A | 5.20 ± 1.57 a,A 5.22 ± 1.37 a,A | 6.43 ± 1.78 ab,A 6.38 ± 1.73 ab,A | 7.61 ± 1.94 bc,A 7.69 ± 1.82 bc,A | 8.64 ± 1.95 c,A 8.52 ± 1.59 c,A | 0.61 0.63 |
| C | D W | 5.04 ± 1.46 a,A 4.74 ± 1.35 a,A | 4.83 ± 0.99 a,A 5.06 ± 0.83 a,A | 6.09 ± 1.10 ab,A 6.33 ± 1.01 ab,A | 7.40 ± 1.31 bc,A 7.45 ± 1.29 bc,A | 8.09 ± 1.12 c,A 8.32 ± 1.17 c,A | 0.69 0.72 | |
| All | 4.90 ± 1.83 a | 5.09 ± 1.24 a | 6.31 ± 1.46 b | 7.54 ± 1.62 c | 8.40 ± 1.51 d | |||
| Glycine (3.57 ppm) | H | D W | 1.19 ± 0.17 a,A 1.14 ± 0.17 a,A | 1.38 ± 0.22 ab,A 1.38 ± 0.21 ab,A | 1.69 ± 0.34 bc,A 1.62 ± 0.25 bc,B | 1.90 ± 0.31 cd,A 1.83 ± 0.28 cd,A | 2.05 ± 0.31 d,A 2.05 ± 0.29 d,A | 0.83 0.83 |
| C | D W | 1.11 ± 0.17 a,A 1.20 ± 0.20 a,B | 1.35 ± 0.26 ab,A 1.32 ± 0.25 a,A | 1.45 ± 0.23 bc,A 1.50 ± 0.21 ab,A | 1.78 ± 0.27 cd,A 1.75 ± 0.20 bc,A | 1.99 ± 0.20 d,A 1.91 ± 0.21 c,A | 0.80 0.80 | |
| All | 1.16 ± 0.18 a | 1.36 ± 0.23 b | 1.57 ± 0.28 c | 1.82 ± 0.27 d | 2.00 ± 0.26 e | |||
| Isoleucine (1.02–1.03 ppm) | H | D W | 0.20 ± 0.06 a,A 0.20 ± 0.05 a,A | 0.34 ± 0.10 ab,A 0.40 ± 0.12 ab,B | 0.55 ± 0.18 bc,A 0.59 ± 0.19 bc,A | 0.76 ± 0.22 cd,A 0.84 ± 0.20 cd,A | 1.02 ± 0.24 d,A 1.12 ± 0.23 d,A | 0.91 0.92 |
| C | D W | 0.20 ± 0.04 a,A 0.23 ± 0.04 a,B | 0.33 ± 0.07 ab,A 0.36 ± 0.09 ab,B | 0.54 ± 0.14 bc,A 0.60 ± 0.15 bc,B | 0.80 ± 0.23 cd,A 0.90 ± 0.25 cd,B | 1.06 ± 0.29 d,A 1.17 ± 0.28 d,B | 0.90 0.92 | |
| All | 0.21 ± 0.05 a | 0.36 ± 0.10 b | 0.57 ± 0.17 c | 0.83 ± 0.23 d | 1.09 ± 0.26 e | |||
| Leucine (0.97–0.98 ppm) | H | D W | 0.33 ± 0.11 a,A 0.31 ± 0.10 a,A | 0.58 ± 0.20 ab,A 0.70 ± 0.25 ab,B | 0.96 ± 0.33 bc,A 1.01 ± 0.33 bc,A | 1.32 ± 0.37 cd,A 1.42 ± 0.33 cd,A | 1.74 ± 0.38 d,A 1.86 ± 0.34 d,A | 0.91 0.92 |
| C | DW | 0.31 ± 0.06 a,A 0.36 ± 0.06 a,B | 0.55 ± 0.14 ab,A 0.60 ± 0.15 ab,B | 0.93 ± 0.27 bc,A 1.02 ± 0.28 bc,B | 1.36 ± 0.40 cd,A 1.50 ± 0.42 cd,B | 1.78 ± 0.47 d,A 1.95 ± 0.48 d,A | 0.90 0.92 | |
| All | 0.32 ± 0.09 a | 0.61 ± 0.20 b | 0.98 ± 0.30 c | 1.40 ± 0.38 d | 1.83 ± 0.42 e | |||
| Methionine (2.64–2.65 ppm) | H | D W | 1.12 ± 0.33 a,A 1.10 ± 0.28 a,A | 1.27 ± 0.28 ab,A 1.37 ± 0.30 ab,B | 1.53 ± 0.27 bc,A 1.54 ± 0.27 bc,A | 1.74 ± 0.32 cd,A 1.76 ± 0.26 cd,A | 1.93 ± 0.32 d,A 1.94 ± 0.28 d,A | 0.74 0.78 |
