Genetic and Probiotic Characteristics of Urolithin A Producing Enterococcus faecium FUA027
Abstract
1. Introduction
2. Materials and Methods
2.1. Bacterial Strain and Growth Conditions
2.2. Whole-Genome Sequencing
2.3. Genome Assembly and Annotation
2.4. Strain Safety Assessment
2.4.1. Identifying Safety-Related Genes from the FUA027 Genome
2.4.2. Antibiotic Susceptibility Testing
2.4.3. Hemolytic Activity Evaluation
2.4.4. Nitrate Reductase and Amino Acid Decarboxylase Activity
2.5. Assessment of Probiotic Properties
2.5.1. Probiotic-Associated Genes in the E. faecium FUA027 Genome
2.5.2. Evaluation of Acid and Bile Salt Tolerance In Vitro
2.5.3. Evaluation of the Antioxidant Activity In Vitro
2.5.4. Hydrophobicity and Auto-Aggregation Tests
2.5.5. Evaluation of Antibacterial Activity
3. Results and Discussion
3.1. Genome Properties
3.2. Evaluation Safety of E. faecium FUA027
3.2.1. Identification of Antibiotic Resistance Gene
3.2.2. Evaluation of Virulence Factor Genes and Toxin-Encoding Genes
3.2.3. Biogenic Amine Production
3.3. Assessment of Probiotic Properties
3.3.1. Acid and Bile Salt Tolerance In Vitro
3.3.2. Antioxidant Ability In Vitro
3.3.3. Evaluation of Adhesion-Related Genes
3.3.4. Antibacterial Test of E. faecium FUA027 against Quality Control Strains
4. Conclusions
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
Appendix A
| Antibiotic | Standard for Judging Diameter of Inhibition Zone Diam (mm) | Drug Contents (µg) | Zone Diam (mm) | Antibacterial Effect | ||
|---|---|---|---|---|---|---|
| R | I | S | ||||
| Amikacin | ≤14 | 15~16 | ≥17 | 30 | 10 ± 0.2 | R |
| Norfloxacin | ≤12 | 13~16 | ≥17 | 10 | 19 ± 0.3 | S |
| Ofloxacin | ≤12 | 13~15 | ≥16 | 5 | 18 ± 0.4 | S |
| Ciprofloxacin | ≤15 | 16~20 | ≥21 | 5 | 15 ± 0.1 | I |
| Levofloxacin | ≤12 | 13~16 | ≥17 | 5 | 25 ± 0.3 | S |
| Erythromycin | ≤13 | 14~22 | ≥23 | 15 | 22 ± 0.1 | I |
| Tetracycline | ≤14 | 15~18 | ≥19 | 30 | 21 ± 0.2 | S |
| Cefuroxime | ≤14 | 15~17 | ≥18 | 30 | 30 ± 0.2 | S |
| Cefazolin | ≤14 | - | ≥15 | 30 | 22 ± 0.3 | S |
| Cephalothin | ≤14 | 15~17 | ≥18 | 30 | 27 ± 0.4 | S |
| Cefotaxime | ≤22 | 23~25 | ≥26 | 30 | 30 ± 0.1 | S |
| Cefatriaxone | ≤13 | 14~20 | ≥21 | 30 | 24 ± 0.4 | S |
| Ceftazidime | ≤14 | 15~17 | ≥18 | 30 | 18 ± 0.3 | S |
| Piperacillin | ≤28 | - | ≥29 | 100 | 21 ± 0.3 | S |
| Ampicillin | ≤16 | 18~24 | ≥25 | 10 | 22 ± 0.2 | S |
| Oxacillin | ≤17 | - | ≥25 | 1 | 14 ± 0.4 | R |
| Penicillin G | ≤28 | - | ≥29 | 10 | 22 ± 0.6 | S |
