Microbial Profiling of Biltong Processing Using Culture-Dependent and Culture-Independent Microbiome Analysis
Abstract
:1. Introduction
2. Materials and Methods
2.1. Beef Sources
2.2. Beef Preparation and Sampling
2.2.1. Preparation of Beef for Biltong Process and Microbiome Analyses
2.2.2. Biltong Beef Processing, Marination, and Drying
2.2.3. Environmental and Marinade Sample Preparation for Microbiome Analysis
2.3. Culture-Dependent Analysis
2.3.1. Bacterial Enumeration
2.3.2. Microbial Profiling (16S PCR, Sequencing, and Identification of Isolates)
2.3.3. Phylogenetic Relationship
2.4. Culture-Independent Analysis
2.4.1. DNA Extraction
2.4.2. 16S rRNA Sequencing
2.4.3. Bioinformatic Analysis
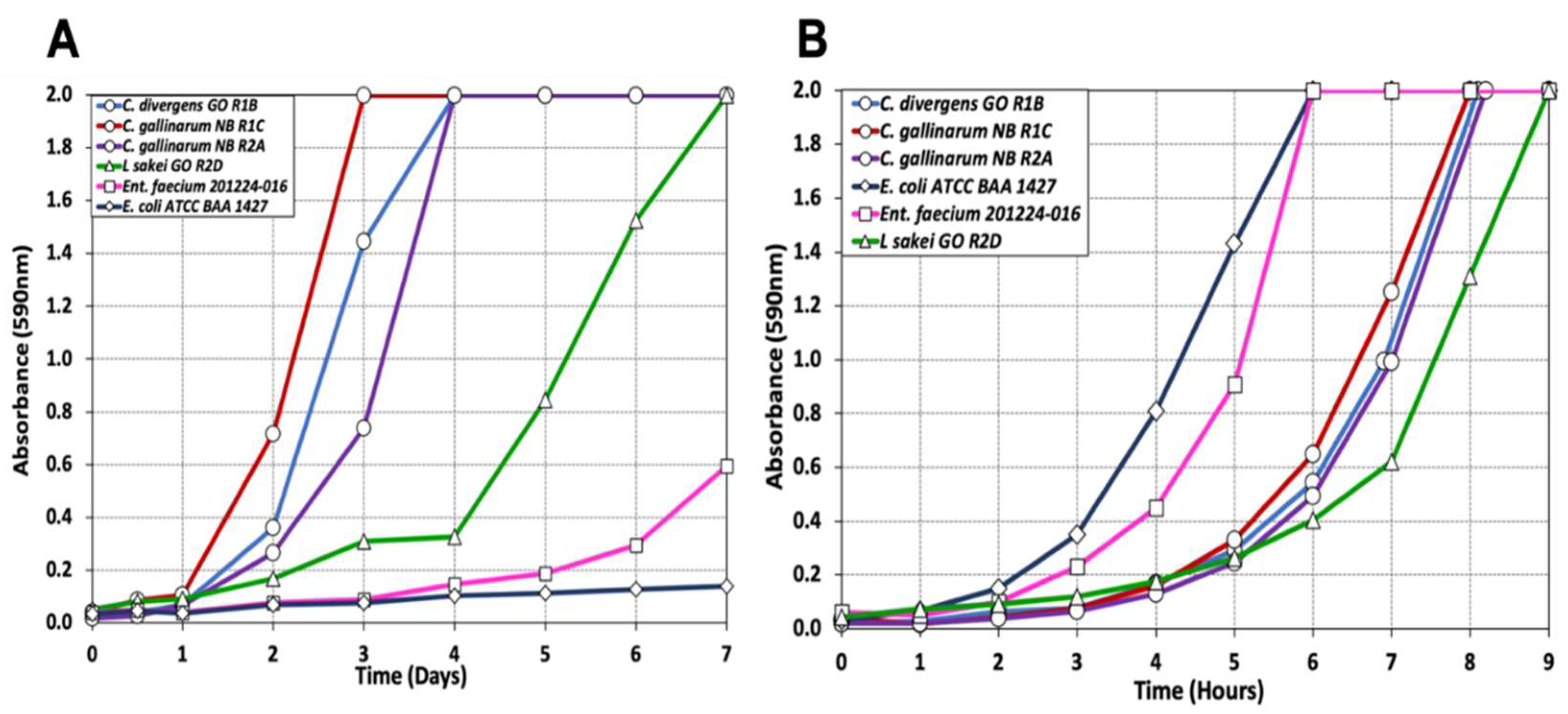
2.5. Growth Assay of Isolates Obtained from Biltong Process
3. Results and Discussion
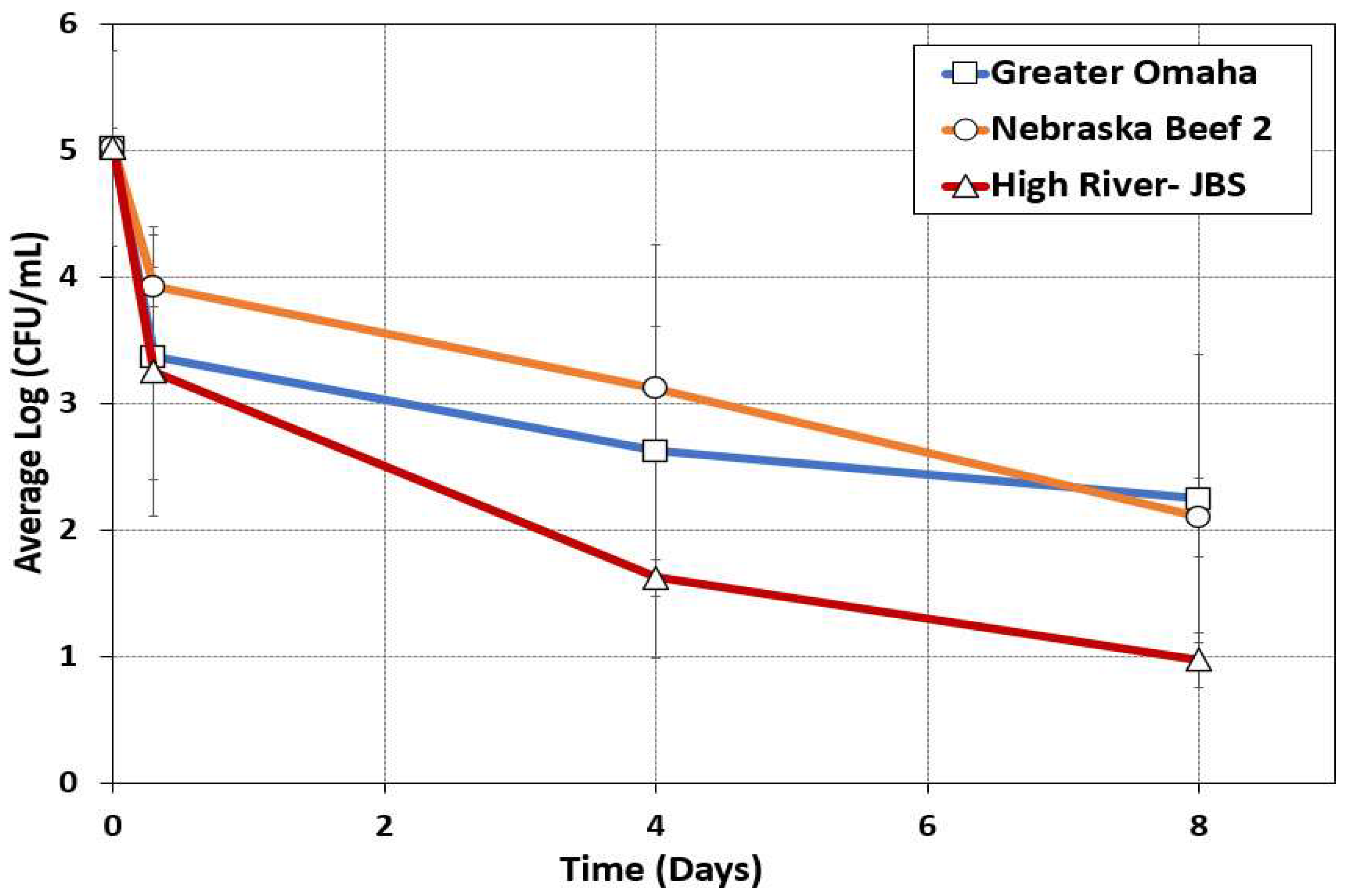
3.1. Culture-Dependent Microbiome Analysis
3.1.1. Bacteria Identified via Culture-Dependent Methodology
3.1.2. Impact of Processing on Culture-Dependent Microbiome
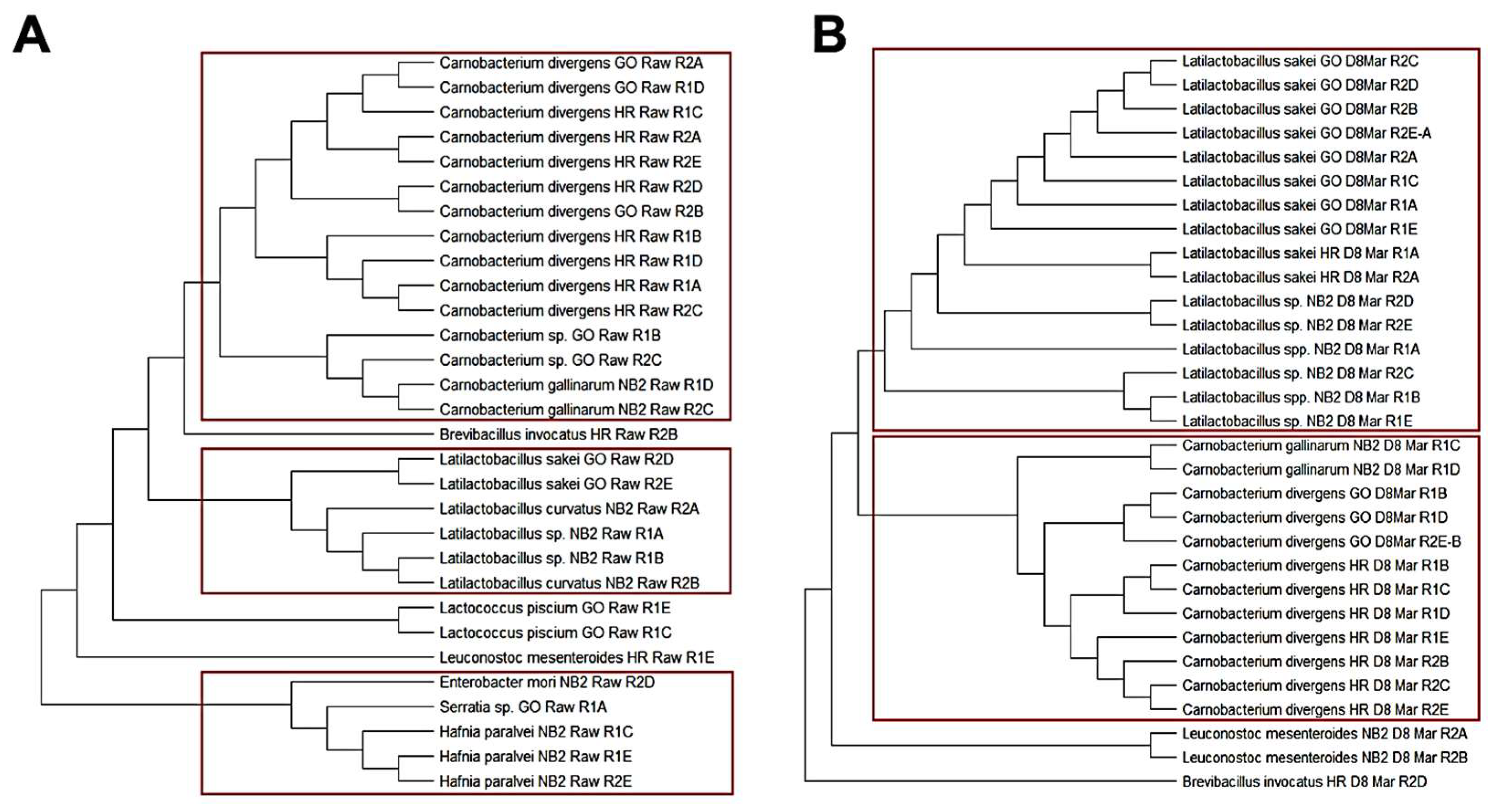
3.1.3. Phylogenetic Relationship among Isolates Obtained from Biltong Processing
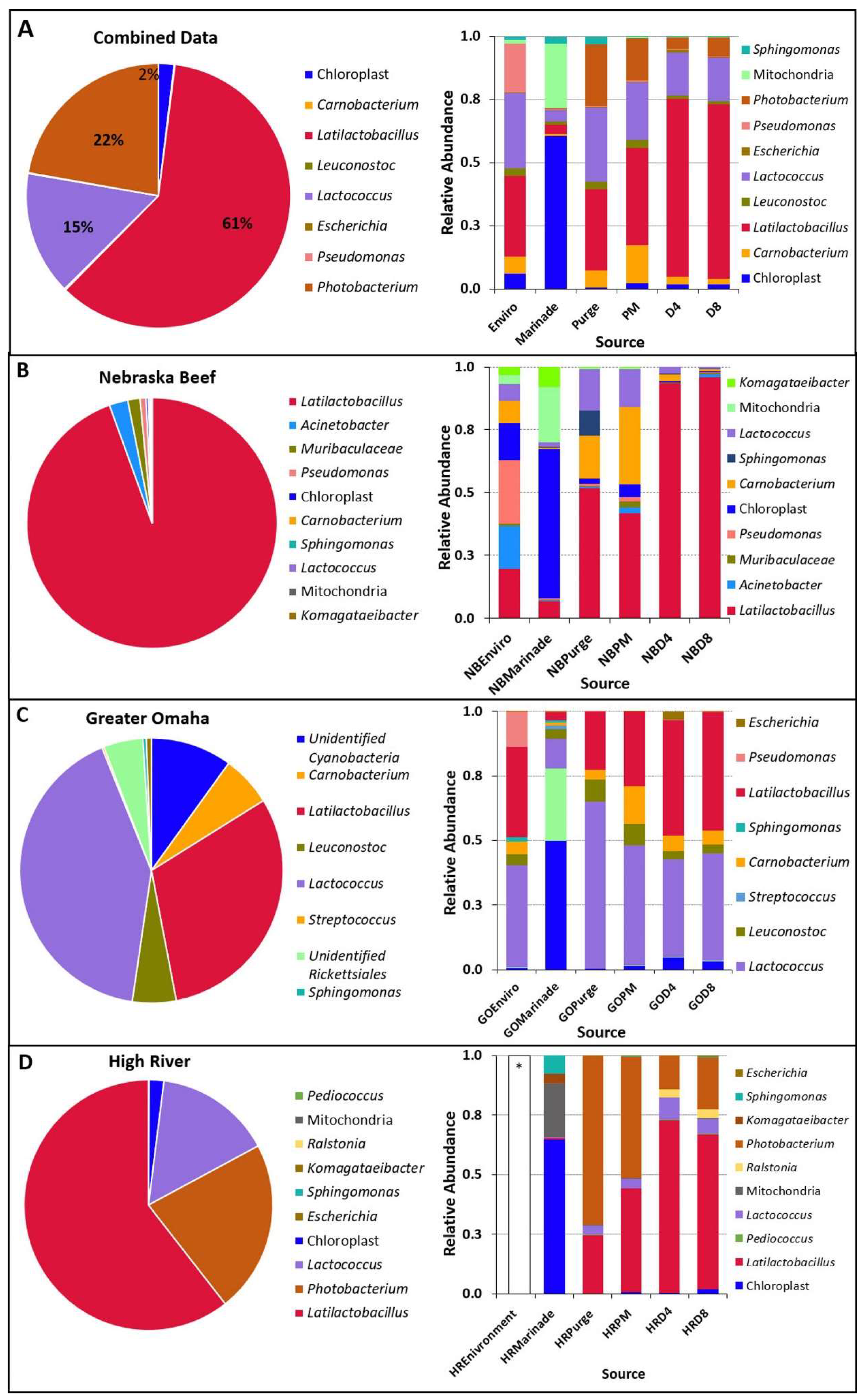
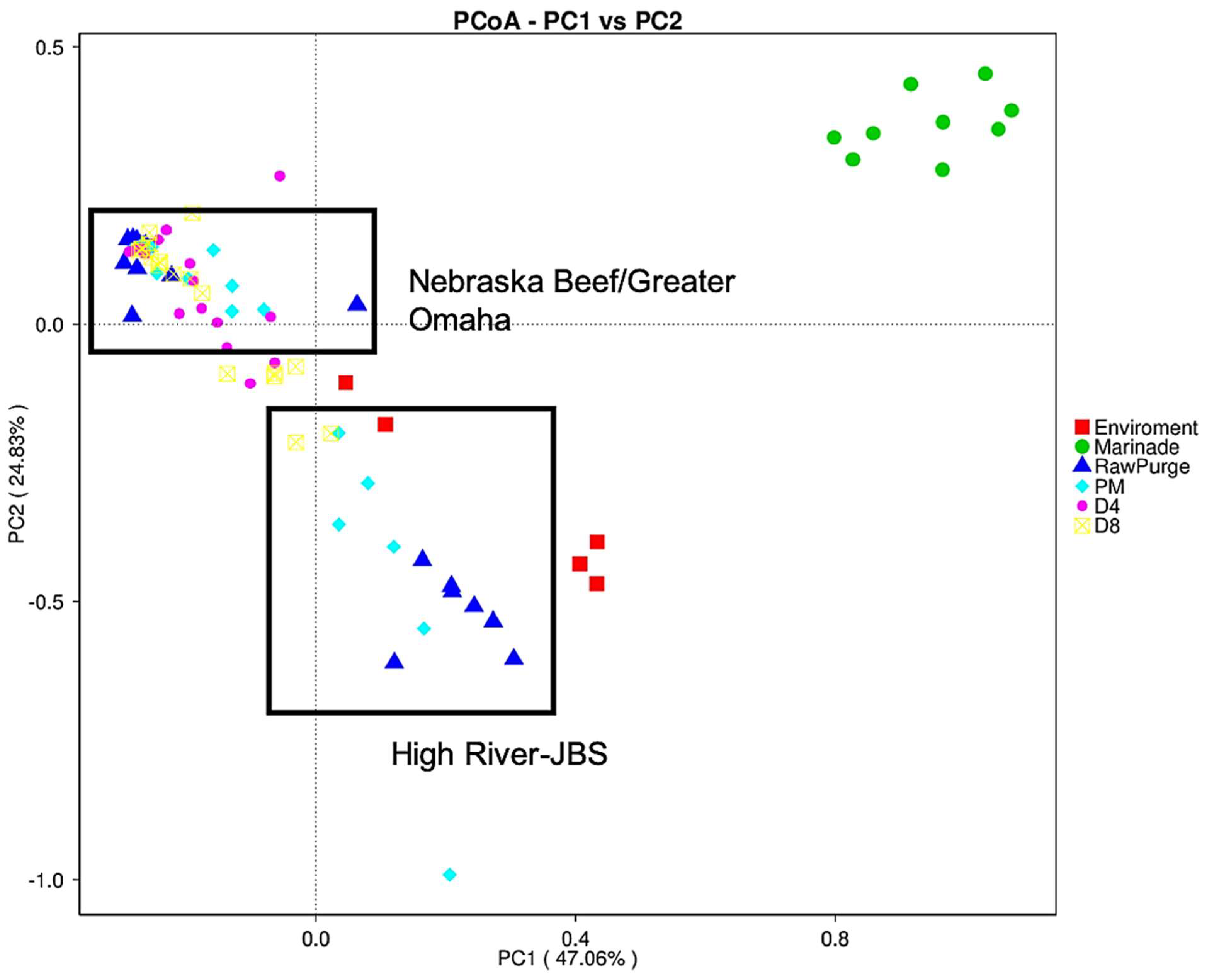
3.2. Culture-Independent Microbiome Analysis
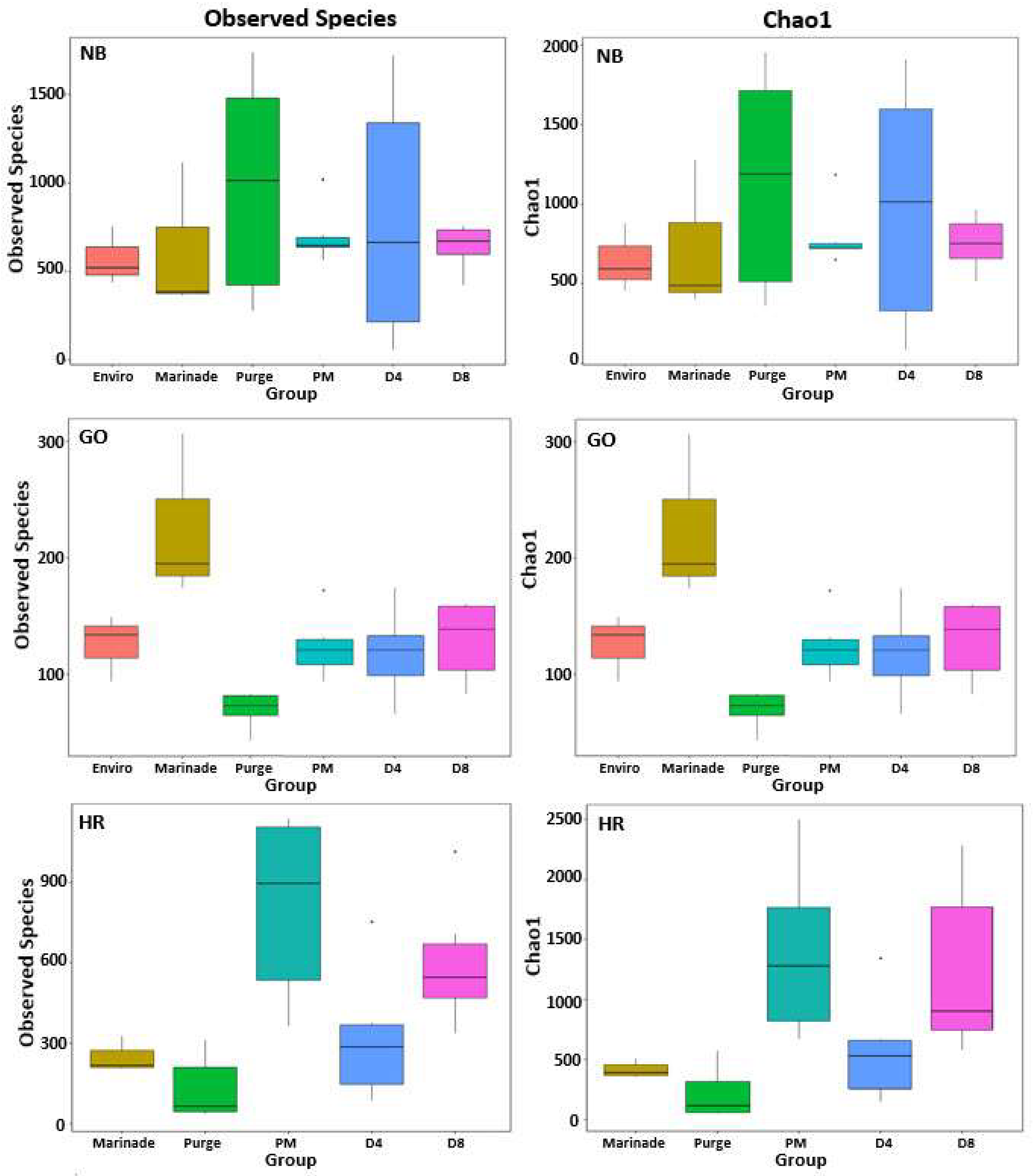
3.2.1. Bacterial Richness and Diversity in Biltong Processing
3.2.2. Changes in the Microbial Community during Processing
4. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Karolenko, C.E.; Bhusal, A.; Nelson, J.L.; Muriana, P.M. Processing of biltong (dried beef) to achieve USDA-FSIS 5-log reduction of Salmonella without a heat lethality step. Microorganisms 2020, 8, 791. [Google Scholar] [CrossRef] [PubMed]
- Gavai, K.; Karolenko, C.; Muriana, P.M. Effect of biltong dried beef processing on the reduction of Listeria monocytogenes, E. coli O157:H7, and Staphylococcus aureus, and the contribution of the major marinade components. Microorganisms 2022, 10, 1308. [Google Scholar] [CrossRef] [PubMed]
- Sitz, B.M.; Calkins, C.R.; Feuz, D.M.; Umberger, W.J.; Eskridge, K.M. Consumer sensory acceptance and value of wet-aged and dry-aged beef steaks. J. Anim. Sci. 2006, 84, 1221–1226. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Barros-Velázquez, J.; Carreira, L.; Franco, C.; Vazquez, B.I.; Fente, C.; Cepeda, A. Microbiological and physicochemical properties of fresh retail cuts of beef packaged under an advanced vacuum skin system and stored at 4C. J. Food Prot. 2003, 66, 2085–2092. [Google Scholar] [CrossRef]
- Venter, P.; Shale, K.; Lues, J.; Buys, E. Microbial proliferation and mathematical indices of vacuum-packed bovine meat. J. Food Process. Preserv. 2006, 30, 433–448. [Google Scholar] [CrossRef]