| C | D W | 1.20 ± 0.13 a,A 1.22 ± 0.14 a,A | 1.39 ± 0.16 ab,A 1.38 ± 0.16 ab,A | 1.59 ± 0.19 bc,A 1.64 ± 0.20 bc,B | 1.88 ± 0.23 cd,A 1.91 ± 0.26 cd,A | 2.12 ± 0.33 d,A 2.11 ± 0.26 d,A | 0.84 0.86 | |
| All | 1.16 ± 0.25 a | 1.35 ± 0.24 b | 1.57 ± 0.24 c | 1.82 ± 0.28 d | 2.02 ± 0.31 e | |||
| Phenylalanine (7.45–7.47 ppm) | H | D W | 0.53 ± 0.04 a,A 0.52 ± 0.04 a,A | 0.64 ± 0.06 ab,A 0.70 ± 0.09 ab,B | 0.82 ± 0.12 bc,A 0.85 ± 0.13 bc,A | 1.00 ± 0.14 cd,A 1.06 ± 0.14 cd,A | 1.19 ± 0.16 d,A 1.28 ± 0.15 d,B | 0.94 0.94 |
| C | D W | 0.51 ± 0.05 a,A 0.55 ± 0.04 a,B | 0.64 ± 0.07 ab,A 0.68 ± 0.05 ab,A | 0.83 ± 0.14 bc,A 0.88 ± 0.11 bc,A | 1.04 ± 0.19 cd,A 1.14 ± 0.19 cd,B | 1.25 ± 0.21 d,A 1.34 ± 0.22 d,B | 0.91 0.94 | |
| All | 0.53 ± 0.04 a | 0.66 ± 0.07 b | 0.84 ± 0.13 c | 1.06 ± 0.17 d | 1.26 ± 0.19 e | |||
| Tryptophan (7.72–7.75 ppm) | H | D W | 0.13 ± 0.01 a,A 0.13 ± 0.01 a,A | 0.15 ± 0.02 ab,A 0.16 ± 0.02 ab,B | 0.19 ± 0.03 bc,A 0.19 ± 0.03 bc,A | 0.22 ± 0.04 cd,A 0.23 ± 0.03 cd,A | 0.25 ± 0.04 d,A 0.27 ± 0.04 d,B | 0.88 0.90 |
| C | D W | 0.13 ± 0.01 a,A 0.13 ± 0.01 a,A | 0.15 ± 0.02 ab,A 0.16 ± 0.02 ab,A | 0.19 ± 0.03 bc,A 0.20 ± 0.03 bc,B | 0.22 ± 0.04 cd,A 0.24 ± 0.04 cd,B | 0.26 ± 0.04 d,A 0.28 ± 0.04 d,B | 0.89 0.91 | |
| All | 0.13 ± 0.01 a | 0.15 ± 0.02 b | 0.19 ± 0.03 c | 0.23 ± 0.04 d | 0.27 ± 0.04 e | |||
| Tyrosine (7.17–7.22 ppm) | H | D W | 0.29 ± 0.06 a,A 0.29 ± 0.06 a,A | 0.43 ± 0.10 ab,A 0.51 ± 0.13 ab,B | 0.64 ± 0.16 bc,A 0.68 ± 0.18 bc,A | 0.82 ± 0.19 cd,A 0.91 ± 0.18 cd,B | 1.04 ± 0.19 d,A 1.14 ± 0.19 d,B | 0.91 0.92 |
| C | D W | 0.30 ± 0.05 a,A 0.33 ± 0.05 a,B | 0.43 ± 0.08 ab,A 0.48 ± 0.08 ab,B | 0.65 ± 0.16 bc,A 0.71 ± 0.16 bc,B | 0.86 ± 0.22 cd,A 0.96 ± 0.23 cd,B | 1.08 ± 0.25 d,A 1.19 ± 0.24 d,B | 0.89 0.92 | |
| All | 0.30 ± 0.06 a | 0.46 ± 0.11 b | 0.67 ± 0.16 c | 0.89 ± 0.21 d | 1.11 ± 0.22 e | |||
| Valine (1.03–1.06 ppm) | H | D W | 0.35 ± 0.08 a,A 0.33 ± 0.08 a,A | 0.55 ± 0.15 ab,A 0.65 ± 0.18 ab,B | 0.89 ± 0.28 bc,A 0.96 ± 0.30 bc,A | 1.24 ± 0.34 cd,A 1.41 ± 0.32 cd,B | 1.67 ± 0.38 d,A 1.89 ± 0.37 d,B | 0.92 0.93 |
| C | D W | 0.36 ± 0.06 a,A 0.39 ± 0.06 a,B | 0.56 ± 0.12 ab,A 0.61 ± 0.14 ab,A | 0.89 ± 0.22 bc,A 1.00 ± 0.24 bc,B | 1.30 ± 0.36 cd,A 1.50 ± 0.39 cd,B | 1.76 ± 0.45 d,A 1.95 ± 0.45 d,B | 0.91 0.93 | |
| All | 0.36 ± 0.08 a | 0.59 ± 0.15 b | 0.93 ± 0.27 c | 1.36 ± 0.36 d | 1.82 ± 0.42 e | |||
| Nucleotides | ||||||||