| Aztreonam | ≤15 | 16~21 | ≥22 | 30 | 0 | R |
| Compound sulfamethoxazole | ≤10 | 11~15 | ≥16 | 23.75 | 0 | R |
| Nitrofurantoin | ≤14 | 15~16 | ≥17 | 300 | 15 ± 0.6 | I |
| Chloramphenicol | ≤12 | 13~17 | ≥18 | 30 | 19 ± 0.3 | S |
| Polymyxin B | ≤11 | 12~14 | ≥15 | 300 | 0 | R |
| Clindamycin | ≤13 | 14~17 | ≥18 | 2 | 8 ± 0.4 | R |
| Kanamycin | ≤12 | 13~14 | ≥15 | 30 | 6 ± 0.4 | R |
| Gentamicin | ≤12 | 13~14 | ≥15 | 10 | 9 ± 0.6 | R |
| Streptomycin | ≤11 | 12~14 | ≥15 | 10 | 7 ± 0.7 | R |
| Vancomycin | ≤14 | 15~16 | ≥17 | 30 | 19 ± 0.3 | S |
| Type of Stress Response | Protein | Gene Locus |
|---|---|---|
| Acid stress response | F0F1-ATPase | GM_000855, GM_000856 |
| GM_000857, GM_000858 | ||
| GM_000859, GM_000860 | ||
| GM_000861, GM_000862 | ||
| Na+/H+ antiporter | GM_000704, GM_001361 | |
| Bile salts stress response | Conjugated bile acid hydrolase | GM_001069 |
| ABC transporter | GM_002271, GM_002452 | |
| GM_000767, GM_002209 | ||
| GM_002135, GM_002210 | ||
| GM_002486, GM_002485 | ||
| GM_002270, GM_000917 | ||
| GM_000916, GM_002136 | ||
| GM_001223, GM_001224 | ||
| Temperature stress response | Cold shock protein | GM_001318 |
| Metal stress response | Divalent metal cation transporter | GM_000913 |
| Cadmium, zinc and cobalt-transporting ATPase | GM_000987 | |
| Potassium/sodium uptake protein | GM_000822 | |
| Oxidative stress response | Alkyl hydroperoxide reductase | GM_001365, GM_001366 |
| Glutathione peroxidase | GM_000501 | |
| Thioredoxin reductases | GM_000926 | |
| Glutathione reductase | GM_002670 | |
| Superoxide dismutase | GM_001908 | |
| Peroxide-responsive repressor | GM_001511 | |
| NADH peroxidase | GM_000518 | |
| Superoxide dismutase | GM_001908 |
References
- García-Villalba, R.; Beltrán, D.; Espín, J.C.; Selma, M.V.; Tomás-Barberán, F.A. Time Course Production of Urolithins from Ellagic Acid by Human Gut Microbiota. J. Agric. Food Chem. 2013, 61, 8797–8806. [Google Scholar] [CrossRef] [PubMed]
- Dao, T.; Green, A.E.; Kim, Y.A.; Bae, S.-J.; Ha, K.-T.; Gariani, K.; Lee, M.-R.; Menzies, K.J.; Ryu, D. Sarcopenia and Muscle Aging: A Brief Overview. Endocrinol. Metab. 2020, 35, 716–732. [Google Scholar] [CrossRef] [PubMed]
- Selma, M.V.; Romo-Vaquero, M.; García-Villalba, R.; González-Sarrías, A.; Tomás-Barberán, F.A.; Espín, J.C. The human gut microbial ecology associated with overweight and obesity determines ellagic acid metabolism. Food Funct. 2016, 7, 1769–1774. [Google Scholar] [CrossRef]
