Untargeted Metabolite Profiling of Adipose Tissue in Rats Exposed to Mepiquat
Abstract
:1. Introduction
2. Materials and Methods
2.1. Chemicals and Reagents
2.2. Animals and Treatments
2.3. Sample Preparation
2.4. GC-MS Analysis
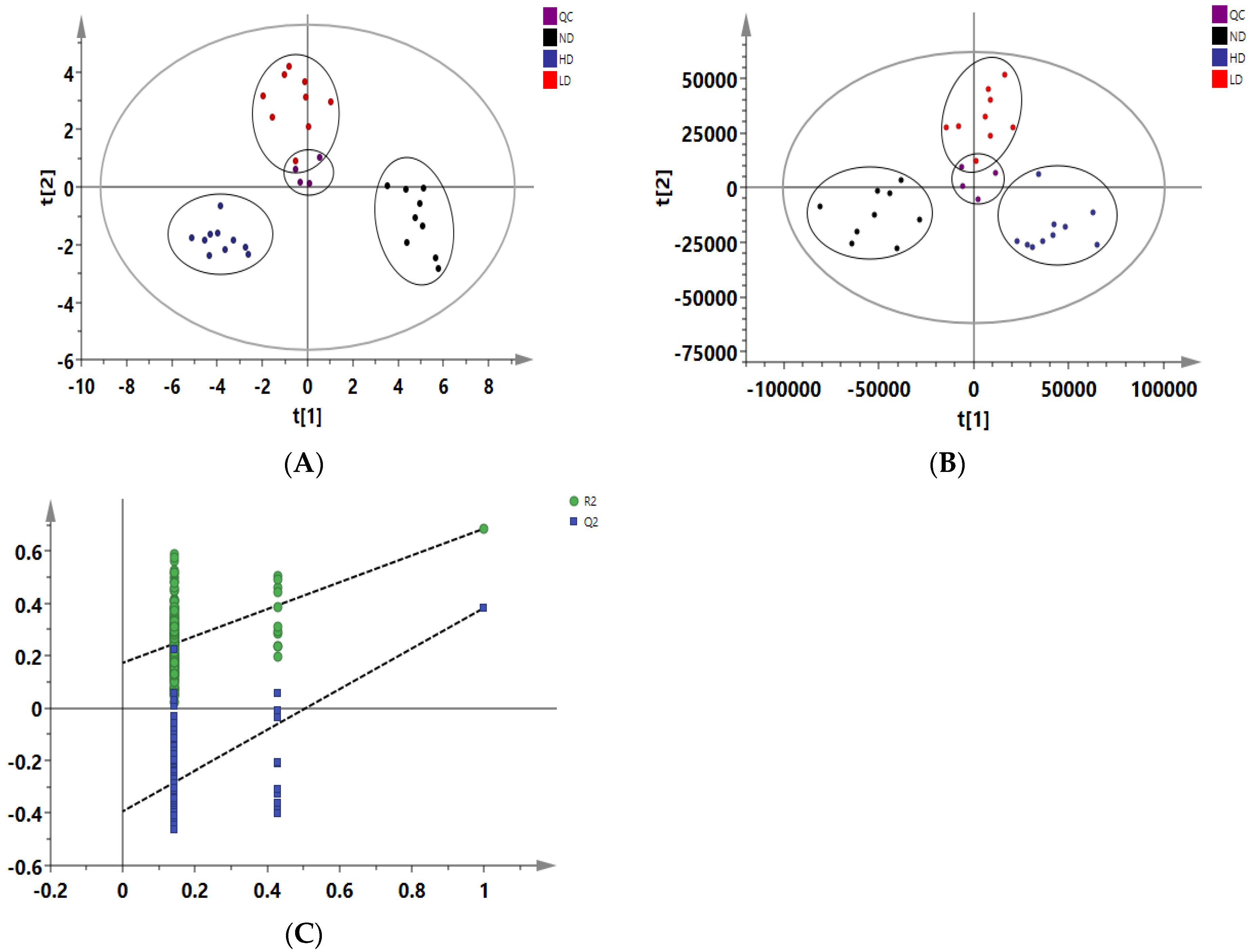
2.5. Data Processing and Multivariate Analysis
2.6. Receiver Operator Characteristic (ROC) Analysis
3. Results and Discussion
3.1. Weight Change
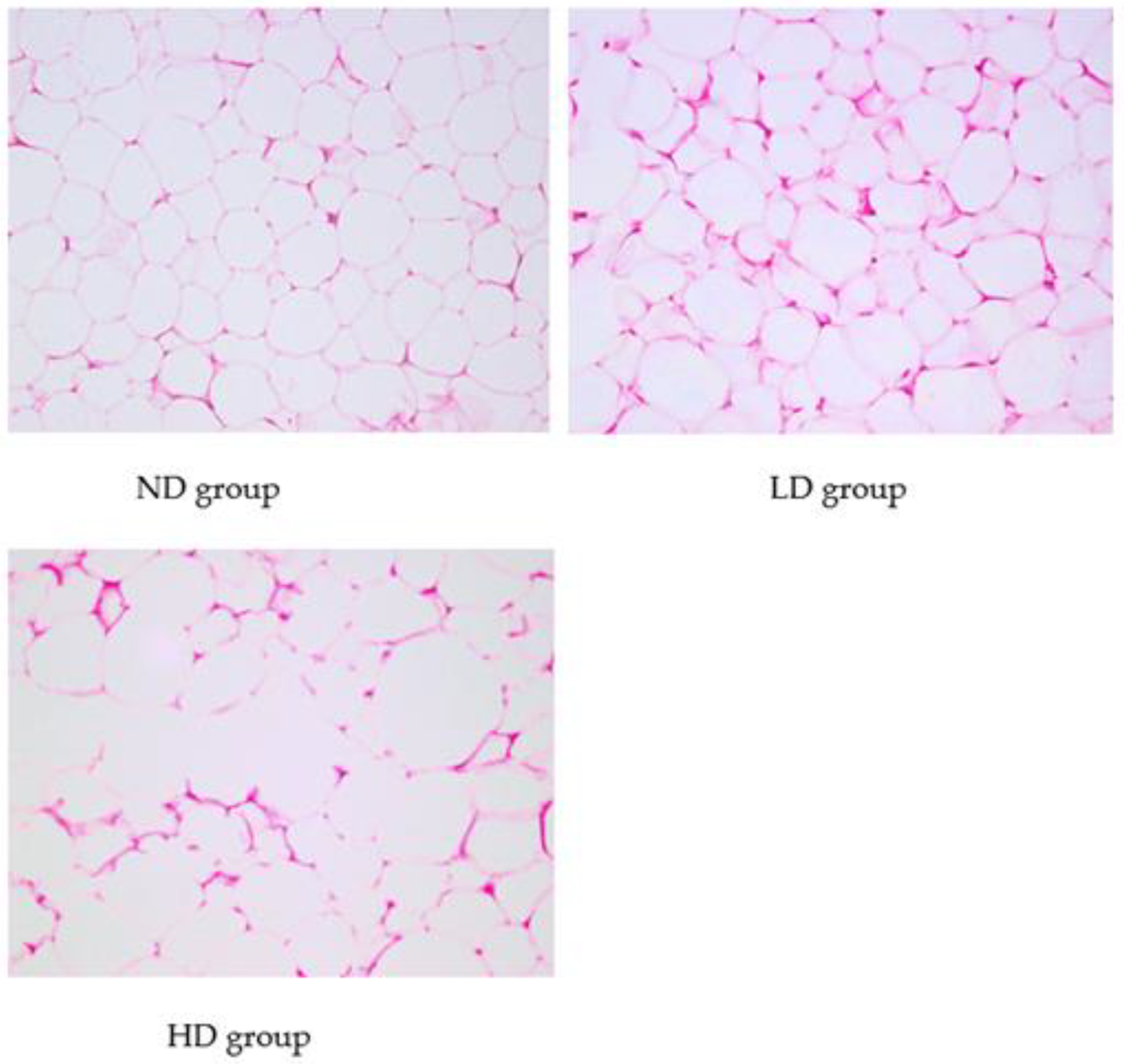
3.2. Histopathological Analysis
3.3. GC-MS Analysis of Adipose Tissue
3.4. Adipose Tissue Metabolomics Analysis
3.5. ROC Curve
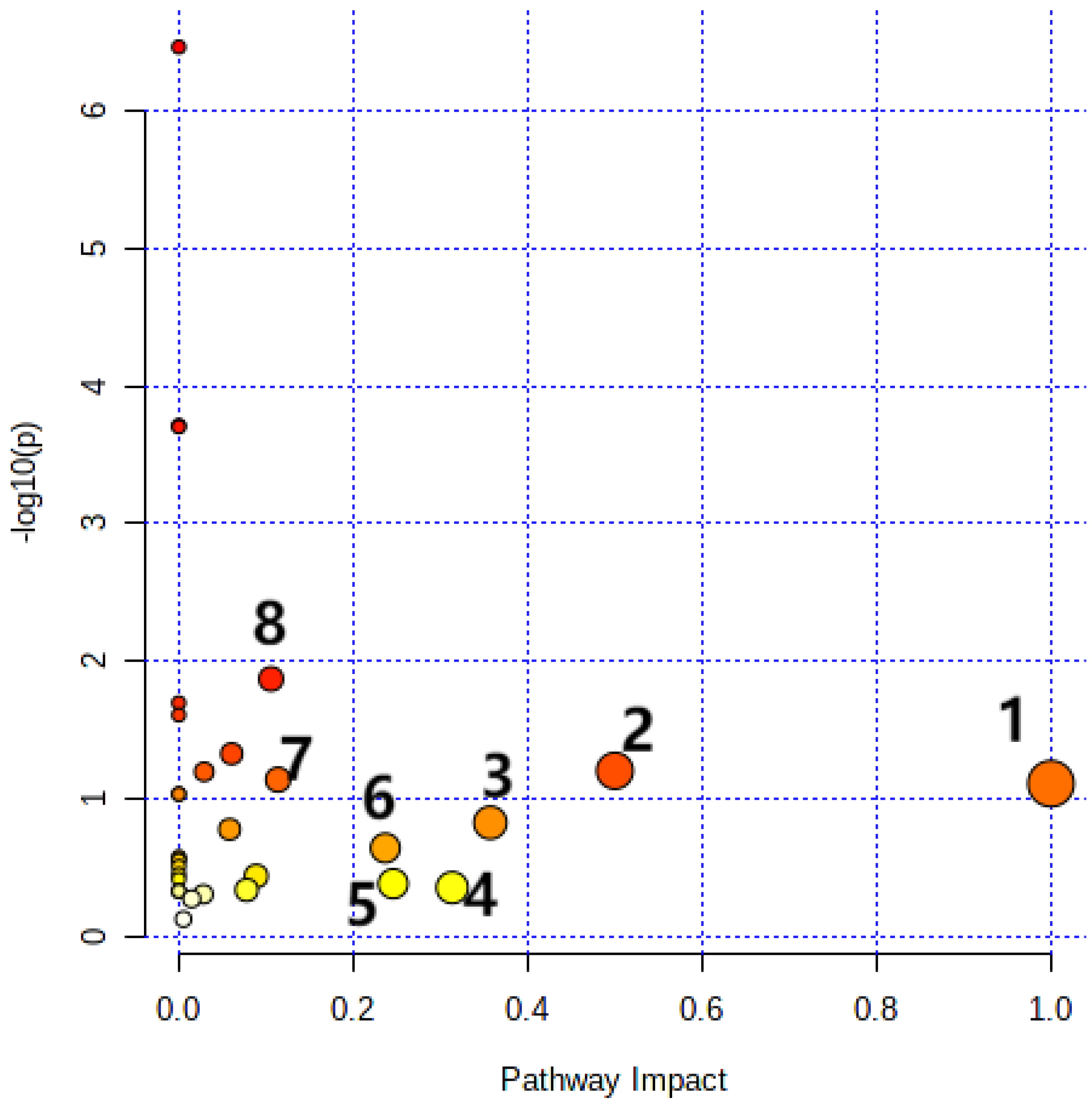
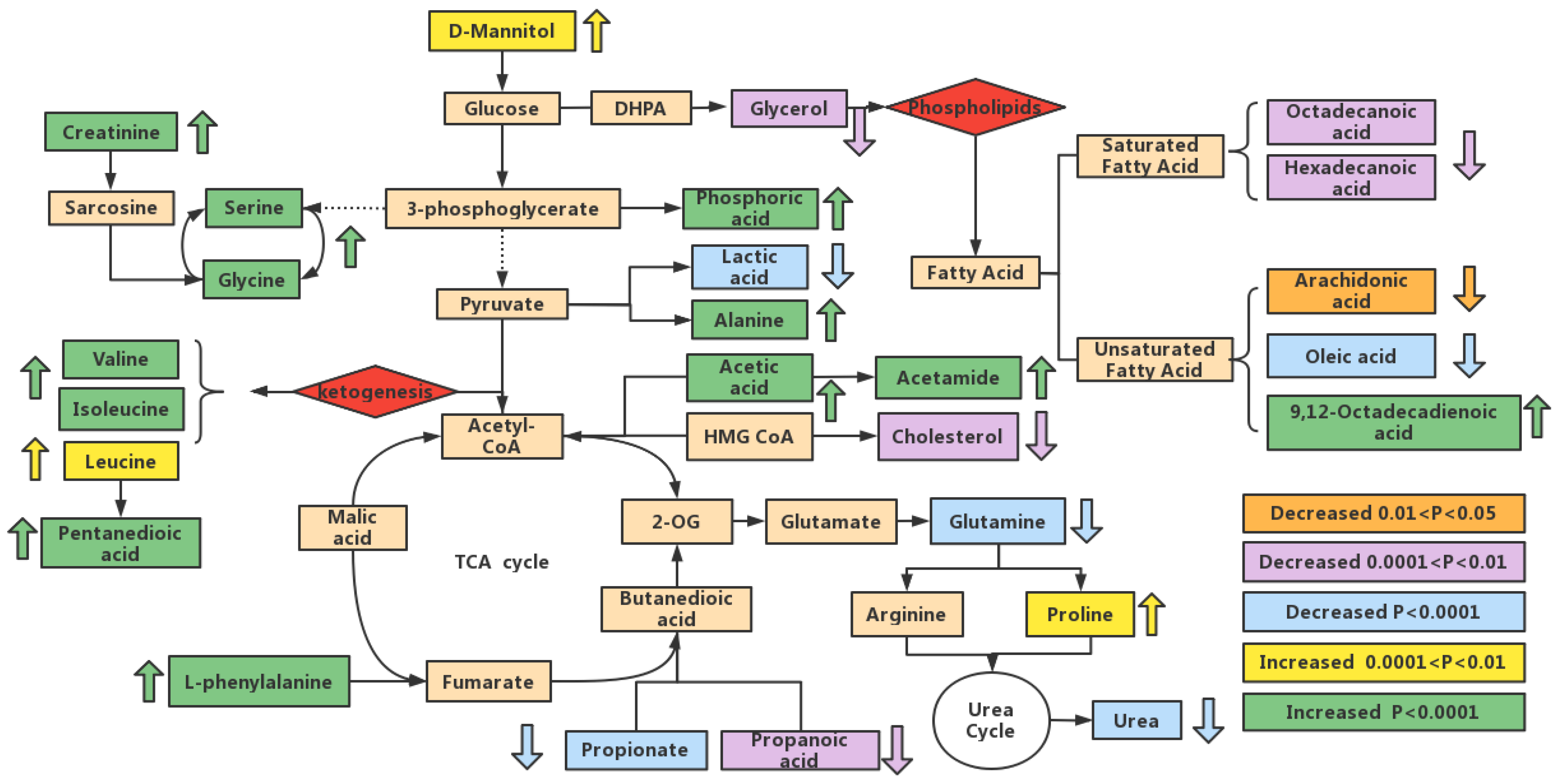
3.6. Metabolic Pathway Analysis
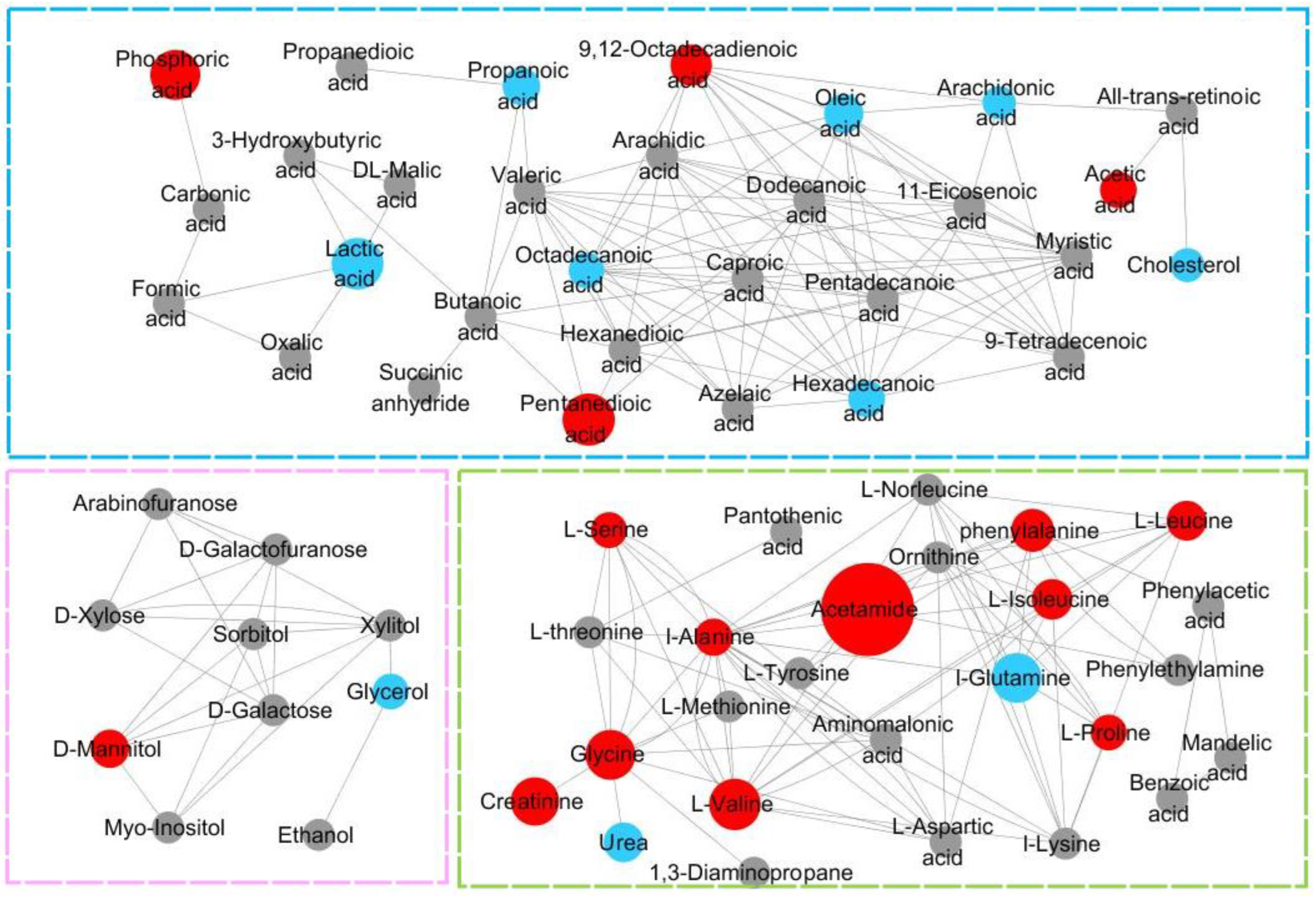
3.7. Analysis of the Metabolic Function of Adipose Tissue in Rats of ND, LD, and HD Groups
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Li, X.; Zhang, X.; Tan, L.; Yan, H.; Yuan, Y. Heat-induced formation of N,N-dimethylpiperidinium (mepiquat) in Arabica and Robusta coffee. J. Food Sci. 2020, 85, 2754–2761. [Google Scholar] [CrossRef] [PubMed]
- Wermann, S.; Theurillat, V.; Verzegnassi, L.; Hofmann, J.; Kuchenbecker, R.; Constable, A.; Delatour, T.; Stadler, R.H. N, N-dimethylpiperidinium (mepiquat) Part 2. Formation in roasted coffee and barley during thermal processing. Food Addit. Contam. Part A 2014, 31, 234–241. [Google Scholar] [CrossRef] [PubMed]
- Li, X.; Pang, Y.; Wang, S.; Liu, H.; Yan, H.; Yuan, Y. Effects of thermal processing on N,N-dimethylpiperidinium (mepiquat) formation in meat and vegetable products. Food Res. Int. 2021, 150, 110771. [Google Scholar] [CrossRef] [PubMed]
- Yuan, Y.; Li, X.; Liu, H.; Qu, Y.; Zhang, W.; Yu, H.; Zhang, J.; Zhuang, H. Carnitine, a New Precursor in the Formation of the Plant Growth Regulator Mepiquat. J. Agric. Food Chem. 2018, 66, 5907–5912. [Google Scholar] [CrossRef]
- European Food Safety Authority. Conclusion regarding the peer review of the pesticide risk assessment of the active substance mepiquat. EFSA J. 2008, 146, 1–73. [Google Scholar]
- Mo, J.; Ma, Z.; Yan, S.; Cheung, N.K.M.; Yang, F.; Yao, X.; Guo, J. Metabolomic profiles in a green alga (Raphidocelis subcapitata) following erythromycin treatment: ABC transporters and energy metabolism. J. Environ. Sci. 2023, 124, 591–601. [Google Scholar] [CrossRef]