| Hypoxanthine (8.22 ppm) | H | D W | 1.21 ± 0.32 a,A 1.32 ± 0.33 a,A | 1.96 ± 0.36 ab,A 2.21 ± 0.50 ab,B | 2.72 ± 0.36 bc,A 2.98 ± 0.43 bc,B | 3.41 ± 0.50 cd,A 3.70 ± 0.44 cd,B | 4.15 ± 0.61 d,A 4.48 ± 0.70 d,B | 0.93 0.94 |
| C | D W | 1.12 ± 0.26 a,A 1.16 ± 0.23 a,A | 1.52 ± 0.20 ab,A 1.81 ± 0.20 ab,B | 2.08 ± 0.27 bc,A 2.43 ± 0.29 bc,B | 2.59 ± 0.21 cd,A 3.07 ± 0.45 cd,B | 3.36 ± 0.44 d,A 3.81 ± 0.77 d,B | 0.94 0.96 | |
| All | 1.20 ± 0.30 a | 1.89 ± 0.42 b | 2.57 ± 0.48 c | 3.22 ± 0.59 d | 3.97 ± 0.76 e | |||
| IMP (6.13–6.16 ppm) | H | D W | 3.63 ± 0.29 a,A 3.45 ± 0.25 a,B | 2.88 ± 0.34 ab,A 2.77 ± 0.34 ab,A | 2.16 ± 0.35 bc,A 2.03 ± 0.41 bc,A | 1.67 ± 0.34 cd,A 1.38 ± 0.32 cd,B | 1.17 ± 0.29 d,A 0.86 ± 0.21 d,B | −0.93 −0.95 |
| C | D W | 3.37 ± 0.35 a,A 3.32 ± 0.32 a,A | 2.92 ± 0.16 ab,A 2.68 ± 0.21 ab,B | 2.43 ± 0.14 bc,A 2.23 ± 0.15 bc,B | 2.07 ± 0.20 cd,A 1.79 ± 0.24 cd,B | 1.65 ± 0.27 d,A 1.39 ± 0.24 d,B | −0.93 −0.94 | |
| All | 3.43 ± 0.33 a | 2.81 ± 0.29 b | 2.20 ± 0.32 c | 1.71 ± 0.38 d | 1.25 ± 0.38 e | |||
| Inosine (6.08–6.12 ppm) | H | D W | 0.85 ± 0.10 a,A 0.86 ± 0.10 a,A | 1.26 ± 0.17 ab,A 1.19 ± 0.08 a,A | 1.52 ± 0.11 bc,A 1.44 ± 0.08 b,B | 1.69 ± 0.15 c,A 1.48 ± 0.16 b,B | 1.66 ± 0.14 c,A 1.49 ± 0.08 b,B | 0.88 0.87 |
| C | D W | 0.79 ± 0.11 a,A 0.83 ± 0.13 a,A | 1.14 ± 0.19 ab,A 1.03 ± 0.16 ab,B | 1.31 ± 0.19 bc,A 1.17 ± 0.21 bc,B | 1.48 ± 0.24 c,A 1.21 ± 0.25 c,B | 1.44 ± 0.28 c,A 1.24 ± 0.25 c,B | 0.80 0.68 | |
| All | 0.84 ± 0.11 a | 1.16 ± 0.17 b | 1.37 ± 0.20 c | 1.47 ± 0.26 c | 1.46 ± 0.25 c | |||
| Acids | ||||||||
| Acetic acid (1.92–1.93 ppm) | H | D W | 0.39 ± 0.10 a,A 0.40 ± 0.09 a,A | 0.49 ± 0.12 ab,A 0.49 ± 0.11 ab,A | 0.61 ± 0.14 bc,A 0.58 ± 0.13 bc,A | 0.74 ± 0.19 cd,A 0.70 ± 0.12 cd,A | 0.88 ± 0.17 d,A 0.83 ± 0.12 d,A | 0.83 0.85 |
| C | D W | 0.47 ± 0.11 a,A 0.49 ± 0.06 a,A | 0.59 ± 0.08 ab,A 0.59 ± 0.06 ab,A | 0.66 ± 0.09 bc,A 0.68 ± 0.10 bc,A | 0.77 ± 0.08 cd,A 0.80 ± 0.12 cd,A | 0.91 ± 0.15 d,A 1.03 ± 0.22 d,A | 0.86 0.87 | |
| All | 0.43 ± 0.10 a | 0.54 ± 0.11 b | 0.63 ± 0.12 c | 0.75 ± 0.14 d | 0.91 ± 0.18 e | |||
| Fumaric Acid (6.52–6.53 ppm) | H | D W | 0.19 ± 0.09 a,A 0.18 ± 0.10 a,A | 0.24 ± 0.08 a,A 0.26 ± 0.12 ab,A | 0.36 ± 0.15 b,A 0.30 ± 0.10 bc,A | 0.42 ± 0.20 b,A 0.34 ± 0.09 bc,A | 0.45 ± 0.12 b,A 0.44 ± 0.15 c,A | 0.68 0.64 |