- Alfei, S.; Marengo, B.; Zuccari, G. Oxidative Stress, Antioxidant Capabilities, and Bioavailability: Ellagic Acid or Urolithins? Antioxidants 2020, 9, 707. [Google Scholar] [CrossRef] [PubMed]
- Al-Harbi, S.A.; Abdulrahman, A.O.; Zamzami, M.A.; Khan, M.I. Urolithins: The Gut Based Polyphenol Metabolites of Ellagitannins in Cancer Prevention, a Review. Front. Nutr. 2021, 8, 647582. [Google Scholar] [CrossRef]
- García-Villalba, R.; Giménez-Bastida, J.A.; Cortés-Martín, A.; Ávila-Gálvez, M.; Tomás-Barberán, F.A.; Selma, M.V.; Espín, J.C.; González-Sarrías, A. Urolithins: A Comprehensive Update on their Metabolism, Bioactivity, and Associated Gut Microbiota. Mol. Nutr. Food Res. 2022, 66, 2101019. [Google Scholar] [CrossRef] [PubMed]
- Xian, W.; Yang, S.; Deng, Y.; Yang, Y.; Chen, C.; Li, W.; Yang, R. Distribution of Urolithins Metabotypes in Healthy Chinese Youth: Difference in Gut Microbiota and Predicted Metabolic Pathways. J. Agric. Food Chem. 2021, 69, 13055–13065. [Google Scholar] [CrossRef]
- D’Amico, D.; Andreux, P.A.; Valdés, P.; Singh, A.; Rinsch, C.; Auwerx, J. Impact of the Natural Compound Urolithin A on Health, Disease, and Aging. Trends Mol. Med. 2021, 27, 687–699. [Google Scholar] [CrossRef]
- Cortés-Martín, A.; García-Villalba, R.; González-Sarrías, A.; Romo-Vaquero, M.; Loria-Kohen, V.; Ramírez-De-Molina, A.; Tomás-Barberán, F.A.; Selma, M.V.; Espín, J.C. The gut microbiota urolithin metabotypes revisited: The human metabolism of ellagic acid is mainly determined by aging. Food Funct. 2018, 9, 4100–4106. [Google Scholar] [CrossRef]
- Kang, I.; Buckner, T.; Shay, N.F.; Gu, L.; Chung, S. Improvements in Metabolic Health with Consumption of Ellagic Acid and Subsequent Conversion into Urolithins: Evidence and Mechanisms. Adv. Nutr. 2016, 7, 961–972. [Google Scholar] [CrossRef]
- Gaya, P.; Peirotén, Á; Medina, M.; Alvaréz, I.; Landete, J.M. Bifidobacterium pseudocatenulatum INIA P815: The first bacterium able to produce urolithins a and b from ellagic acid. J. Funct. Foods 2018, 45, 95–99. [Google Scholar]
- Liu, Q.; Liu, S.; Ye, Q.; Hou, X.; Yang, G.; Lu, J.; Hai, Y.; Shen, J.; Fang, Y. A Novel Streptococcus thermophilus FUA329 Isolated from Human Breast Milk Capable of Producing Urolithin A from Ellagic Acid. Foods 2022, 11, 3280. [Google Scholar] [CrossRef]