- Ercolini, D.; Russo, F.; Torrieri, E.; Masi, P.; Villani, F. Changes in the spoilage-related microbiota of beef during refrigerated storage under different packaging conditions. Appl. Environ. Microbiol. 2006, 72, 4663–4671. [Google Scholar] [CrossRef] [Green Version]
- Dainty, R.H.; Mackey, B.M. The relationship between the phenotypic properties of bacteria from chill-stored meat and spoilage processes. J. Appl. Microbiol. 1992, 73, 103s–114s. [Google Scholar] [CrossRef]
- Gram, L.; Ravn, L.; Rasch, M.; Bruhn, J.B.; Christensen, A.B.; Givskov, M. Food spoilage--interactions between food spoilage bacteria. Int. J. Food Microbiol. 2002, 78, 79–97. [Google Scholar] [CrossRef]
- Johansson, P.; Jaaskelainen, E.; Nieminen, T.T.; Hultman, J.; Auvinen, P.; Björkroth, J. Microbiomes in the context of refrigerated raw meat spoilage. MMB 2020, 4, 2. [Google Scholar] [CrossRef]
- Nieminen, T.T.; Välitalo, H.; Säde, E.; Paloranta, A.; Koskinen, K.; Björkroth, J. The effect of marination on lactic acid bacteria communities in raw broiler fillet strips. Front. Microbiol. 2012, 3, 376. [Google Scholar] [CrossRef]
- Chaillou, S.; Chaulot-Talmon, A.; Caekebeke, H.; Cardinal, M.; Christieans, S.; Denis, C.; Desmonts, M.H.; Dousset, X.; Feurer, C.; Hamon, E.; et al. Origin and ecological selection of core and food-specific bacterial communities associated with meat and seafood spoilage. ISME J. 2015, 9, 1105–1118. [Google Scholar] [CrossRef] [Green Version]
- De Filippis, F.; La Storia, A.; Villani, F.; Ercolini, D. Exploring the sources of bacterial spoilers in beefsteaks by culture-independent high-throughput sequencing. PLoS ONE 2013, 8, e70222. [Google Scholar] [CrossRef] [Green Version]
- Pini, F.; Aquilani, C.; Giovannetti, L.; Viti, C.; Pugliese, C. Characterization of the microbial community composition in Italian Cinta Senese sausages dry-fermented with natural extracts as alternatives to sodium nitrite. Food Microbiol. 2020, 89, 103417. [Google Scholar] [CrossRef]
- Cao, Y.; Fanning, S.; Proos, S.; Jordan, K.; Srikumar, S. A Review on the applications of next generation sequencing technologies as applied to food-related microbiome studies. Front. Microbiol. 2017, 8, 1829. [Google Scholar] [CrossRef] [Green Version]
- Jarvis, K.G.; Daquigan, N.; White, J.R.; Morin, P.M.; Howard, L.M.; Manetas, J.E.; Ottesen, A.; Ramachandran, P.; Grim, C.J. Microbiomes associated with foods from plant and animal sources. Front. Microbiol. 2018, 9, 2540. [Google Scholar] [CrossRef] [Green Version]
- Säde, E.; Penttinen, K.; Björkroth, J.; Hultman, J. Exploring lot-to-lot variation in spoilage bacterial communities on commercial modified atmosphere packaged beef. Food Microbiol. 2017, 62, 147–152. [Google Scholar] [CrossRef] [Green Version]