| C | D W | 0.22 ± 0.08 a,A 0.26 ± 0.06 a,A | 0.29 ± 0.11 ab,A 0.29 ± 0.09 ab,A | 0.35 ± 0.08 b,A 0.39 ± 0.09 c,A | 0.31 ± 0.09 ab,A 0.34 ± 0.10 abc,A | 0.42 ± 0.15 b,A 0.37 ± 0.12 bc,A | 0.50 0.51 | |
| All | 0.21 ± 0.09 a | 0.27 ± 0.10 b | 0.35 ± 0.11 c | 0.35 ± 0.14 c | 0.42 ± 0.14 d | |||
| Lactic Acid (1.30–1.37 ppm) | H | D W | 70.22 ± 5.02 a,A 66.64 ± 8.95 a,A | 72.84 ± 5.94 ab,A 70.39 ± 5.28 a,A | 76.92 ± 8.19 abc,A 69.15 ± 5.04 a,B | 81.32 ± 8.77 bc,A 70.39 ± 4.57 a,B | 83.27 ± 9.49 c,A 70.71 ± 4.40 a,B | 0.62 0.08 |
| C | D W | 70.02 ± 4.00 a,A 70.89 ± 4.12 a,A | 75.51 ± 4.60 ab,A 72.29 ± 3.91 ab,A | 78.07 ± 4.64 bc,A 74.85 ± 2.11 bc,A | 87.89 ± 8.84 c,A 76.01 ± 4.07 c,B | 90.45 ± 11.62 c,A 77.37 ± 3.16 c,B | 0.79 0.66 | |
| All | 69.37 ± 6.15 a | 72.68 ± 5.30 b | 74.63 ± 6.50 bc | 78.70 ± 9.43 cd | 80.22 ± 10.71 d | |||
| Succinic Acid (2.41–2.42 ppm) | H | D W | 0.37 ± 0.12 a,A 0.36 ± 0.11 a,A | 0.62 ± 0.36 ab,A 0.51 ± 0.21 ab,A | 0.68 ± 0.29 bc,A 0.70 ± 0.37 bc,A | 0.86 ± 0.34 c,A 1.00 ± 0.52 c,A | 0.97 ± 0.60 c,A 0.86 ± 0.31 c,A | 0.69 0.71 |
| C | D W | 0.59 ± 0.19 a,A 0.82 ± 0.21 a,A | 0.96 ± 0.22 b,A 1.02 ± 0.20 ab,A | 0.98 ± 0.23 b,A 1.00 ± 0.17 ab,A | 1.14 ± 0.28 b,A 1.11 ± 0.24 b,A | 1.05 ± 0.31 b,A 1.13 ± 0.25 b,B | 0.49 0.57 | |
| All | 0.52 ± 0.25 a | 0.76 ± 0.34 b | 0.83 ± 0.31 bc | 1.02 ± 0.38 d | 1.00 ± 0.40 cd | |||
| Sugars | ||||||||
| α Glucose (5.25–5.27 ppm) | H | D W | 2.12 ± 0.39 a,A 1.88 ± 0.53 a,A | 2.67 ± 0.49 ab,A 2.90 ± 0.56 ab,A | 3.89 ± 0.69 bc,A 3.90 ± 0.59 bc,A | 4.48 ± 0.60 c,A 4.61 ± 0.85 c,A | 4.70 ± 0.73 c,A 4.71 ± 0.70 c,A | 0.88 0.88 |
| C | D W | 2.10 ± 0.50 a,A 2.19 ± 0.37 a,A | 2.90 ± 0.45 ab,A 2.87 ± 0.38 ab,A | 3.59 ± 0.84 bc,A 3.69 ± 0.46 bc,A | 4.53 ± 0.70 cd,A 4.42 ± 0.73 c,A | 5.07 ± 0.88 d,A 4.58 ± 0.77 c,A | 0.90 0.89 | |
| All | 2.07 ± 0.46 a | 2.83 ± 0.48 b | 3.77 ± 0.66 c | 4.51 ± 0.72 d | 4.76 ± 0.78 d | |||
| β Glucose (4.64–4.67 ppm) | H | D W | 3.61 ± 0.79 a,A 3.47 ± 1.68 a,A | 4.43 ± 0.96 a,A 4.78 ± 1.11 ab,A | 6.00 ± 1.01 b,A 6.04 ± 0.95 bc,A | 6.63 ± 0.88 b,A 6.69 ± 1.16 c,A | 6.49 ± 0.96 b,A 6.51 ± 1.02 c,A | 0.80 0.76 |
| C | D W | 3.45 ± 1.09 a,A 3.71 ± 0.78 a,A | 4.59 ± 0.97 ab,A 4.53 ± 0.77 ab,A | 5.68 ± 2.33 bc,A 5.50 ± 0.92 bc,A | 6.66 ± 1.23 cd,A 6.38 ± 1.24 c,A | 6.92 ± 1.26 d,A 6.32 ± 1.18 c,A | 0.80 0.78 | |