- Mi, H.; Liu, S.; Hai, Y.; Yang, G.; Lu, J.; He, F.; Zhao, Y.; Xia, M.; Hou, X.; Fang, Y. Lactococcus garvieae FUA009, a Novel Intestinal Bacterium Capable of Producing the Bioactive Metabolite Urolithin A from Ellagic Acid. Foods 2022, 11, 2621. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.; Fang, Y.; Yang, G.; Hou, X.; Hai, Y.; Xia, M.; He, F.; Zhao, Y.; Liu, S. Isolation and characterization of a novel human intestinal Enterococcus faecium FUA027 capable of producing urolithin A from ellagic acid. Front. Nutr. 2022, 9, 1039697. [Google Scholar] [CrossRef] [PubMed]
- Koirala, S.; Anal, A.K. Probiotics-based foods and beverages as future foods and their overall safety and regulatory claims. Future Foods 2021, 3, 100013. [Google Scholar] [CrossRef]
- Zhang, C.; Ma, K.; Nie, K.; Deng, M.; Luo, W.; Wu, X.; Huang, Y.; Wang, X. Assessment of the safety and probiotic properties of Roseburia intestinalis: A potential “Next Generation Probiotic”. Front. Microbiol. 2022, 13, 973046. [Google Scholar] [CrossRef]
- Tatusova, T.; DiCuccio, M.; Badretdin, A.; Chetvernin, V.; Nawrocki, E.P.; Zaslavsky, L.; Lomsadze, A.; Pruitt, K.D.; Borodovsky, M.; Ostell, J. NCBI prokaryotic genome annotation pipeline. Nucleic Acids Res. 2016, 44, 6614–6624. [Google Scholar] [CrossRef] [PubMed]
- Liu, B.; Zheng, D.; Zhou, S.; Chen, L.; Yang, J. VFDB 2022: A general classification scheme for bacterial virulence factors. Nucleic Acids Res. 2022, 50, D912–D917. [Google Scholar] [CrossRef]
- Zankari, E.; Hasman, H.; Cosentino, S.; Vestergaard, M.; Rasmussen, S.; Lund, O.; Aarestrup, F.M.; Larsen, M.V. Identification of acquired antimicrobial resistance genes. J. Antimicrob. Chemother. 2012, 67, 2640–2644. [Google Scholar] [CrossRef]
- Alcock, B.P.; Raphenya, A.R.; Lau, T.T.Y.; Tsang, K.K.; Bouchard, M.; Edalatmand, A.; Huynh, W.; Nguyen, A.-L.V.; Cheng, A.A.; Liu, S.; et al. CARD 2020: Antibiotic resistome surveillance with the comprehensive antibiotic resistance database. Nucleic Acids Res. 2020, 48, D517–D525. [Google Scholar] [CrossRef]
- Hombach, M.; Maurer, F.; Pfiffner, T.; Böttger, E.C.; Furrer, R. Standardization of Operator-Dependent Variables Affecting Precision and Accuracy of the Disk Diffusion Method for Antibiotic Susceptibility Testing. J. Clin. Microbiol. 2015, 53, 3864–3869. [Google Scholar] [CrossRef] [PubMed]
- Kim, Y.; Choi, S.I.; Jeong, Y.; Kang, C.H. Evaluation of Safety and Probiotic Potential of Enterococcus faecalis MG5206 and Enterococcus faecium MG5232 Isolated from Kimchi, a Korean Fermented Cabbage. Microorganisms 2022, 10, 2070. [Google Scholar] [CrossRef] [PubMed]