- Yang, X.; Noyes, N.R.; Doster, E.; Martin, J.N.; Linke, L.M.; Magnuson, R.J.; Yang, H.; Geornaras, I.; Woerner, D.R.; Jones, K.L.; et al. Use of metagenomic shotgun sequencing technology to detect foodborne pathogens within the microbiome of the beef production chain. Appl. Environ. Microbiol. 2016, 82, 2433–2443. [Google Scholar] [CrossRef] [Green Version]
- Clark, D.; Clark, D.; Bass, P.; Capouya, R.; Mitchell, T. A survey of microbial communities on dry-aged beef in commercial meat processing facilities. Meat Muscle Biol. 2020, 4. [Google Scholar] [CrossRef]
- Ojo-Okunola, A.; Claassen-Weitz, S.; Mwaikono, K.S.; Gardner-Lubbe, S.; Zar, H.J.; Nicol, M.P.; du Toit, E. The influence of DNA extraction and lipid removal on human milk bacterial profiles. Methods Protoc. 2020, 3, 39. [Google Scholar] [CrossRef]
- Hanlon, K.; Miller, J.; Brashears, M.M.; Smith, M.; Brooks, C.; Legako, J. Microbial profile evaluation of beef steaks from different packaging and retail lighting display conditions. Meat Muscle Biol. 2021, 5, 1–9. [Google Scholar] [CrossRef]
- Karolenko, C.; Muriana, P. Quantification of process lethality (5-Log reduction) of Salmonella and salt concentration during sodium replacement in biltong marinade. Foods 2020, 9, 1570. [Google Scholar] [CrossRef]
- Coton, E.; Coton, M. Multiplex PCR for colony direct detection of Gram-positive histamine- and tyramine-producing bacteria. J. Microbiol. Methods 2005, 63, 296–304. [Google Scholar] [CrossRef] [PubMed]
- Kumar, S.; Stecher, G.; Li, M.; Knyaz, C.; Tamura, K. MEGA X: Molecular evolutionary genetics analysis across computing platforms. Mol. Biol. Evol. 2018, 35, 1547–1549. [Google Scholar] [CrossRef] [PubMed]
- Desjardins, P.; Conklin, D. NanoDrop microvolume quantitation of nucleic acids. JoVE 2010, 45, e2565. [Google Scholar] [CrossRef] [Green Version]
- Edgar, R.C. Updating the 97% identity threshold for 16S ribosomal RNA OTUs. Bioinformatics 2018, 34, 2371–2375. [Google Scholar] [CrossRef] [Green Version]
- Janda, J.M.; Abbott, S.L. 16S rRNA gene sequencing for bacterial identification in the diagnostic laboratory: Pluses, perils, and pitfalls. J. Clin. Microbiol. 2007, 45, 2761. [Google Scholar] [CrossRef] [Green Version]
- Petti, C.A. Detection and identification of microorganisms by gene amplification and sequencing. Clin. Infect. Dis. 2007, 44, 1108–1114. [Google Scholar] [CrossRef]
- Rossi-Tamisier, M.; Benamar, S.; Raoult, D.; Fournier, P.E. Cautionary tale of using 16S rRNA gene sequence similarity values in identification of human-associated bacterial species. Int. J. Syst. Evol. Microbiol. 2015, 65, 1929–1934. [Google Scholar] [CrossRef]
- Pennacchia, C.; Ercolini, D.; Villani, F. Spoilage-related microbiota associated with chilled beef stored in air or vacuum pack. Food Microbiol. 2011, 28, 84–93. [Google Scholar] [CrossRef]