- Grech, O.; Seneviratne, S.Y.; Alimajstorovic, Z.; Yiangou, A.; Mitchell, J.L.; Smith, T.B.; Mollan, S.P.; Lavery, G.G.; Ludwig, C.; Sinclair, A.J. Nuclear Magnetic Resonance Spectroscopy Metabolomics in Idiopathic Intracranial Hypertension to Identify Markers of Disease and Headache. Neurology 2022, 99, e1702–e1714. [Google Scholar] [CrossRef]
- Li, J.; Luu, L.D.W.; Wang, X.; Cui, X.; Huang, X.; Fu, J.; Zhu, X.; Li, Z.; Wang, Y.; Tai, J. Metabolomic analysis reveals potential biomarkers and the underlying pathogenesis involved in Mycoplasma pneumoniae pneumonia. Emerg. Microbes Infect. 2022, 11, 593–605. [Google Scholar] [CrossRef]
- Wang, Y.; Wang, Q.; Zhou, L.; Zeng, Z.; Zhao, C.; You, L.; Lu, X.; Liu, X.; Ouyang, R.; Wang, Y.; et al. Metabolomics insights into the prenatal exposure effects of polybrominated diphenyl ethers on neonatal birth outcomes. Sci. Total Environ. 2022, 836, 155601. [Google Scholar] [CrossRef]
- Zhang, Q.; Deng, X.; Wang, J. The effects of mepiquat chloride (DPC) on the soluble protein content and the activities of protective enzymes in cotton in response to aphid feeding and on the activities of detoxifying enzymes in aphids. BMC Plant Biol. 2022, 22, 213. [Google Scholar] [CrossRef]
- van Dam, A.D.; Boon, M.R.; Berbee, J.F.P.; Rensen, P.C.N.; van Harmelen, V. Targeting white, brown and perivascular adipose tissue in atherosclerosis development. Eur. J. Pharmacol. 2017, 816, 82–92. [Google Scholar] [CrossRef]
- Jones, O.A.H.; Murfitt, S.; Svendsen, C.; Turk, A.; Turk, H.; Spurgeon, D.J.; Walker, L.A.; Shore, R.F.; Long, S.M.; Griffin, J.L. Comparisons of metabolic and physiological changes in rats following short term oral dosing with pesticides commonly found in food. Food Chem. Toxicol. 2013, 59, 438–445. [Google Scholar] [CrossRef] [Green Version]
- Hu, C.; Zhang, Y.; Liu, G.; Liu, Y.; Wang, J.; Sun, B. Untargeted metabolite profiling of adipose tissue in hyperlipidemia rats exposed to hawthorn ethanol extracts. J. Food Sci. 2019, 84, 717–725. [Google Scholar] [CrossRef]
- Hu, C.; Li, R.; Wang, J.; Liu, Y.; Wang, J.; Sun, B. Untargeted metabolite profiling of liver in mice exposed to 2-methylfuran. J. Food Sci. 2021, 86, 242–250. [Google Scholar] [CrossRef]