- De Aguero, N.L.; Frizzo, L.S.; Ouwehand, A.C.; Aleu, G.; Rosmini, M.R. Technological Characterisation of Probiotic Lactic Acid Bacteria as Starter Cultures for Dry Fermented Sausages. Foods 2020, 9, 596. [Google Scholar] [CrossRef] [PubMed]
- Eddy, S.R. Accelerated Profile HMM Searches. PLoS Comput. Biol. 2011, 7, e1002195. [Google Scholar] [CrossRef] [PubMed]
- Wu, Y.-P.; Liu, D.-M.; Zhao, S.; Huang, Y.-Y.; Yu, J.-J.; Zhou, Q.-Y. Assessing the safety and probiotic characteristics of Bacillus coagulans 13002 based on complete genome and phenotype analysis. LWT 2022, 155, 112847. [Google Scholar] [CrossRef]
- Blin, K.; Shaw, S.; Steinke, K.; Villebro, R.; Ziemert, N.; Lee, S.Y.; Medema, M.H.; Weber, T. antiSMASH 5.0: Updates to the secondary metabolite genome mining pipeline. Nucleic Acids Res. 2019, 47, W81–W87. [Google Scholar] [CrossRef] [PubMed]
- Pieniz, S.; Andreazza, R.; Anghinoni, T.; Camargo, F.; Brandelli, A. Probiotic potential, antimicrobial and antioxidant activities of Enterococcus durans strain LAB18s. Food Control 2014, 37, 251–256. [Google Scholar] [CrossRef]
- Wu, Y.; Li, S.; Tao, Y.; Li, D.; Han, Y.; Show, P.L.; Wen, G.; Zhou, J. Fermentation of blueberry and blackberry juices using Lactobacillus plantarum, Streptococcus thermophilus and Bifidobacterium bifidum: Growth of probiotics, metabolism of phenolics, antioxidant capacity in vitro and sensory evaluation. Food Chem. 2021, 348, 129083. [Google Scholar] [CrossRef]
- Mu, G.; Gao, Y.; Tuo, Y.; Li, H.; Zhang, Y.; Qian, F.; Jiang, S. Assessing and comparing antioxidant activities of lactobacilli strains by using different chemical and cellular antioxidant methods. J. Dairy Sci. 2018, 101, 10792–10806. [Google Scholar] [CrossRef]
- Azat, R.; Liu, Y.; Li, W.; Kayir, A.; Lin, D.-B.; Zhou, W.-W.; Zheng, X.-D. Probiotic properties of lactic acid bacteria isolated from traditionally fermented Xinjiang cheese. J. Zhejiang Univ. Sci. B 2016, 17, 597–609. [Google Scholar] [CrossRef]
- Maragkoudakis, P.A.; Zoumpopoulou, G.; Miaris, C.; Kalantzopoulos, G.; Pot, B.; Tsakalidou, E. Probiotic potential of Lactobacillus strains isolated from dairy products. Int. Dairy J. 2006, 16, 189–199. [Google Scholar] [CrossRef]
- Humphries, R.; Bobenchik, A.M.; Hindler, J.A.; Schuetz, A.N. Overview of Changes to the Clinical and Laboratory Standards Institute Performance Standards for Antimicrobial Susceptibility Testing, M100, 31st Edition. J. Clin. Microbiol. 2021, 59, e00213-21. [Google Scholar] [CrossRef] [PubMed]