- Laursen, B.G.; Bay, L.; Cleenwerck, I.; Vancanneyt, M.; Swings, J.; Dalgaard, P.; Leisner, J.J. Carnobacterium divergens and Carnobacterium maltaromaticum as spoilers or protective cultures in meat and seafood: Phenotypic and genotypic characterization. Syst. Appl. Microbiol. 2005, 28, 151–164. [Google Scholar] [CrossRef]
- Aznar, R.; Chenoll, E. Intraspecific diversity of Lactobacillus curvatus, Lactobacillus plantarum, Lactobacillus sakei, and Leuconostoc mesenteroides associated with vacuum-packed meat product spoilage analyzed by Randomly Amplified Polymorphic DNA PCR. J. Food Prot. 2006, 69, 2403–2410. [Google Scholar] [CrossRef]
- Iskandar, C.F.; Borges, F.; Taminiau, B.; Daube, G.; Zagorec, M.; Remenant, B.; Leisner, J.J.; Hansen, M.A.; Sørensen, S.J.; Mangavel, C.; et al. Comparative genomic analysis reveals ecological differentiation in the genus Carnobacterium. Front. Microbiol. 2017, 8, 357. [Google Scholar] [CrossRef] [Green Version]
- Chaillou, S.; Champomier-Vergès, M.-C.; Cornet, M.; Crutz-Le Coq, A.-M.; Dudez, A.-M.; Martin, V.; Beaufils, S.; Darbon-Rongère, E.; Bossy, R.; Loux, V.; et al. The complete genome sequence of the meat-borne lactic acid bacterium Lactobacillus sakei 23K. Nat. Biotech. 2005, 23, 1527–1533. [Google Scholar] [CrossRef] [Green Version]
- Chiaramonte, F.; Blugeon, S.; Chaillou, S.; Langella, P.; Zagorec, M. Behavior of the meat-borne bacterium Lactobacillus sakei during Its transit through the gastrointestinal tracts of axenic and conventional mice. Appl. Environ. Microbiol. 2009, 75, 4498–4505. [Google Scholar] [CrossRef] [Green Version]
- Minks, D.; Stringer, W.C. The influence of aging beef in vacuum. J. Food Sci. 1972, 37, 736–738. [Google Scholar] [CrossRef]
- Terjung, N.; Witte, F.; Heinz, V. The dry aged beef paradox: Why dry aging is sometimes not better than wet aging. Meat Sci. 2021, 172, 108355. [Google Scholar] [CrossRef]
- Leisner, J.J.; Laursen, B.G.; Prévost, H.; Drider, D.; Dalgaard, P. Carnobacterium: Positive and negative effects in the environment and in foods. FEMS Microbiol. Rev. 2007, 31, 592–613. [Google Scholar] [CrossRef] [Green Version]
- Chen, Y.; Li, N.; Zhao, S.; Zhang, C.; Qiao, N.; Duan, H.; Xiao, Y.; Yan, B.; Zhao, J.; Tian, F.; et al. Integrated phenotypic–genotypic analysis of Latilactobacillus sakei from different niches. Foods 2021, 10, 1717. [Google Scholar] [CrossRef]
- Doulgeraki, A.I.; Ercolini, D.; Villani, F.; Nychas, G.J. Spoilage microbiota associated to the storage of raw meat in different conditions. Int. J. Food Microbiol. 2012, 157, 130–141. [Google Scholar] [CrossRef]
- Nychas, G.-J.E.; Skandamis, P.N.; Tassou, C.C.; Koutsoumanis, K.P. Meat spoilage during distribution. Meat Sci. 2008, 78, 77–89. [Google Scholar] [CrossRef]