- Rossmeislova, L.; Gojda, J.; Smolkova, K. Pancreatic cancer: Branched-chain amino acids as putative key metabolic regulators? Cancer Metastasis Rev. 2021, 40, 1115–1139. [Google Scholar] [CrossRef]
- Shou, J.; Chen, P.; Xiao, W. The effects of BCAAs on insulin resistance in athletes. J. Nutr. Sci. Vitaminol. 2019, 65, 383–389. [Google Scholar] [CrossRef] [Green Version]
- Zhenyukh, O.; Gonzalez-Amor, M.; Rodrigues-Diez, R.R.; Esteban, V.; Ruiz-Ortega, M.; Salaices, M.; Mas, S.; Briones, A.M.; Egido, J. Branched-chain amino acids promote endothelial dysfunction through increased reactive oxygen species generation and inflammation. J. Cell. Mol. Med. 2018, 22, 4948–4962. [Google Scholar] [CrossRef]
- Bispo, D.S.C.; Jesus, C.S.H.; Correia, M.; Ferreira, F.; Bonifazio, G.; Goodfellow, B.J.; Oliveira, M.B.; Mano, J.F.; Gil, A.M. NMR Metabolomics Assessment of Osteogenic Differentiation of Adipose-Tissue-Derived Mesenchymal Stem Cells. J. Proteome Res. 2022, 21, 654–670. [Google Scholar] [CrossRef]
- Parihar, R.; Shukla, R.; Baishya, B.; Kalita, J.; Haldar, R.; Misra, U.K. NMR based CSF metabolomics in tuberculous meningitis: Correlation with clinical and MRI findings. Metab. Brain Dis. 2022, 37, 773–785. [Google Scholar] [CrossRef]
- Altman, B.J.; Stine, Z.E.; Dang, C.V. From Krebs to clinic: Glutamine metabolism to cancer therapy. Nat. Rev. Cancer 2016, 16, 619–634. [Google Scholar] [CrossRef] [Green Version]
- McFadden, J.W.; Girard, C.L.; Tao, S.; Zhou, Z.; Bernard, J.K.; Duplessis, M.; White, H.M. Symposium review: One-carbon metabolism and methyl donor nutrition in the dairy cow. J. Dairy Sci. 2020, 103, 5668–5683. [Google Scholar] [CrossRef]
- Metcalf, J.S.; Dunlop, R.A.; Powell, J.T.; Banack, S.A.; Cox, P.A. L-Serine: A Naturally-Occurring Amino Acid with Therapeutic Potential. Neurotox. Res. 2018, 33, 213–221. [Google Scholar] [CrossRef] [PubMed]
- Ovrehus, M.A.; Bruheim, P.; Ju, W.; Zelnick, L.R.; Langlo, K.A.; Sharma, K.; de Boer, I.H.; Hallan, S.I. Gene Expression Studies and Targeted Metabolomics Reveal Disturbed Serine, Methionine, and Tyrosine Metabolism in Early Hypertensive Nephrosclerosis. Kidney Int. 2019, 4, 321–333. [Google Scholar]
- Imenshahidi, M.; Hossenzadeh, H. Effects of glycine on metabolic syndrome components: A review. J. Endocrinol. Investig. 2022, 45, 927–939. [Google Scholar] [CrossRef] [PubMed]