| All | 3.56 ± 1.15 a | 4.58 ± 0.96 b | 5.82 ± 1.41 c | 6.59 ± 1.12 d | 6.55 ± 1.11 d | |||
| Glucose-6-phosphate (5.22–5.24 ppm) | H | D W | 1.53 ± 0.44 a,A 1.35 ± 0.58 a,A | 1.75 ± 0.47 ab,A 1.87 ± 0.45 abc,A | 2.19 ± 0.40 c,A 2.16 ± 0.39 b,A | 2.12 ± 0.41 bc,A 2.07 ± 0.43 bc,A | 1.81 ± 0.35 ab,A 1.75 ± 0.31 ac,A | 0.40 0.34 |
| C | D W | 1.47 ± 0.54 a,A 1.61 ± 0.39 a,A | 1.75 ± 0.53 ab,A 1.70 ± 0.35 a,A | 1.93 ± 0.65 ab,A 1.89 ± 0.42 a,A | 2.08 ± 0.46 b,A 1.97 ± 0.50 a,A | 1.94 ± 0.40 b,A 1.76 ± 0.41 a,A | 0.35 0.23 | |
| All | 1.49 ± 0.50 a | 1.77 ± 0.46 b | 2.05 ± 0.48 c | 2.06 ± 0.45 c | 1.81 ± 0.37 d | |||
| Further compounds | ||||||||
| Anserine (7.08–7.10 ppm) | H | D W | 1.61 ± 0.66 a,A 1.72 ± 0.70 a,A | 1.96 ± 0.68 ab,A 2.03 ± 0.76 ab,A | 2.40 ± 0.91 bc,A 2.39 ± 0.89 bc,A | 2.65 ± 0.95 c,A 2.63 ± 1.00 c,A | 2.89 ± 1.10 c,A 2.59 ± 0.99 c,A | 0.51 0.45 |
| C | D W | 2.12 ± 0.46 a,A 2.16 ± 0.38 a,A | 2.24 ± 0.51 ab,A 2.18 ± 0.47 a,A | 2.58 ± 0.46 abc,A 2.56 ± 0.44 ab,A | 3.03 ± 0.53 c,A 2.57 ± 0.51 ab,B | 2.76 ± 0.88 bc,A 2.93 ± 0.48 b,A | 0.44 0.51 | |
| All | 1.89 ± 0.62 a | 2.09 ± 0.63 a | 2.48 ± 0.72 b | 2.71 ± 0.81 b | 2.79 ± 0.91 b | |||
| Betaine (3.27–3.28 ppm) | H | D W | 4.50 ± 0.81 a,A 4.33 ± 0.87 a,A | 5.38 ± 0.97 ab,A 5.60 ± 1.16 ab,A | 6.64 ± 0.86 bc,A 6.54 ± 0.91 bc,A | 7.37 ± 1.30 c,A 7.23 ± 1.02 cd,A | 7.95 ± 1.37 c,A 7.66 ± 0.81 d,A | 0.82 0.81 |
| C | D W | 4.47 ± 0.66 a,A 4.56 ± 0.67 a,A | 5.16 ± 0.79 ab,A 5.19 ± 0.64 ab,A | 6.06 ± 0.94 bc,A 6.11 ± 0.80 bc,A | 7.12 ± 1.07 cd,A 6.87 ± 0.98 cd,B | 8.12 ± 1.29 d,A 7.56 ± 1.08 d,A | 0.83 0.84 | |
| All | 4.46 ± 0.76 a | 5.34 ± 0.93 b | 6.35 ± 0.91 c | 7.16 ± 1.11 d | 7.82 ± 1.16 e | |||
| Carnitine (3.22–3.23 ppm) | H | D W | 4.57 ± 0.55 ab,A 4.45 ± 0.71 a,A | 4.19 ± 0.42 a,A 4.20 ± 0.42 ab,A | 4.31 ± 0.33 ab,A 4.10 ± 0.37 b,A | 4.66 ± 0.61 ab,A 4.23 ± 0.35 ab,B | 4.65 ± 0.47 b,A 4.11 ± 0.42 b,B | 0.09 −0.31 |
| C | D W | 2.19 ± 0.27 a,A 2.21 ± 0.23 a,A | 2.29 ± 0.31 a,A 2.20 ± 0.20 a,A | 2.29 ± 0.23 a,A 2.26 ± 0.20 a,A | 2.61 ± 0.36 b,A 2.37 ± 0.24 ab,B | 2.65 ± 0.44 b,A 2.41 ± 0.23 b,A | 0.45 0.31 | |
| All | 3.43 ± 1.26 a | 3.28 ± 1.04 a | 3.30 ± 1.01 a | 3.53 ± 1.08 a | 3.52 ± 1.03 a | |||
| Carnosine (7.10–7.14 ppm) | H | D W | 16.12 ± 5.86 a,A 15.92 ± 5.76 a,A | 16.77 ± 5.79 a,A 17.56 ± 5.93 b,B | 18.02 ± 6.30 ab,A 17.90 ± 6.16 b,A | 18.93 ± 6.53 b,A 18.24 ± 6.26 b,A | 18.76 ± 6.24 b,A 18.10 ± 6.01 b,A | 0.37 0.30 |