- Zhang, Y.; Zhu, L.; Dong, P.; Liang, R.; Mao, Y.; Qiu, S.; Luo, X. Bio-protective potential of lactic acid bacteria: Effect of Lactobacillus sakei and Lactobacillus curvatus on changes of the microbial community in vacuum-packaged chilled beef. Asian-Australas J. Anim. Sci. 2018, 31, 585–594. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Casaburi, A.; Nasi, A.; Ferrocino, I.; Di Monaco, R.; Mauriello, G.; Villani, F.; Ercolini, D. Spoilage-related activity of Carnobacterium maltaromaticum strains in air-stored and vacuum-packed meat. Appl. Environ. Microbiol. 2011, 77, 7382–7393. [Google Scholar] [CrossRef] [Green Version]
- Borch, E.; Kant-Muermans, M.L.; Blixt, Y. Bacterial spoilage of meat and cured meat products. Int. J. Food Microbiol. 1996, 33, 103–120. [Google Scholar] [CrossRef] [PubMed]
- Pothakos, V.; Devlieghere, F.; Villani, F.; Björkroth, J.; Ercolini, D. Lactic acid bacteria and their controversial role in fresh meat spoilage. Meat Sci. 2015, 109, 66–74. [Google Scholar] [CrossRef] [PubMed]
- Karolenko, C.E.; Bhusal, A.; Gautam, D.; Muriana, P.M. Selenite cystine agar for enumeration of inoculated Salmonella serovars recovered from stressful conditions during antimicrobial validation studies. Microorganisms 2020, 8, 338. [Google Scholar] [CrossRef] [Green Version]
- Koutsoumanis, K.P.; Kendall, P.A.; Sofos, J.N. Effect of food processing-related stresses on acid tolerance of Listeria monocytogenes. Appl. Environ. Microbiol. 2003, 69, 7514–7516. [Google Scholar] [CrossRef] [Green Version]
- Capozzi, V.; Fiocco, D.; Amodio, M.L.; Gallone, A.; Spano, G. Bacterial stressors in minimally processed food. Int. J. Mol. Sci. 2009, 10, 3076–3105. [Google Scholar] [CrossRef] [Green Version]
- Rodriguez-Romo, L.; Yousef, A.; Griffiths, M. Cross-protective effects of bacterial stress. In Understanding Pathogen Behaviour; Elsevier: Amsterdam, The Netherlands, 2005; pp. 128–151. [Google Scholar]
- Karolenko, C.E.; Wilkinson, J.; Muriana, P.M. Evaluation of various lactic acid bacteria and generic E. coli as potential nonpathogenic surrogates for in-plant validation of biltong dried beef processing. Microorganisms 2022, 10, 1648. [Google Scholar] [CrossRef]
- Woese, C.R. Bacterial evolution. Microbiol. Rev. 1987, 51, 221–271. [Google Scholar] [CrossRef]
- Li, Y.; Zhuang, S.; Liu, Y.; Zhang, L.; Liu, X.; Cheng, H.; Liu, J.; Shu, R.; Luo, Y. Effect of grape seed extract on quality and microbiota community of container-cultured snakehead (Channa argus) fillets during chilled storage. Food Microbiol. 2020, 91, 103492. [Google Scholar] [CrossRef]
- Teng, F.; Darveekaran Nair, S.S.; Zhu, P.; Li, S.; Huang, S.; Li, X.; Xu, J.; Yang, F. Impact of DNA extraction method and targeted 16S-rRNA hypervariable region on oral microbiota profiling. Sci. Rep. 2018, 8, 16321. [Google Scholar] [CrossRef] [Green Version]