- Wang, T.; Li, S.; Wu, Y.; Yan, X.; Zhu, Y.; Jiang, Y.; Jiang, F.; Liu, W. Mechanistic Investigation of Xuebijing for Treatment of Paraquat-Induced Pulmonary Fibrosis by Metabolomics and Network Pharmacology. ACS Omega 2021, 6, 19717–19730. [Google Scholar] [CrossRef]
- Ye, J.; Lin, X.; Cheng, Z.; Su, Y.; Li, W.; Ali, F.; Zheng, J.; Peng, B. Identification and efficacy of glycine, serine and threonine metabolism in potentiating kanamycin-mediated killing of Edwardsiella piscicida. J. Proteom. 2018, 183, 34–44. [Google Scholar] [CrossRef]
- Sawicka, M.M.; Sawicki, K.; Lyson, T.; Politynska, B.; Miltyk, W. Proline Metabolism in Malignant Gliomas: A Systematic Literature Review. Cancers 2022, 14, 2030. [Google Scholar] [CrossRef]
- Xie, Y.; Feng, W.; Liu, M.; Xie, J.; Yu, L.; Gong, X.; Li, Y.; Peng, C. Investigation of Efficacy Enhancing and Toxicity Reducing Mechanism of Combination of Aconiti Lateralis Radix Praeparata and Paeoniae Radix Alba in Adjuvant-Induced Arthritis Rats by Metabolomics. Evid.-Based Complement. Alternat. Med. 2019, 2019, 9864841. [Google Scholar] [CrossRef] [Green Version]
- Dong, M.; Du, H.; Li, X.; Zhang, L.; Wang, X.; Wang, Z.; Jiang, H. Discovery of Biomarkers and Potential Mechanisms of Agarwood Incense Smoke Intervention by Untargeted Metabolomics and Network Pharmacology. Drug Des. Dev. Ther. 2022, 16, 265–278. [Google Scholar] [CrossRef]
- Liu, Y.; Mei, B.; Chen, D.; Cai, L. GC-MS metabolomics identifies novel biomarkers to distinguish tuberculosis pleural effusion from malignant pleural effusion. J. Clin. Lab. Anal. 2021, 35, e23706. [Google Scholar] [CrossRef]
- Guo, B.; Li, Y.; Huang, G.; Yu, L. Fatty acid esters of 3-monochloropropanediol: A review. Annu. Rev. Food Sci. Technol. 2019, 10, 259–284. [Google Scholar] [CrossRef]
- Stanimirova, I.; Banasik, M.; Zabek, A.; Dawiskiba, T.; Koscielska-Kasprzak, K.; Wojtowicz, W.; Krajewska, M.; Janczak, D.; Mlynarz, P. Serum metabolomics approach to monitor the changes in metabolite profiles following renal transplantation. Sci. Rep. 2020, 10, 17223. [Google Scholar] [CrossRef]
- Gill, S.S.; Kavanagh, M.; Cherry, W.; Barker, M.; Weld, M.; Cooke, G.M. A 28-day Gavage Toxicity Study in Male Fischer 344 Rats with 2-methylfuran. Toxicol. Pathol. 2014, 42, 352–360. [Google Scholar] [CrossRef] [Green Version]