- Klare, I.; Konstabel, C.; Badstubner, D.; Werner, G.; Witte, W. Occurrence and spread of antibiotic resistances in Enterococcus faecium. Int. J. Food Microbiol. 2003, 88, 269–290. [Google Scholar] [CrossRef] [PubMed]
- Natarajan, P.; Parani, M. First Complete Genome Sequence of a Probiotic Enterococcus faecium Strain T-110 and Its Comparative Genome Analysis with Pathogenic and Non-pathogenic Enterococcus faecium Genomes. J. Genet. Genom. 2015, 42, 43–46. [Google Scholar] [CrossRef]
- Graham, K.; Stack, H.; Rea, R. Safety, beneficial and technological properties of enterococci for use in functional food applications—A review. Crit. Rev. Food Sci. Nutr. 2020, 60, 3836–3861. [Google Scholar] [CrossRef] [PubMed]
- Terai, T.; Kato, K.; Ishikawa, E.; Nakao, M.; Ito, M.; Miyazaki, K.; Kushiro, A.; Imai, S.; Nomura, Y.; Hanada, N.; et al. Safety assessment of the candidate oral probiotic Lactobacillus crispatus YIT 12319: Analysis of antibiotic resistance and virulence-associated genes. Food Chem. Toxicol. 2020, 140, 111278. [Google Scholar] [CrossRef]
- Linares, D.M.; Martín, M.; Ladero, V.; Alvarez, M.A.; Fernández, M. Biogenic Amines in Dairy Products. Crit. Rev. Food Sci. Nutr. 2011, 51, 691–703. [Google Scholar] [CrossRef]
- Sarantinopoulos, P.; Andrighetto, C.; Georgalaki, M.D.; Rea, M.; Lombardi, A.; Cogan, T.M.; Kalantzopoulos, G.; Tsakalidou, E. Biochemical properties of enterococci relevant to their technological performance. Int. Dairy J. 2001, 11, 621–647. [Google Scholar] [CrossRef]
- Ladero, V.; Fernandez, M.; Calles-Enriquez, M.; Sanchez-Llana, E.; Canedo, E.; Martin, M.C.; Alvarez, M.A. Is the production of the biogenic amines tyramine and putrescine a species-level trait in enterococci? Food Microbiol. 2012, 30, 132–138. [Google Scholar] [CrossRef]
- Wang, J.; Da, R.; Tuo, X.; Cheng, Y.; Wei, J.; Jiang, K.; Lv, J.; Adediji, O.M.; Han, B. Probiotic and Safety Properties Screening of Enterococcus faecalis from Healthy Chinese Infants. Probiotics Antimicrob. Proteins 2020, 12, 1115–1125. [Google Scholar] [CrossRef]
- Succi, M.; Tremonte, P.; Reale, A.; Sorrentino, E.; Grazia, L.; Pacifico, S.; Coppola, R. Bile salt and acid tolerance of Lactobacillus rhamnosus strains isolated from Parmigiano Reggiano cheese. FEMS Microbiol. Lett. 2005, 244, 129–137. [Google Scholar] [CrossRef]