| C | D W | 18.56 ± 2.46 a,A 18.70 ± 2.74 a,A | 19.53 ± 2.52 ab,A 19.18 ± 2.03 a,A | 20.35 ± 1.96 abc,A 20.35 ± 1.95 ab,A | 22.39 ± 1.68 c,A 20.87 ± 2.06 bc,B | 21.57 ± 3.60 bc,A 22.03 ± 1.85 c,A | 0.46 0.47 | |
| All | 17.24 ± 4.73 a | 18.19 ± 4.64 a | 19.08 ± 4.84 b | 20.01 ± 5.06 b | 20.00 ± 5.12 b | |||
| Creatine (3.92–3.94 ppm) | H | D W | 32.40 ± 2.45 a,A 30.92 ± 1.09 a,B | 32.64 ± 3.36 a,A 32.31 ± 2.51 a,A | 34.23 ± 4.35 ab,A 32.59 ± 2.72 a,B | 35.52 ± 3.07 b,A 32.54 ± 2.20 a,B | 34.66 ± 2.88 ab,A 31.76 ± 2.40 a,B | 0.48 0.05 |
| C | D W | 30.21 ± 1.61 a,A 30.18 ± 1.73 a,A | 31.52 ± 1.65 a,A 30.37 ± 2.10 a,B | 31.84 ± 1.84 ab,A 31.40 ± 1.38 ab,A | 34.74 ± 3.01 b,A 31.88 ± 1.68 b,B | 34.71 ± 3.01 b,A 31.32 ± 1.49 ab,B | 0.64 0.31 | |
| All | 30.97 ± 1.99 a | 31.76 ± 2.64 ab | 32.58 ± 3.04 bc | 33.69 ± 2.95 d | 33.12 ± 2.95 cd | |||
| Creatinine (4.06–4.07 ppm) | H | D W | 1.23 ± 0.31 a,A 1.16 ± 0.29 a,A | 1.69 ± 0.42 ab,A 1.91 ± 0.50 b,A | 2.07 ± 0.42 bc,A 2.06 ± 0.40 b,A | 2.16 ± 0.39 bc,A 2.05 ± 0.30 b,A | 2.41 ± 0.40 c,A 2.20 ± 0.22 b,A | 0.79 0.74 |
| C | D W | 1.39 ± 0.44 a,A 1.40 ± 0.35 a,A | 1.67 ± 0.40 ab,A 1.78 ± 0.47 ab,A | 2.12 ± 0.44 bc,A 2.05 ± 0.27 b,A | 2.17 ± 0.29 c,A 2.09 ± 0.24 bc,A | 2.44 ± 0.39 c,A 2.38 ± 0.28 c,A | 0.73 0.72 | |
| All | 1.29 ± 0.36 a | 1.76 ± 0.45 b | 2.08 ± 0.39 c | 2.12 ± 0.31 c | 2.35 ± 0.34 d | |||
| Niacinamide (8.71–8.73 ppm) | H | D W | 0.42 ± 0.03 a,A 0.40 ± 0.04 a,A | 0.43 ± 0.04 a,A 0.48 ± 0.03 ab,B | 0.50 ± 0.05 b,A 0.50 ± 0.03 bc,A | 0.51 ± 0.06 b,A 0.53 ± 0.03 c,A | 0.54 ± 0.06 b,A 0.53 ± 0.03 c,A | 0.74 0.86 |
| C | D W | 0.36 ± 0.04 a,A 0.37 ± 0.03 a,A | 0.41 ± 0.05 ab,A 0.41 ± 0.04 ab,A | 0.47 ± 0.05 bc,A 0.50 ± 0.03 bc,B | 0.52 ± 0.07 c,A 0.54 ± 0.04 c,A | 0.55 ± 0.08 c,A 0.54 ± 0.04 c,A | 0.81 0.87 | |
| All | 0.39 ± 0.04 a | 0.43 ± 0.05 b | 0.49 ± 0.04 c | 0.53 ± 0.05 d | 0.54 ± 0.05 d | |||
| O-Acetyl-L-carnitine (5.57–5.65 ppm) | H | D W | 1.50 ± 0.36 a,A 1.62 ± 0.50 a,A | 1.95 ± 0.25 b,A 1.79 ± 0.26 ab,B | 2.04 ± 0.22 b,A 1.91 ± 0.19 b,A | 2.04 ± 0.28 b,A 1.84 ± 0.20 ab,B | 1.97 ± 0.28 ab,A 1.81 ± 0.24 ab,B | 0.49 0.34 |
| C | D W | 0.93 ± 0.16 a,A 0.98 ± 0.22 a,A | 0.98 ± 0.17 a,A 0.97 ± 0.19 a,A | 0.99 ± 0.28 a,A 0.94 ± 0.15 ab,A | 0.91 ± 0.13 a,A 0.86 ± 0.13 bc,A | 0.89 ± 0.14 a,A 0.82 ± 0.12 c,B | −0.15 −0.31 | |