- Deng, F.; Pan, J.; Zheng, P.; Xia, J.; Yin, B.; Liang, W.; Li, Y.; Wu, J.; Xu, F.; Wu, Q.; et al. Metabonomics reveals peripheral and central short-chain fatty acid and amino acid dysfunction in a naturally occurring depressive model of macaques. Neuropsychiatr. Dis. Treat. 2019, 15, 1077–1088. [Google Scholar] [CrossRef] [Green Version]
- Yan, X.; Li, L.; Liu, P.; Xu, J.; Wang, Z.; Ding, L.; Yang, L. Targeted metabolomics profiles serum fatty acids by HFD induced non-alcoholic fatty liver in mice based on GC-MS. J. Pharm. Biomed. Anal. 2022, 211, 114620. [Google Scholar] [CrossRef]
- Li, X.; Shen, Y.; Wu, G.; Qi, X.; Zhang, H.; Wang, L.; Qian, H. Determination of key active components in different edible oils affecting lipid accumulation and reactive oxygen species production in HepG2 cells. J. Agric. Food Chem. 2018, 66, 11943–11956. [Google Scholar] [CrossRef]
- Annie, M.A.S.; Prem, S.S.; Jayasuriya, R.; Ganesh, G.; Ramkumar, K.M.; Sarada, D.V.L. The pivotal role of Nrf2 activators in adipocyte biology. Pharmacol. Res. 2021, 173, 105853. [Google Scholar] [CrossRef]
- Li, Y.; Liu, J.; Zhou, H.; Liu, J.; Xue, X.; Wang, L.; Ren, S. Liquid chromatography-mass spectrometry method for discovering the metabolic markers to reveal the potential therapeutic effects of naringin on osteoporosis. J. Chromatogr. B Anal. Technol. Biomed. Life Sci. 2022, 1194, 123170. [Google Scholar] [CrossRef]
- Nguyen, H.T.; Li, L.; Eguchi, A.; Agusa, T.; Yamamoto, K.; Kannan, K.; Kim, E.Y.; Iwata, H. Effects of gestational exposure to bisphenol A on the hepatic transcriptome and lipidome of rat dams: Intergenerational comparison of effects in the offspring. Sci. Total Environ. 2022, 826, 153990. [Google Scholar] [CrossRef]
- Kakisaka, K.; Cazanave, S.C.; Fingas, C.D.; Guicciardi, M.E.; Bronk, S.F.; Werneburg, N.W.; Mott, J.L.; Gores, G.J. Mechanisms of lysophosphatidylcholine-induced hepatocyte lipoapoptosis. Am. J. Physiol. 2012, 302, G77–G84. [Google Scholar] [CrossRef] [Green Version]
- Banan, A.; Harvey, K.A.; Whitley, P.; Xu, Z.; Walker, C.; Pavlina, T.M.; Zaloga, G.P.; Siddiqui, R.A. Oleic acid prevents stearic acid-induced inhibition of cell growth and pro-inflammatory responses in human aortic endothelial cells. FASEB J. 2010, 24, 3470–3480. [Google Scholar] [CrossRef]