- Zago, M.; Fornasari, M.E.; Carminati, D.; Burns, P.; Suàrez, V.; Vinderola, G.; Reinheimer, J.; Giraffa, G. Characterization and probiotic potential of Lactobacillus plantarum strains isolated from cheeses. Food Microbiol. 2011, 28, 1033–1040. [Google Scholar] [CrossRef]
- Rahmeh, R.; Akbar, A.; Alonaizi, T.; Kishk, M.; Shajan, A.; Akbar, B. Characterization and application of antimicrobials produced by Enterococcus faecium S6 isolated from raw camel milk. J. Dairy Sci. 2020, 103, 11106–11115. [Google Scholar] [CrossRef] [PubMed]
- Valenzuela, A.S.; Benomar, N.; Abriouel, H.; Cañamero, M.M.; Gálvez, A. Isolation and identification of Enterococcus faecium from seafoods: Antimicrobial resistance and production of bacteriocin-like substances. Food Microbiol. 2010, 27, 955–961. [Google Scholar] [CrossRef] [PubMed]
- Basanta, A.; Sánchez, J.; Gómez-Sala, B.; Herranz, C.; Hernández, P.E.; Cintas, L.M. Antimicrobial activity of Enterococcus faecium L50, a strain producing enterocins L50 (L50A and L50B), P and Q, against beer-spoilage lactic acid bacteria in broth, wort (hopped and unhopped), and alcoholic and non-alcoholic lager beers. Int. J. Food Microbiol. 2008, 125, 293–307. [Google Scholar] [CrossRef] [PubMed]
- Lee, H.; van der Donk, W. Macrocyclization and Backbone Modification in RiPP Biosynthesis. Annu. Rev. Biochem. 2022, 91, 269–294. [Google Scholar] [CrossRef] [PubMed]

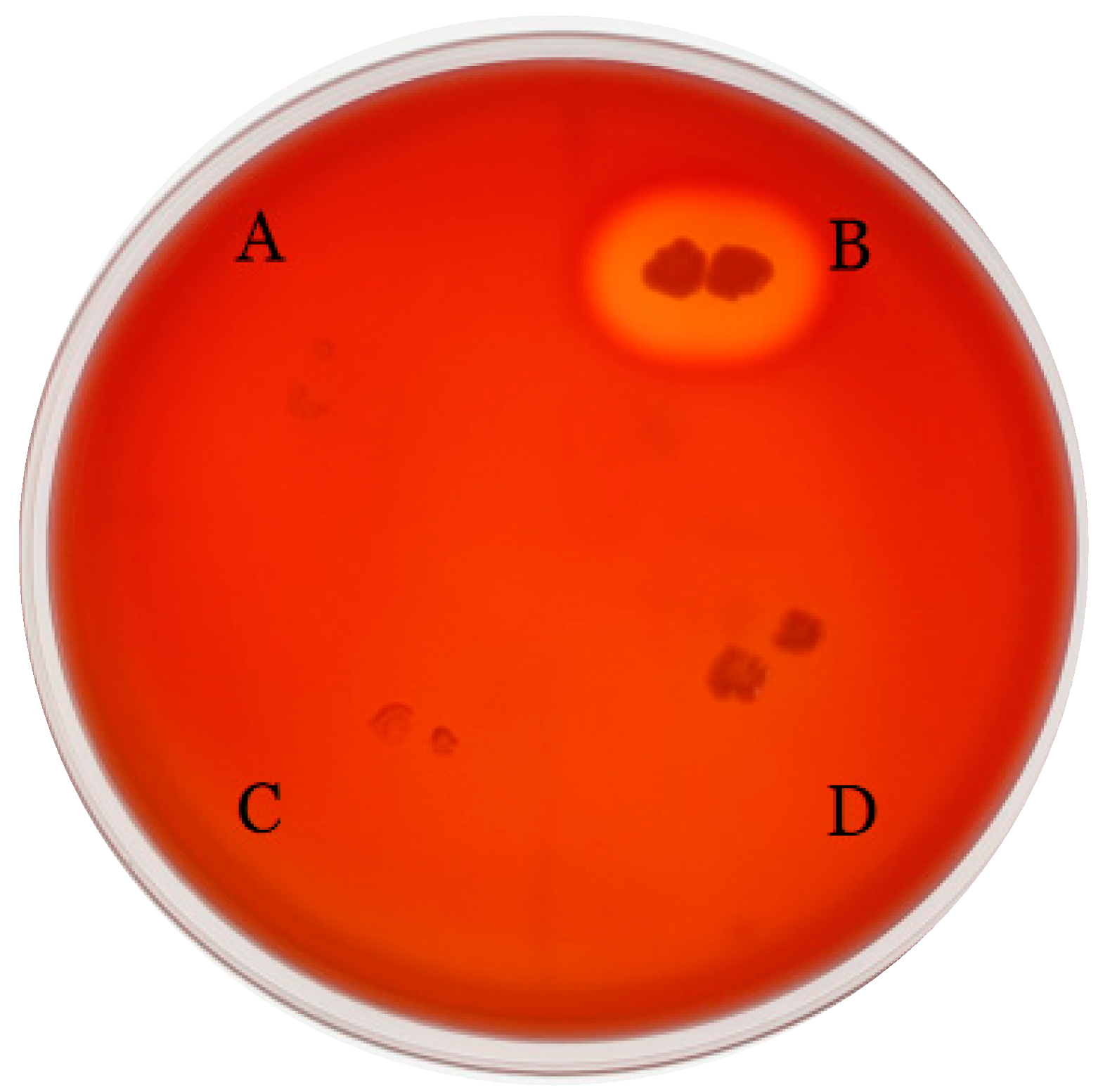
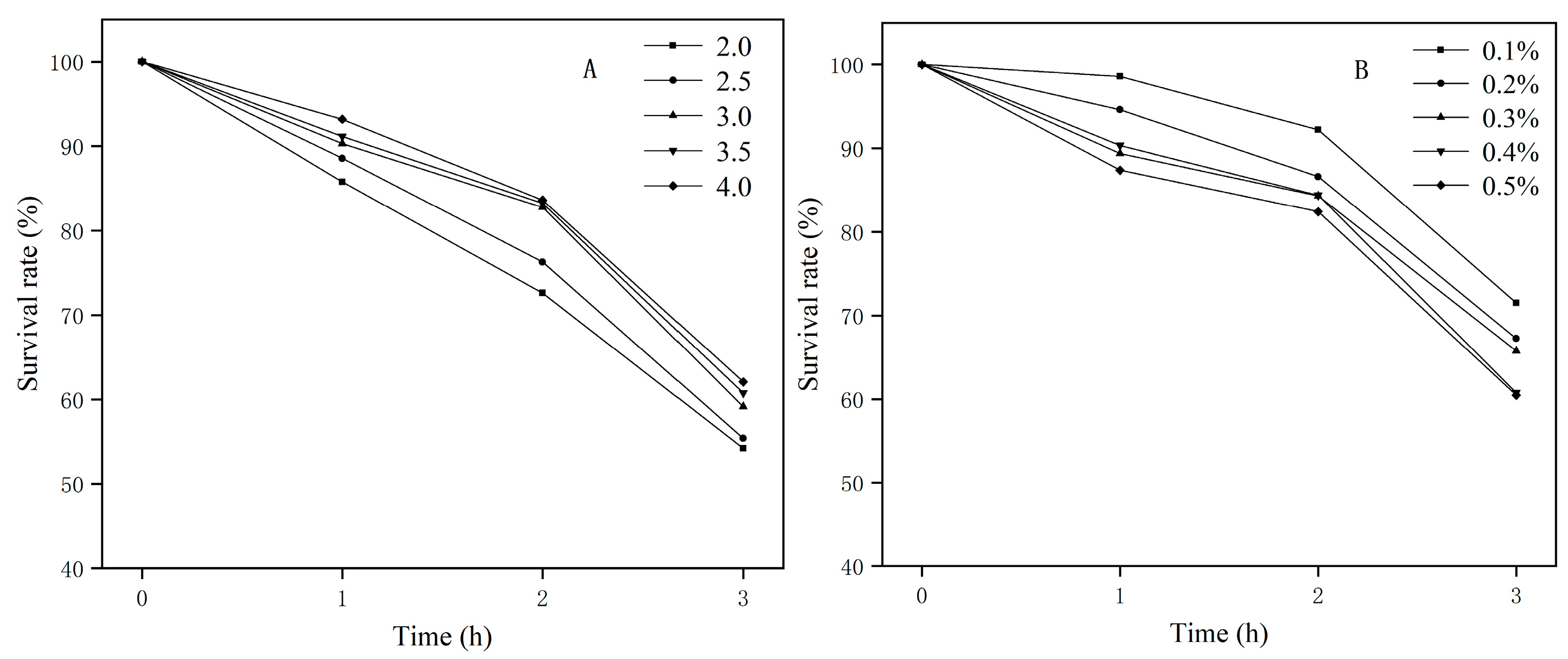
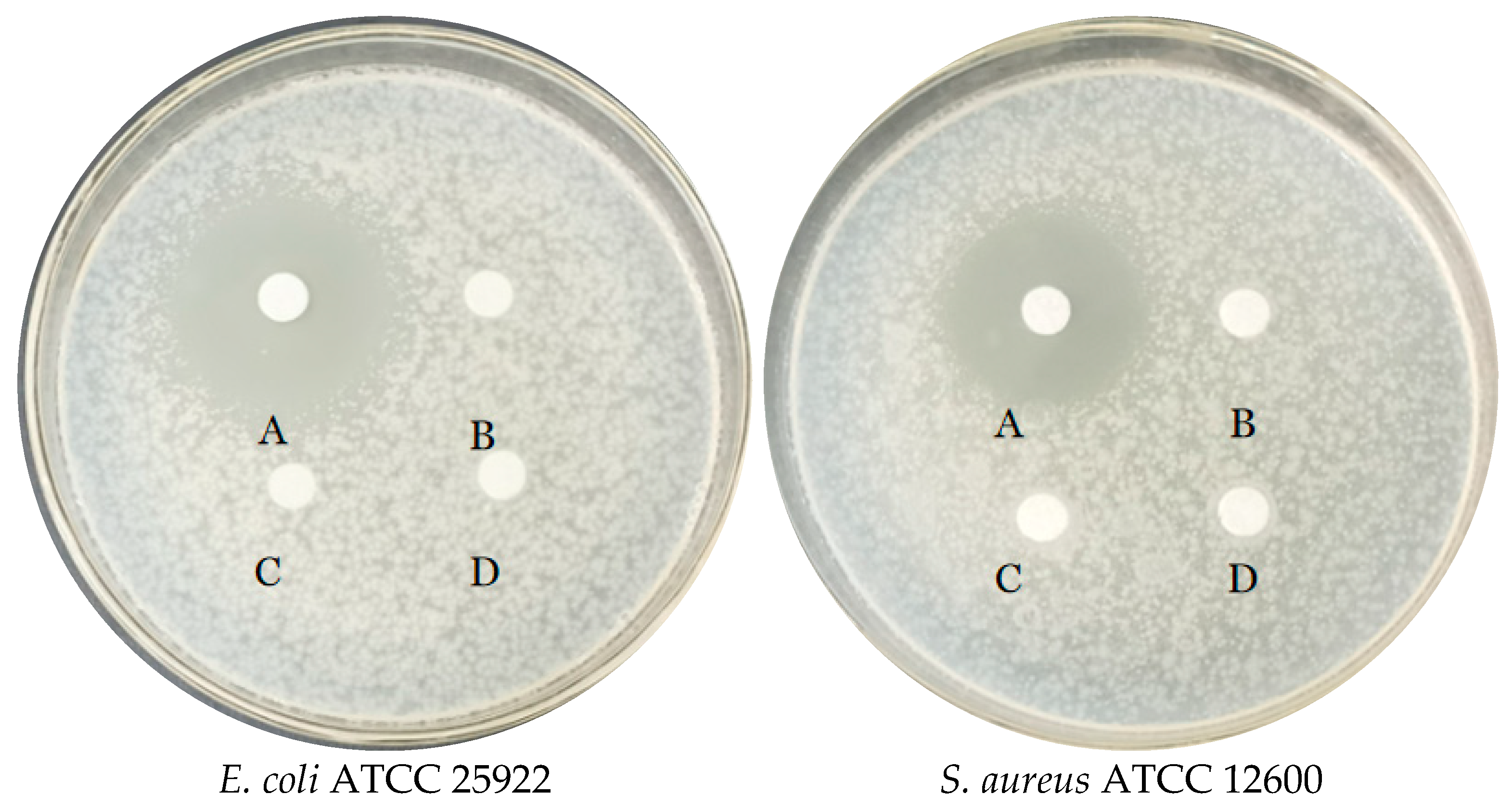
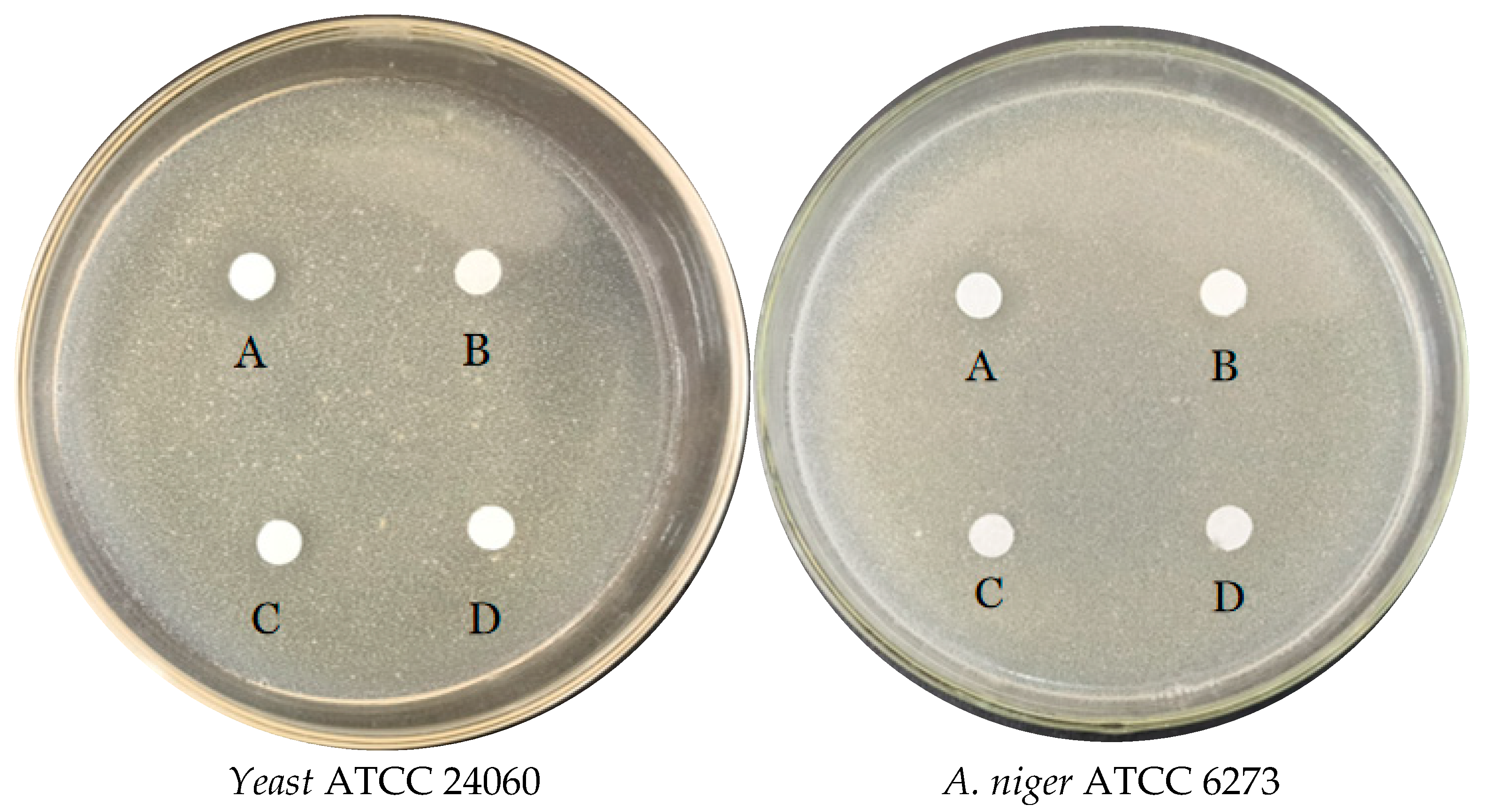
| Attribute | Value | % of Total |
|---|---|---|
| Total size, bp | 2,718,096 | 100 |
| Overall GC content | 1,040,215 | 38.27 |
| Total length of genes | 2,367,573 | 87.1 |
| Total genes | 2700 | 100 |
| Number of protein-coding genes | 2617 | 96.93 |
| Number of tRNA genes | 64 | 2.37 |
| Number of rRNA genes | 17 | 0.63 |
| Number of sRNA genes | 2 | 0.07 |
| Resistance Type | ARO_Name | Identity (%) | Gene Locus |
|---|---|---|---|
| Aminoglycoside antibiotic | AAC(6’)-Ii | 100 | GM_002230 |
| AAC(3)-IIb | 59.1 | GM_002310 | |
| ANT(4’)-Ib | 54.4 | GM_001125 | |
| Lincosamide antibiotic | msrC | 99.8 | GM_002574 |
| lsaA | 68.3 | GM_000711 | |
| Fluoroquinolone antibiotic; | efrA | 82.7 | GM_002136 |
| efrB | 81.7 | GM_002135 | |
| efmA | 74.8 | GM_000589 | |
| patB | 63.8 | GM_002485 | |
| patA | 57.7 | GM_002486 | |
| pmrA | 53 | GM_001845 | |
| adeH | 60.4 | GM_002244 | |
| arlR | 57.3 | GM_001836 | |
| Diaminopyrimidine antibiotic | dfrE | 62.6 | GM_001700 |
