Benchmarking and Validation of a Bioinformatics Workflow for Meat Species Identification Using 16S rDNA Metabarcoding
Abstract
1. Introduction
2. Materials and Methods
2.1. Reference Material
2.2. In Silico Barcode Analysis
2.3. Metabarcoding Analysis
2.3.1. Reads Preprocessing
2.3.2. De Novo Identity Clustering
2.3.3. Denoising
2.3.4. Taxonomic Assignment
2.4. Performance Analysis
2.5. Figure Preparation and Statistical Analysis
3. Results
3.1. Barcode Specificity
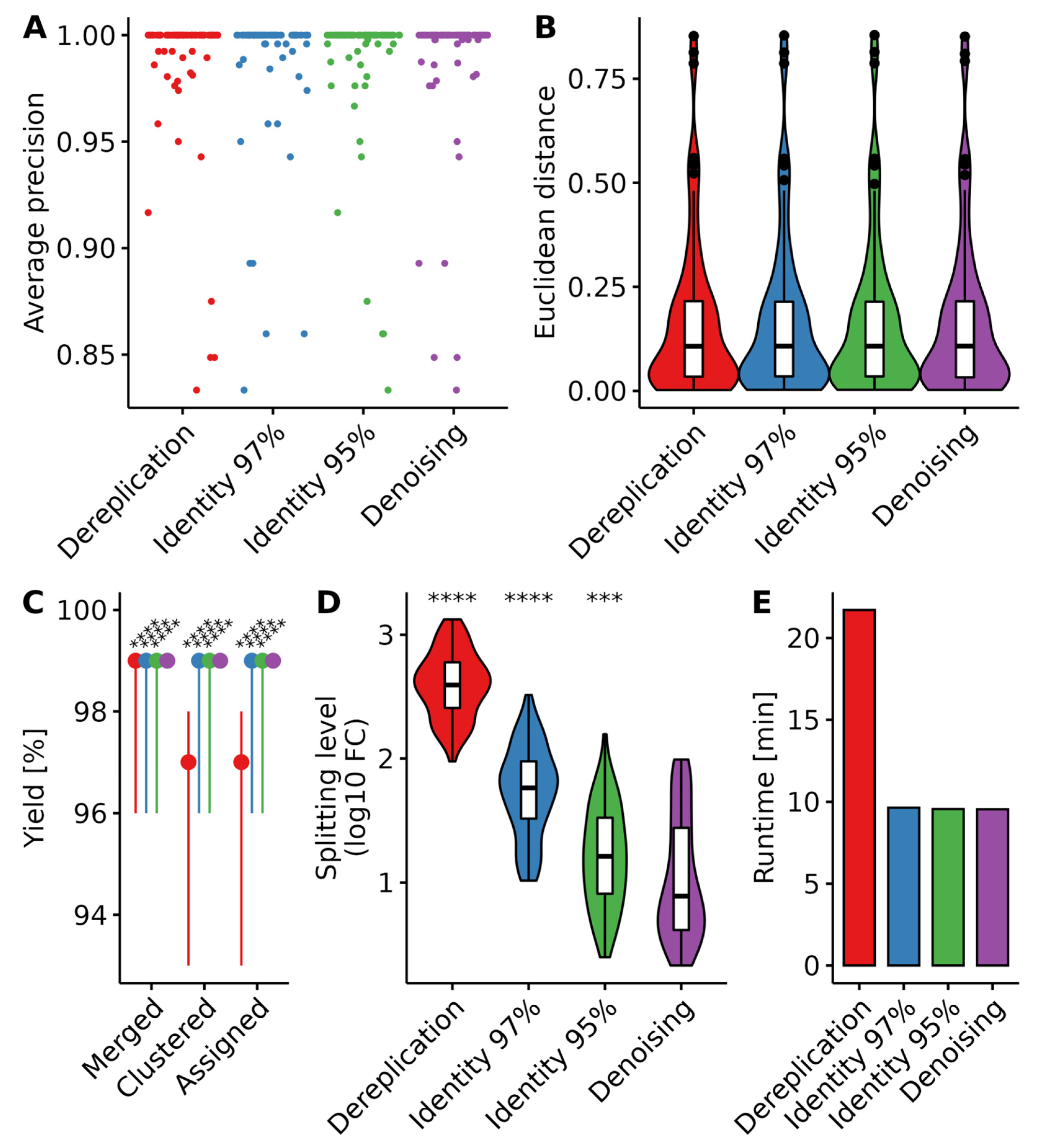
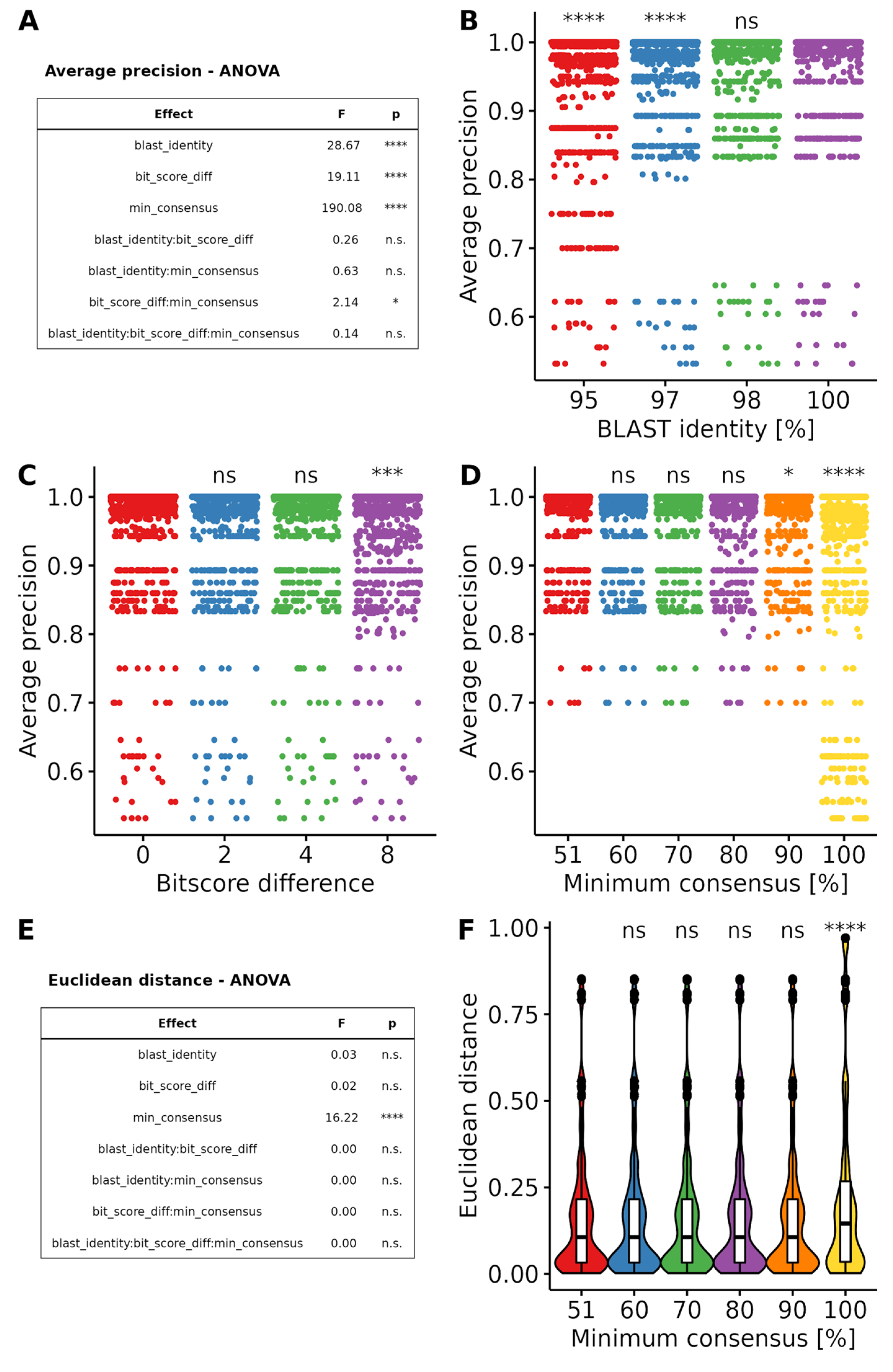
3.2. Workflow Benchmarking and Optimization
- Reads preprocessing, where primer sequences are removed, and bad quality bases are trimmed;
- Clustering, where reads satisfying a given identity level are grouped together;
- Taxonomic assignment, where clusters of reads are assigned to taxonomic nodes.