- Subczynski, W.K.; Pasenkiewicz, G.M.; Widomska, J.; Mainali, L.; Raguz, M. High Cholesterol/Low Cholesterol: Effects in Biological Membranes: A Review. Cell Biochem. Biophys. 2017, 75, 369–385. [Google Scholar] [CrossRef] [PubMed]
- Su, Z.; Ling, X.; Ji, K.; Huang, H.; Liu, X.; Yin, C.; Zhu, H.; Guo, Y.; Mo, Y.; Lu, Y.; et al. H-1 NMR-based urinary metabonomic study of the antidiabetic effects of Rubus Suavissimus, S. Lee in STZ-induced T1DM rats. J. Chromatogr. B Anal. Technol. Biomed. Life Sci. 2020, 1158, 122347. [Google Scholar] [CrossRef] [PubMed]
- Polderman, K.H.; van de Kraats, G.; Dixon, J.M.; Vandertop, W.P.; Girbes, A.R.J. Increases in spinal fluid osmolarity induced by mannitol. Crit. Care Med. 2003, 31, 584–590. [Google Scholar] [CrossRef]
- Al-Lahham, S.H.; Peppelenbosch, M.P.; Roelofsen, H.; Vonk, R.J.; Venema, K. Biological effects of propionic acid in humans; metabolism, potential applications and underlying mechanisms. Biochim. Biophys. Acta (BBA)-Mol. Cell Biol. Lipids 2010, 1801, 1175–1183. [Google Scholar] [CrossRef] [PubMed]
- Chen, R.; Liao, C.; Guo, Q.; Wu, L.; Zhang, L.; Wang, X. Combined systems pharmacology and fecal metabonomics to study the biomarkers and therapeutic mechanism of type 2 diabetic nephropathy treated with Astragalus and Leech. RSC Adv. 2018, 8, 27448–27463. [Google Scholar] [CrossRef] [Green Version]
- Al Mahri, S.; Malik, S.S.; Al Ibrahim, M.; Haji, E.; Dairi, G.; Mohammad, S. Free Fatty Acid Receptors (FFARs) in Adipose: Physiological Role and Therapeutic Outlook. Cells 2022, 11, 750. [Google Scholar] [CrossRef]
- Alasmari, F.; Assiri, M.A.; Ahamad, S.R.; Aljumayi, S.R.; Alotaibi, W.H.; Alhamdan, M.M.; Alhazzani, K.; Alharbi, M.; Alqahtani, F.; Alasmari, A.F. Serum Metabolomic Analysis of Male Patients with Cannabis or Amphetamine Use Disorder. Metabolites 2022, 12, 179. [Google Scholar] [CrossRef]
- Wen, C.; Wang, Z.; Zhang, M.; Wang, S.; Geng, P.; Sun, F.; Chen, M.; Lin, G.; Hu, L.; Ma, J.; et al. Metabolic changes in rat urine after acute paraquat poisoning and discriminated by support vector machine. Biomed. Chromatogr. 2016, 30, 75–80. [Google Scholar] [CrossRef]
- Dybing, E.; Soderlund, E.J.; Gordon, W.P.; Holme, J.A.; Christensen, T.; Becher, G.; Rivedal, E.; Thorgeirsson, S.S. Studies on the mechanism of acetamide hepatocarcinogenicity. Pharmacol. Toxicol. 1987, 60, 9–16. [Google Scholar] [CrossRef]