- Hilgarth, M.; Behr, J.; Vogel, R.F. Monitoring of spoilage-associated microbiota on modified atmosphere packaged beef and differentiation of psychrophilic and psychrotrophic strains. J. Appl. Microbiol. 2018, 124, 740–753. [Google Scholar] [CrossRef]
- Claesson, M.J.; Wang, Q.; O’Sullivan, O.; Greene-Diniz, R.; Cole, J.R.; Ross, R.P.; O’Toole, P.W. Comparison of two next-generation sequencing technologies for resolving highly complex microbiota composition using tandem variable 16S rRNA gene regions. Nucleic Acids Res. 2010, 38, e200. [Google Scholar] [CrossRef]
- Braz, V.S.; Melchior, K.; Moreira, C.G. Escherichia coli as a multifaceted pathogenic and versatile bacterium. Front. Cell. Infect. Microbiol. 2020, 10, 548492. [Google Scholar] [CrossRef]
- Nickelson, R.; Luchansky, J.B.; Kaspar, C.; Johnson, E. Dry fermented sausage and Escherichia coli O157:H7 validation research. In An executive summary prepared by the Blue Ribbon Task Force of the National Cattlemen’s Beef Association; Research Report No. 11-316; National Cattlemen’s Beef Association: Chicago, IL, USA, 1996. [Google Scholar]
- Fuertes-Perez, S.; Hauschild, P.; Hilgarth, M.; Vogel, R.F. Biodiversity of photobacterium spp. isolated from meats. Front. Microbiol. 2019, 10, 2399. [Google Scholar] [CrossRef]
- Fougy, L.; Desmonts, M.; Coeuret, G.; Fassel, C.; Harmon, E.; Hezard, B.; Champomier-Vergès, M.; Chaillou, S.; Besser, T.E. Reducing salt in raw pork sausages increases spoilage and correlates with reduced bacterial diversity. Appl. Environ. Microbiol. 2016, 82, 3928–3939. [Google Scholar] [CrossRef] [Green Version]
- Champomier-Vergès, M.C.; Chaillou, S.; Cornet, M.; Zagorec, M. Lactobacillus sakei: Recent developments and future prospects. Res. Microbiol. 2001, 152, 839–848. [Google Scholar] [CrossRef]
- Hanshew, A.S.; Mason, C.J.; Raffa, K.F.; Currie, C.R. Minimization of chloroplast contamination in 16S rRNA gene pyrosequencing of insect herbivore bacterial communities. J. Microbiol. Methods 2013, 95, 149–155. [Google Scholar] [CrossRef] [Green Version]
- Rastogi, G.; Tech, J.J.; Coaker, G.L.; Leveau, J.H. A PCR-based toolbox for the culture-independent quantification of total bacterial abundances in plant environments. J. Microbiol. Methods 2010, 83, 127–132. [Google Scholar] [CrossRef]
- Anguita-Maeso, M.; Olivares-García, C.; Haro, C.; Imperial, J.; Navas-Cortés, J.A.; Landa, B.B. Culture-dependent and culture-independent characterization of the olive xylem microbiota: Effect of sap extraction methods. Front. Plant Sci. 2019, 10, 1708. [Google Scholar] [CrossRef] [Green Version]
- Hinsu, A.; Dumadiya, A.; Joshi, A.; Kotadiya, R.; Andharia, K.; Koringa, P.; Kothari, R. To culture or not to culture: A snapshot of culture-dependent and culture-independent bacterial diversity from peanut rhizosphere. PeerJ 2021, 9, e12035. [Google Scholar] [CrossRef] [PubMed]