| All | 1.28 ± 0.46 a | 1.45 ± 0.50 ab | 1.50 ± 0.55 b | 1.45 ± 0.57 ab | 1.41 ± 0.56 ab | |||
References
- Liu, J.; Ellies-Oury, M.-P.; Stoyanchev, T.; Hocquette, J.-F. Consumer Perception of Beef Quality and How to Control, Improve and Predict It? Focus on Eating Quality. Foods 2022, 11, 1732. [Google Scholar] [CrossRef] [PubMed]
- Xu, L.; Liu, S.; Cheng, Y.; Qian, H. The effect of aging on beef taste, aroma and texture, and the role of microorganisms: A review. Crit. Rev. Food Sci. Nutr. 2021, 1–12. [Google Scholar] [CrossRef] [PubMed]
- Bischof, G.; Witte, F.; Terjung, N.; Heinz, V.; Juadjur, A.; Gibis, M. Metabolic, proteomic and microbial changes postmortem and during beef aging. Crit. Rev. Food Sci. Nutr. 2022, 1–34. [Google Scholar] [CrossRef] [PubMed]
- Kodani, Y.; Miyakawa, T.; Komatsu, T.; Tanokura, M. NMR-based metabolomics for simultaneously evaluating multiple determinants of primary beef quality in Japanese Black cattle. Sci. Rep. 2017, 7, 1297. [Google Scholar] [CrossRef] [PubMed]
- Kim, Y.H.B.; Kemp, R.; Samuelsson, L.M. Effects of dry-aging on meat quality attributes and metabolite profiles of beef loins. Meat Sci. 2016, 111, 168–176. [Google Scholar] [CrossRef] [PubMed]
- Bischof, G.; Witte, F.; Terjung, N.; Januschewski, E.; Heinz, V.; Juadjur, A.; Gibis, M. Effect of sampling position in fresh, dry-aged and wet-aged beef from M. Longissimus dorsi of Simmental cattle analyzed by 1H NMR spectroscopy. Food Res. Int. 2022, 156, 111334. [Google Scholar] [CrossRef] [PubMed]
- Muroya, S.; Nomura, R.; Nagai, H.; Ojima, K.; Matsukawa, K. Metabolomic profiling of postmortem aged muscle in Japanese Brown beef cattle revealed an interbreed difference from Japanese Black beef. Anim. Biosci. 2022; Epub ahead of print. [Google Scholar] [CrossRef] [PubMed]
- Cho, S.; Seol, K.; Kang, S.; Kim, Y.; Seo, H.; Lee, W.; Kim, J.; Van Ba, H. Comparison of Tastes-Related Components and Eating Quality between Hanwoo Steer and Cow Longissimus thoracis Muscles. Food Sci. Anim. Resour. 2020, 40, 908. [Google Scholar] [CrossRef] [PubMed]
- EU. EU Regulation No. 1308/2013 Appendix IV 2013. Available online: https://eur-lex.europa.eu/legal-content/EN/TXT/?uri=celex%3A32013R1308#d2125e32-825-1 (accessed on 20 December 2022).