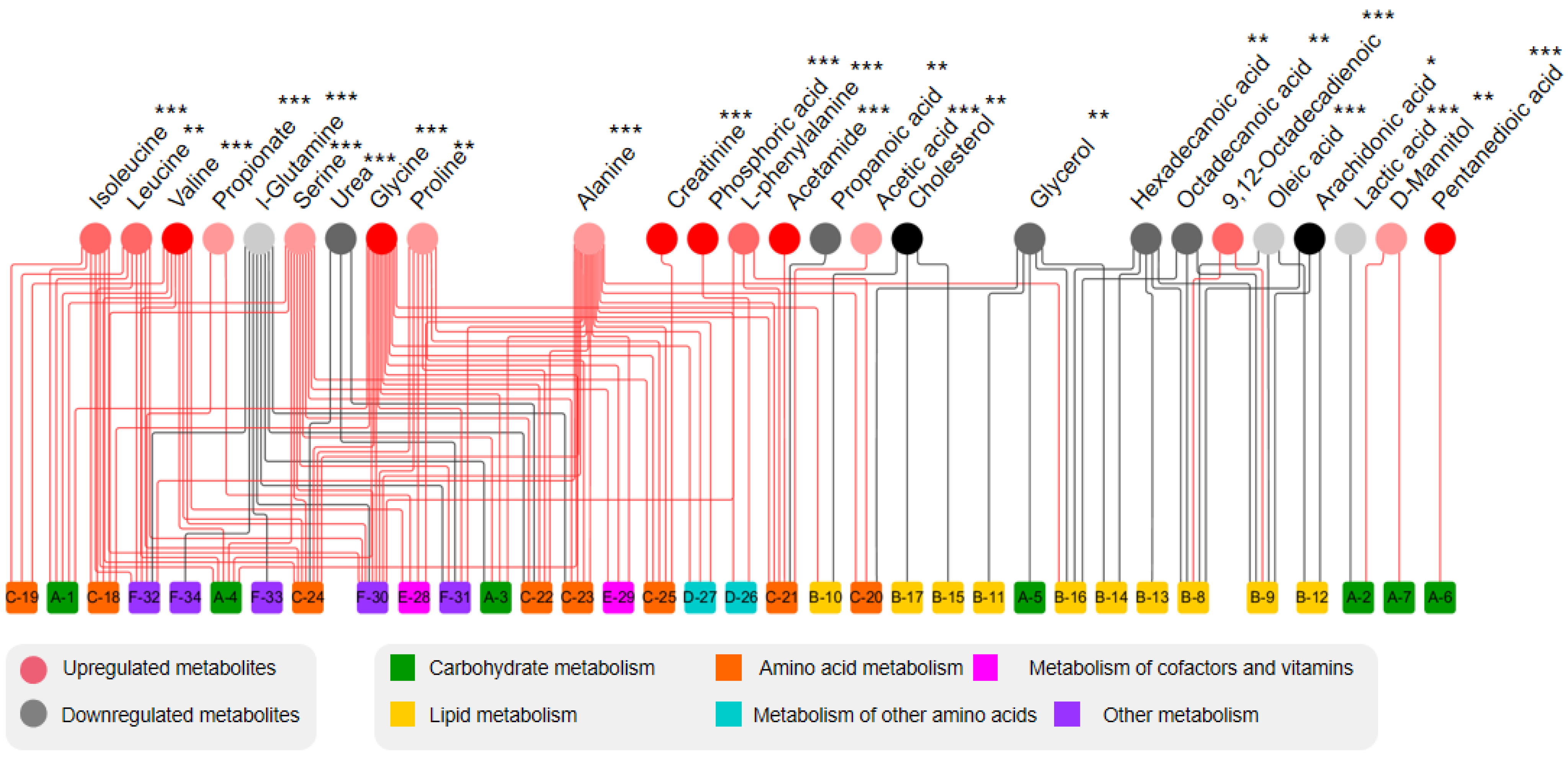
| Metabolite | RT (min) | p-Value | VIP | Fold Change (HD-ND) | FDR | Trend | Metabolic Pathways |
|---|---|---|---|---|---|---|---|
| Lactic acid | 5.30 | 1.90 × 10−5 | 2.13 | 0.25 | 3.53 × 10−5 | ↓ | Glycolysis/Gluconeogenesis |
| Propanoic acid | 8.94 | 7.00 × 10−3 | 3.31 | 0.69 | 7.28 × 10−3 | ↓ | Propionic acid metabolism |
| Alanine | 10.03 | 1.30 × 10−5 | 1.11 | 1.72 | 2.60 × 10−5 | ↑ | Alanine, aspartate, and glutamate metabolism |
| Glycine | 10.51 | 9.80 × 10−9 | 1.07 | 3.28 | 3.64 × 10−8 | ↑ | Glycine, serine, and threonine metabolism |
| Valine | 13.16 | 4.00 × 10−9 | 1.03 | 3.48 | 1.73 × 10−8 | ↑ | Valine, leucine and isoleucine biosynthesis |
| Urea | 14.19 | 1.20 × 10−8 | 1.99 | 0.50 | 3.90 × 10−8 | ↓ | Urea cycle |
| Leucine | 14.71 | 1.50 × 10−4 | 1.90 | 2.05 | 2.17 × 10−4 | ↑ | Valine, leucine and isoleucine biosynthesis |
| Glycerol | 14.9 | 3.00 × 10−3 | 2.29 | 0.67 | 3.39 × 10−3 | ↓ | Glycerolipid metabolism |
| Isoleucine | 15.28 | 3.50 × 10−9 | 1.19 | 2.15 | 1.82 × 10−8 | ↑ | Valine, leucine and isoleucine biosynthesis |
| Serine | 17.16 | 5.40 × 10−5 | 1.12 | 1.52 | 8.78 × 10−5 | ↑ | Glycine, serine, and threonine metabolism |
| Propionate | 18.88 | 1.70 × 10−9 | 1.05 | 1.69 | 1.11 × 10−8 | ↑ | Pantothenate and CoA biosynthesis |
| Acetic acid | 20.70 | 1.70 × 10−9 | 1.04 | 1.58 | 2.74 × 10−4 | ↑ | TCA cycle |
| Proline | 21.07 | 1.40 × 10−4 | 1.70 | 1.52 | 2.14 × 10−4 | ↑ | Arginine and proline metabolism |
| Pentanedioic acid | 21.30 | 3.60 × 10−10 | 1.67 | 3.64 | 4.68 × 10−9 | ↑ | Pentose and glucuronate interconversions |
| Creatinine | 22.87 | 3.70 × 10−8 | 1.11 | 3.15 | 9.62 × 10−8 | ↑ | Arginine and proline metabolism |
| Phenylalanine | 23.49 | 1.10 × 10−9 | 1.16 | 2.44 | 9.53 × 10−9 | ↑ | Phenylalanine, tyrosine, and tryptophan biosynthesis |
| Acetamide | 26.14 | 2.30 × 10−10 | 1.82 | 8.90 | 5.98 × 10−9 | ↑ | Phenylalanine metabolism |
| Phosphoric acid | 26.82 | 3.30 × 10−6 | 3.27 | 3.29 | 7.15 × 10−6 | ↑ | Propionic acid metabolism |
| Glutamine | 27.14 | 2.30 × 10−8 | 1.01 | 0.30 | 6.64 × 10−8 | ↓ | Alanine, aspartate, and glutamate metabolism |
| D-Mannitol | 30.44 | 2.70 × 10−4 | 1.16 | 1.84 | 3.51 × 10−4 | ↑ | Fructose and mannose metabolism |