- Tidjani Alou, M.; Naud, S.; Khelaifia, S.; Bonnet, M.; Lagier, J.C.; Raoult, D. State of the art in the culture of the human microbiota: New interests and strategies. Clin. Microbiol. Rev. 2020, 34, e00129-19. [Google Scholar] [CrossRef] [PubMed]


| Sample | Genus (Species) | Sequence Length (bp) | Query Coverage (%) | Percent ID |
|---|---|---|---|---|
| GO Raw R1A 1,2 | Serratia sp. | 1477 | 99 | 97.6 |
| GO Raw R1B 1,2 | Carnobacterium sp. | 1411 | 100 | 98.1 |
| GO Raw R1C 1,2 | Lactococcus piscium | 1449 | 98 | 99.7 |
| GO Raw R1D 1,2 | Carnobacterium divergens | 1421 | 100 | 99.8 |
| GO Raw R1E 1,2 | Lactococcus piscium | 1449 | 98 | 99.7 |
| GO Raw R2A 1,2 | Carnobacterium divergens | 1407 | 100 | 99.9 |
| GO Raw R2B 1,2 | Carnobacterium divergens | 1467 | 98 | 99.7 |
| GO Raw R2C 1,2 | Carnobacterium sp. | 1420 | 99 | 98.1 |
| GO Raw R2D 1,2 | Latilactobacillus sakei | 1382 | 100 | 99.9 |
| GO Raw R2E 1,2 | Latilactobacillus sakei | 1408 | 100 | 100.0 |
| GO D8Mar R1A 1,3 | Latilactobacillus sakei | 1489 | 99 | 99.7 |
| GO D8Mar R1B 1,3 | Carnobacterium divergens | 1469 | 98 | 99.7 |
| GO D8Mar R1C 1,3 | Latilactobacillus sakei | 1494 | 99 | 99.7 |
| GO D8Mar R1D 1,3 | Carnobacterium divergens | 1468 | 98 | 99.8 |
| GO D8Mar R1E 1,3 | Latilactobacillus sakei | 1489 | 99 | 99.7 |
| GO D8Mar R2A 1,3 | Latilactobacillus sakei | 1490 | 98 | 99.9 |
| GO D8Mar R2B 1,3 | Latilactobacillus sakei | 1493 | 98 | 99.9 |
| GO D8Mar R2C 1,3 | Latilactobacillus sakei | 1489 | 98 | 99.8 |
| GO D8Mar R2D 1,3 | Latilactobacillus sakei | 1475 | 98 | 99.8 |
| GO D8Mar R2E-A 1,3 | Latilactobacillus sakei | 1489 | 98 | 99.7 |
| GO D8Mar R2E-B 1,3 | Carnobacterium divergens | 1467 | 98 | 99.6 |
| Sample | Genus (Species) | Sequence Length (bp) | Query Coverage (%) | Percent ID |
|---|---|---|---|---|
| NB2 Raw R1A 1,2 | Latilactobacillus sp. (sakei, graminis, or curvatus) | 1501 | 100 | 99.7 |
| NB2 Raw R1B 1,2 | Latilactobacillus sp. (graminis, or curvatus) | 1461 | 100 | 99.8 |
| NB2 Raw R1C 1,2 | Hafnia paralvei | 1453 | 99 | 99.7 |
| NB2 Raw R1D 1,2 | Carnobacterium gallinarum | 1436 | 99 | 98.5 |
| NB2 Raw R1E 1,2 | Hafnia paralvei | 1472 | 99 | 99.6 |
| NB2 Raw R2A 1,2 | Latilactobacillus curvatus | 1467 | 100 | 99.5 |
| NB2 Raw R2B 1,2 | Latilactobacillus curvatus | 1467 | 100 | 99.7 |
| NB2 Raw R2C 1,2 | Carnobacterium gallinarum | 1461 | 99 | 98.5 |
| NB2 Raw R2D 1,2 | Enterobacter mori | 1472 | 100 | 99.3 |
| NB2 Raw R2E 1,2 | Hafnia paralvei | 1473 | 99 | 99.6 |
| NB2 D8Mar R1A 1,3 | Latilactobacillus sp. (sakei or curvatus) | 1473 | 100 | 99.6 |
| NB2 D8Mar R1B 1,3 | Latilactobacillus sp. (sakei or curvatus) | 1503 | 100 | 99.7 |
| NB2 D8Mar R1C 1,3 | Carnobacterium gallinarum | 1500 | 99 | 98.2 |
| NB2 D8Mar R1D 1,3 | Carnobacterium gallinarum | 1472 | 100 | 98.2 |
| NB2 D8Mar R1E 1,3 | Latilactobacillus sp. (sakei or curvatus) | 1498 | 100 | 99.7 |