- Pabst, W. Die Aufzucht. In Tierproduktion; Weiß, J.W., Pabst, W., Strack, K.E., Granz, S., Eds.; Parey Verlag: Stuttgart, Germany, 2005. [Google Scholar]
- Pabst, W. Rindfleischproduktion. In Tierproduktion; Weiß, J.W., Pabst, W., Strack, K.E., Granz, S., Eds.; Parey Verlag: Stuttgart, Germany, 2005. [Google Scholar]
- Watanabe, A.; Ueda, Y.; Higuchi, M. Effects of slaughter age on the levels of free amino acids and dipeptides in fattening cattle. Anim. Sci. J. 2004, 75, 361–367. [Google Scholar] [CrossRef]
- Cho, S.; Kang, S.; Kang, G.; Seong, P.; Park, K.; Chang, S.; Lee, S.; Cho, Y.; Park, B. Physicochemical meat quality, fatty acid and free amino acid composition of strip loin, chuck tender, and eye of round produced by different age groups of Hanwoo cow. Food Sci. Anim. Resour. 2013, 33, 708–714. [Google Scholar] [CrossRef] [Green Version]
- Bischof, G.; Witte, F.; Terjung, N.; Januschewski, E.; Heinz, V.; Juadjur, A.; Gibis, M. Analysis of aging type-and aging time-related changes in the polar fraction of metabolome of beef by 1H NMR spectroscopy. Food Chem. 2020, 342, 128353. [Google Scholar] [CrossRef] [PubMed]
- Castejón, D.; García-Segura, J.M.; Escudero, R.; Herrera, A.; Cambero, M.I. Metabolomics of meat exudate: Its potential to evaluate beef meat conservation and aging. Anal. Chim. Acta 2015, 901, 1–11. [Google Scholar] [CrossRef] [PubMed]
- Muroya, S.; Oe, M.; Ojima, K.; Watanabe, A. Metabolomic approach to key metabolites characterizing postmortem aged loin muscle of Japanese Black (Wagyu) cattle. Asian-Australas. J. Anim. Sci. 2019, 32, 1172. [Google Scholar] [CrossRef] [PubMed]
- Cônsolo, N.R.; Silva, J.; Buarque, V.L.; Samuelsson, L.M.; Miller, P.; Maclean, P.H.; Moraes, T.B.; Barbosa, L.C.G.S.; Higuera-Padilla, A.; Colnago, L.A.; et al. Using TD-NMR relaxometry and 1D 1H NMR spectroscopy to evaluate aging of Nellore beef. Meat Sci. 2021, 181, 108606. [Google Scholar] [CrossRef] [PubMed]
- Oh, J.; Lee, H.J.; Kim, H.C.; Kim, H.J.; Yun, Y.G.; Kim, K.T.; Choi, Y.I.; Jo, C. The effects of dry or wet aging on the quality of the longissimus muscle from 4-year-old Hanwoo cows and 28-month-old Hanwoo steers. Anim. Prod. Sci. 2018, 58, 2344–2351. [Google Scholar] [CrossRef]
- Feidt, C.; Petit, A.; Bruas-Reignier, F.; Brun-Bellut, J. Release of free amino-acids during ageing in bovine meat. Meat Sci. 1996, 44, 19–25. [Google Scholar] [CrossRef] [PubMed]
- Dashdorj, D.; Amna, T.; Hwang, I. Influence of specific taste-active components on meat flavor as affected by intrinsic and extrinsic factors: An overview. Eur. Food Res. Technol. 2015, 241, 157–171. [Google Scholar] [CrossRef]
- Pabst, W. Die Milchproduktion. In Tierproduktion; Weiß, J.W., Pabst, W., Strack, K.E., Granz, S., Eds.; Parey Verlag: Stuttgart, Germany, 2005. [Google Scholar]
- Decker, C.; Krapf, R.; Kuballa, T.; Bunzel, M. Nontargeted Analysis of Lipid Extracts Using 1H NMR Spectroscopy Combined with Multivariate Statistical Analysis to Discriminate between the Animal Species of Raw and Processed Meat. J. Agric. Food Chem. 2022, 70, 7230–7239. [Google Scholar] [CrossRef] [PubMed]
- Kim, H.C.; Baek, K.H.; Ko, Y.-J.; Lee, H.J.; Yim, D.-G.; Jo, C. Characteristic metabolic changes of the crust from dry-aged beef using 2D NMR spectroscopy. Molecules 2020, 25, 3087. [Google Scholar] [CrossRef] [PubMed]
- Setyabrata, D.; Vierck, K.; Sheets, T.R.; Legako, J.F.; Cooper, B.R.; Johnson, T.A.; Kim, Y.H.B. Characterizing the flavor precursors and liberation mechanisms of various dry-aging methods in cull beef loins using metabolomics and microbiome approaches. Metabolites 2022, 12, 472. [Google Scholar] [CrossRef] [PubMed]


Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Bischof, G.; Januschewski, E.; Witte, F.; Terjung, N.; Heinz, V.; Juadjur, A.; Gibis, M. Variations in the Metabolome of Unaged and Aged Beef from Black-and-White Cows and Heifers by 1H NMR Spectroscopy. Foods 2023, 12, 785. https://doi.org/10.3390/foods12040785
Bischof G, Januschewski E, Witte F, Terjung N, Heinz V, Juadjur A, Gibis M. Variations in the Metabolome of Unaged and Aged Beef from Black-and-White Cows and Heifers by 1H NMR Spectroscopy. Foods. 2023; 12(4):785. https://doi.org/10.3390/foods12040785
Chicago/Turabian StyleBischof, Greta, Edwin Januschewski, Franziska Witte, Nino Terjung, Volker Heinz, Andreas Juadjur, and Monika Gibis. 2023. "Variations in the Metabolome of Unaged and Aged Beef from Black-and-White Cows and Heifers by 1H NMR Spectroscopy" Foods 12, no. 4: 785. https://doi.org/10.3390/foods12040785
APA StyleBischof, G., Januschewski, E., Witte, F., Terjung, N., Heinz, V., Juadjur, A., & Gibis, M. (2023). Variations in the Metabolome of Unaged and Aged Beef from Black-and-White Cows and Heifers by 1H NMR Spectroscopy. Foods, 12(4), 785. https://doi.org/10.3390/foods12040785