3.2.1. Benchmarking Clustering Parameters
3.2.2. Optimization of Taxonomic Assignment
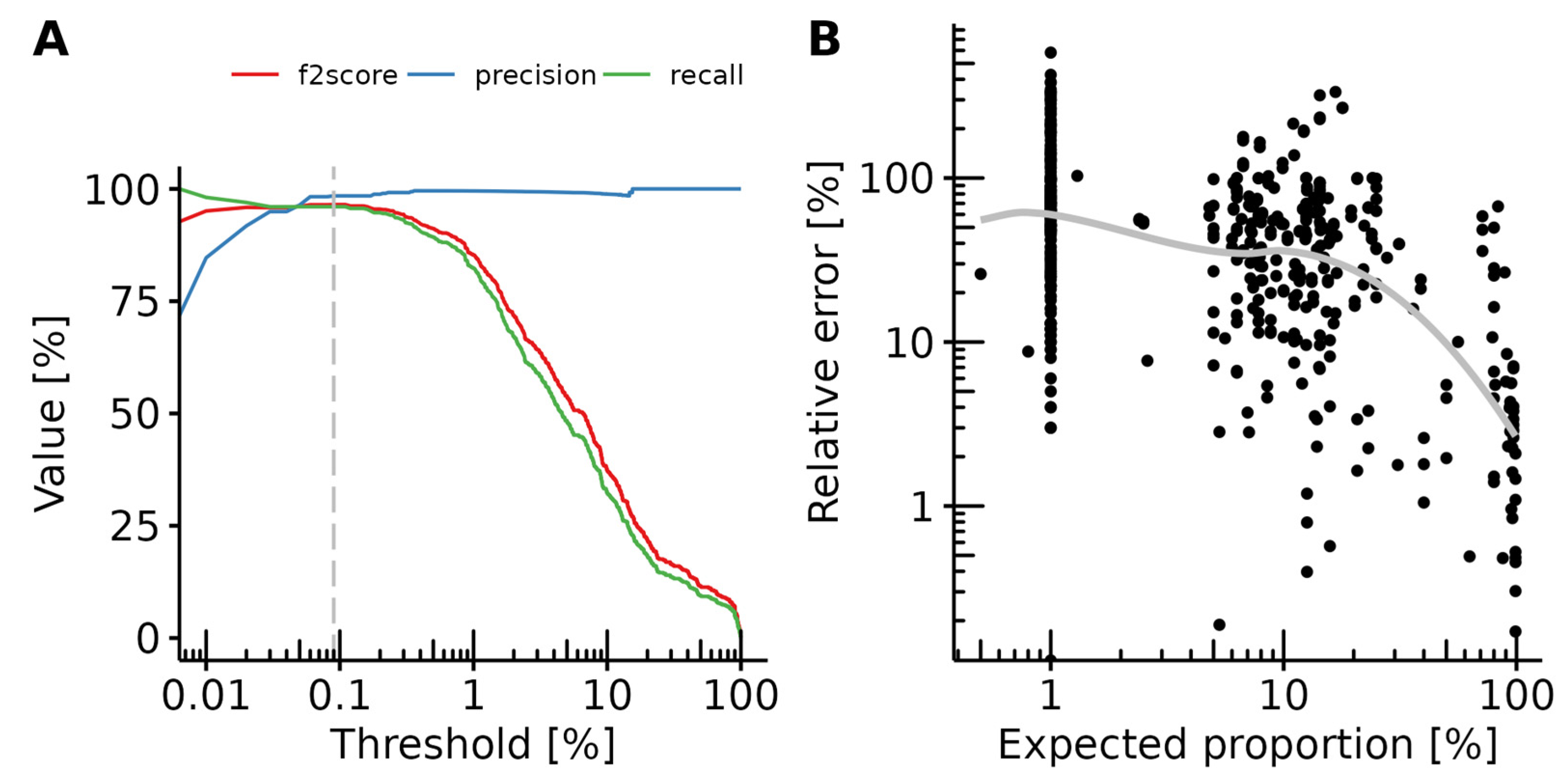
3.3. Detection Limit
3.4. Performance Evaluation
3.5. Effects of Sequencing Depth on Prediction Recall and Variance
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Ballin, N.Z. Authentication of Meat and Meat Products. Meat Sci. 2010, 86, 577–587. [Google Scholar] [CrossRef] [PubMed]
- Montowska, M.; Pospiech, E. Authenticity Determination of Meat and Meat Products on the Protein and DNA Basis. Food Rev. Int. 2010, 27, 84–100. [Google Scholar] [CrossRef]
- Rau, J.; Hiller, E.; Männig, A.; Dyk, M.; Wenninger, O.; Stoll, P.; Wibbelt, G.; Schreiter, P. Animal Species Identification of Meat Using MALDI-TOF Mass Spectrometry. ChemRxiv 2021. [Google Scholar] [CrossRef]
- NCBI Resource Coordinators Database Resources of the National Center for Biotechnology Information. Nucleic Acids Res. 2016, 44, D7–D19. [CrossRef]
- Ratnasingham, S.; Hebert, P.D.N. Bold: The Barcode of Life Data System. Mol. Ecol. Notes 2007, 7, 355–364. [Google Scholar] [CrossRef]