| Hexadecanoic acid | 32.01 | 7.40 × 10−4 | 4.06 | 0.61 | 8.75 × 10−4 | ↓ | Fatty acid biosynthesis |
| 9,12-Octadecadienoic acid | 35.16 | 9.70 × 10−8 | 3.28 | 2.20 | 2.29 × 10−7 | ↑ | Fatty acid biosynthesis |
| Octadecanoic acid | 35.48 | 3.30 × 10−4 | 1.79 | 0.62 | 4.09 × 10−4 | ↓ | Fatty acid biosynthesis |
| Arachidonic acid | 39.52 | 1.20 × 10−2 | 1.31 | 0.77 | 1.20 × 10−2 | ↓ | Arachidonic acid metabolism |
| Oleic acid | 41.94 | 2.80 × 10−5 | 1.93 | 0.46 | 4.85 × 10−5 | ↓ | Fatty acid biosynthesis |
| Cholesterol | 44.48 | 5.00 × 10−3 | 1.12 | 0.71 | 5.42 × 10−3 | ↓ | Steroid hormone biosynthesis |
| Total | Expected | Hits | Raw p | −log(p) | Holm p | FDR | |
|---|---|---|---|---|---|---|---|
| Glyoxylate and dicarboxylate metabolism | 32 | 0.51613 | 3 | 0.013508 | 1.8694 | 1 | 0.28367 |
| Phenylalanine, tyrosine, and tryptophan biosynthesis | 4 | 0.064516 | 1 | 0.063032 | 1.2004 | 1 | 0.59703 |
| Alanine, aspartate, and glutamate metabolism | 28 | 0.45161 | 2 | 0.073113 | 1.136 | 1 | 0.59703 |
| Linoleic acid metabolism | 5 | 0.080645 | 1 | 0.078183 | 1.1069 | 1 | 0.59703 |
| Phenylalanine metabolism | 10 | 0.16129 | 1 | 0.15048 | 0.82252 | 1 | 0.90288 |
| Glycerolipid metabolism | 16 | 0.25806 | 1 | 0.23006 | 0.63815 | 1 | 1 |
| Glycine, serine, and threonine metabolism | 33 | 0.53226 | 1 | 0.41857 | 0.37823 | 1 | 1 |
| Arachidonic acid metabolism | 36 | 0.58065 | 1 | 0.44686 | 0.34983 | 1 | 1 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Hu, C.; Song, X.; Shao, Z.; Liu, Y.; Wang, J.; Sun, B. Untargeted Metabolite Profiling of Adipose Tissue in Rats Exposed to Mepiquat. Foods 2023, 12, 867. https://doi.org/10.3390/foods12040867
Hu C, Song X, Shao Z, Liu Y, Wang J, Sun B. Untargeted Metabolite Profiling of Adipose Tissue in Rats Exposed to Mepiquat. Foods. 2023; 12(4):867. https://doi.org/10.3390/foods12040867
Chicago/Turabian StyleHu, Chuanqin, Xinyu Song, Zhenzhen Shao, Yingli Liu, Jing Wang, and Baoguo Sun. 2023. "Untargeted Metabolite Profiling of Adipose Tissue in Rats Exposed to Mepiquat" Foods 12, no. 4: 867. https://doi.org/10.3390/foods12040867
APA StyleHu, C., Song, X., Shao, Z., Liu, Y., Wang, J., & Sun, B. (2023). Untargeted Metabolite Profiling of Adipose Tissue in Rats Exposed to Mepiquat. Foods, 12(4), 867. https://doi.org/10.3390/foods12040867