| NB2 D8Mar R2A 1,3 | Leuconostoc mesenteroides | 1441 | 100 | 99.4 |
| NB2 D8Mar R2B 1,3 | Leuconostoc mesenteroides | 1457 | 100 | 99.9 |
| NB2 D8Mar R2C 1,3 | Latilactobacillus sp. (sakei or curvatus) | 1524 | 100 | 99.7 |
| NB2 D8Mar R2D 1,3 | Latilactobacillus sp. (sakei or curvatus) | 1500 | 100 | 99.6 |
| NB2 D8Mar R2E 1,3 | Latilactobacillus sp. (sakei or curvatus) | 1467 | 100 | 99.7 |
| Sample | Genus (Species) | Sequence Length (bp) | Query Coverage (%) | Percent ID |
|---|---|---|---|---|
| HR Raw R1A 1,2 | Carnobacterium divergens | 1472 | 98 | 99.5 |
| HR Raw R1B 1,2 | Carnobacterium divergens | 1519 | 96 | 98.6 |
| HR Raw R1C 1,2 | Carnobacterium divergens | 1440 | 96 | 100.0 |
| HR Raw R1D 1,2 | Carnobacterium divergens | 1481 | 97 | 99.2 |
| HR Raw R1E 1,2 | Leuconostoc mesenteroides | 1477 | 99 | 99.7 |
| HR Raw R2A 1,2 | Carnobacterium divergens | 1504 | 96 | 99.3 |
| HR Raw R2B 1,2 | Brevibacillus invocatus | 1467 | 99 | 98.8 |
| HR Raw R2C 1,2 | Carnobacterium divergens | 1477 | 96 | 99.6 |
| HR Raw R2D 1,2 | Carnobacterium divergens | 1510 | 96 | 98.2 |
| HR Raw R2E 1,2 | Carnobacterium divergens | 1506 | 96 | 98.4 |
| HR D8Mar R1A 1,3 | Lactilactobacillus sakei | 1494 | 100 | 99.5 |
| HR D8Mar R1B 1,3 | Carnobacterium divergens | 1485 | 98 | 98.5 |
| HR D8Mar R1C 1,3 | Carnobacterium divergens | 1478 | 98 | 98.8 |
| HR D8Mar R1D 1,3 | Carnobacterium divergens | 1448 | 98 | 99.2 |
| HR D8Mar R1E 1,3 | Carnobacterium divergens | 1441 | 99 | 99.3 |
| HR D8Mar R2A 1,3 | Latilactobacillus sakei | 1512 | 100 | 99.3 |
| HR D8Mar R2B 1,3 | Carnobacterium divergens | 1476 | 98 | 99.3 |
| HR D8Mar R2C 1,3 | Carnobacterium divergens | 1546 | 96 | 98.9 |
| HR D8Mar R2D 1,3 | Brevibacillus invocatus | 1471 | 99 | 98.9 |
| HR D8Mar R2E 1,3 | Carnobacterium divergens | 1502 | 96 | 99.4 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Karolenko, C.; DeSilva, U.; Muriana, P.M. Microbial Profiling of Biltong Processing Using Culture-Dependent and Culture-Independent Microbiome Analysis. Foods 2023, 12, 844. https://doi.org/10.3390/foods12040844
Karolenko C, DeSilva U, Muriana PM. Microbial Profiling of Biltong Processing Using Culture-Dependent and Culture-Independent Microbiome Analysis. Foods. 2023; 12(4):844. https://doi.org/10.3390/foods12040844
Chicago/Turabian StyleKarolenko, Caitlin, Udaya DeSilva, and Peter M. Muriana. 2023. "Microbial Profiling of Biltong Processing Using Culture-Dependent and Culture-Independent Microbiome Analysis" Foods 12, no. 4: 844. https://doi.org/10.3390/foods12040844
APA StyleKarolenko, C., DeSilva, U., & Muriana, P. M. (2023). Microbial Profiling of Biltong Processing Using Culture-Dependent and Culture-Independent Microbiome Analysis. Foods, 12(4), 844. https://doi.org/10.3390/foods12040844