| Peptide antibiotic | ugd | 58.7 | GM_000959 |
| Glycopeptide antibiotic | vanRF | 51.5 | GM_002680 |
| Phosphonic acid antibiotic | mdtG | 53.3 | GM_000601 |
| Rifamycin antibiotic | Bado_rpoB_RIF | 52.8 | GM_002629 |
| Role | Virulence Factor | Related Genes | Identity (%) | Gene Locus |
|---|---|---|---|---|
| Adherence | Periplasmic solute binding protein | efaA | 94.6 | GM_000532 |
| Cell wall anchored protein | SgrA | 77.8 | GM_001311 | |
| Collagen adhesin precursor | Acm | 95.9 | GM_002239 | |
| Collagen adhesin protein | Scm | 93 | GM_002646 | |
| Immune modulation | Capsule | capD | 56.4 | GM_000949 |
| Exoenzyme | Hyaluronidase | EF0818 | 60.8 | GM_002331 |
| Biofilm | Bopd | EFAU085_00344 | 97.6 | GM_000438 |
| Strain | DPPH | OH | O2− |
|---|---|---|---|
| Fermentation Supernatant (%) | Fermentation Supernatant (%) | Fermentation Supernatant (%) | |
| FUA027 | 57.62 ± 0.58 | 30.12 ± 0.76 | 36.23 ± 0.32 |
| LP001 | 62.78 ± 0.72 | 28.54 ± 0.62 | 40.78 ± 0.24 |
| FUA004 | 26.36 ± 0.54 | 9.56 ± 0.12 | 19.32 ± 0.58 |
| Protein/Domain | Gene | Gene Locus |
|---|---|---|
| Segregation and condensation protein B | scpB | GM_001728 |
| Segregation and condensation protein A | scpA | GM_001729 |
| Sortase A | strA | GM_000664 |
| S-ribosylhomocysteine lyase | luxS | GM_000502 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Xia, M.; Mu, S.; Fang, Y.; Zhang, X.; Yang, G.; Hou, X.; He, F.; Zhao, Y.; Huang, Y.; Zhang, W.; et al. Genetic and Probiotic Characteristics of Urolithin A Producing Enterococcus faecium FUA027. Foods 2023, 12, 1021. https://doi.org/10.3390/foods12051021
Xia M, Mu S, Fang Y, Zhang X, Yang G, Hou X, He F, Zhao Y, Huang Y, Zhang W, et al. Genetic and Probiotic Characteristics of Urolithin A Producing Enterococcus faecium FUA027. Foods. 2023; 12(5):1021. https://doi.org/10.3390/foods12051021
Chicago/Turabian StyleXia, Mengjie, Shuting Mu, Yaowei Fang, Xiaomeng Zhang, Guang Yang, Xiaoyue Hou, Fuxiang He, Yaling Zhao, Yichen Huang, Wei Zhang, and et al. 2023. "Genetic and Probiotic Characteristics of Urolithin A Producing Enterococcus faecium FUA027" Foods 12, no. 5: 1021. https://doi.org/10.3390/foods12051021
APA StyleXia, M., Mu, S., Fang, Y., Zhang, X., Yang, G., Hou, X., He, F., Zhao, Y., Huang, Y., Zhang, W., Shen, J., & Liu, S. (2023). Genetic and Probiotic Characteristics of Urolithin A Producing Enterococcus faecium FUA027. Foods, 12(5), 1021. https://doi.org/10.3390/foods12051021