- Haiminen, N.; Edlund, S.; Chambliss, D.; Kunitomi, M.; Weimer, B.C.; Ganesan, B.; Baker, R.; Markwell, P.; Davis, M.; Huang, B.C.; et al. Food Authentication from Shotgun Sequencing Reads with an Application on High Protein Powders. Npj Sci. Food 2019, 3, 24. [Google Scholar] [CrossRef]
- Ripp, F.; Krombholz, C.F.; Liu, Y.; Weber, M.; Schäfer, A.; Schmidt, B.; Köppel, R.; Hankeln, T. All-Food-Seq (AFS): A Quantifiable Screen for Species in Biological Samples by Deep DNA Sequencing. BMC Genom. 2014, 15, 639. [Google Scholar] [CrossRef]
- Xing, R.-R.; Wang, N.; Hu, R.-R.; Zhang, J.-K.; Han, J.-X.; Chen, Y. Application of next Generation Sequencing for Species Identification in Meat and Poultry Products: A DNA Metabarcoding Approach. Food Control 2019, 101, 173–179. [Google Scholar] [CrossRef]
- Preckel, L.; Brünen-Nieweler, C.; Denay, G.; Petersen, H.; Cichna-Markl, M.; Dobrovolny, S.; Hochegger, R. Identification of Mammalian and Poultry Species in Food and Pet Food Samples Using 16S RDNA Metabarcoding. Foods 2021, 10, 2875. [Google Scholar] [CrossRef]
- Dobrovolny, S.; Uhlig, S.; Frost, K.; Schlierf, A.; Nichani, K.; Simon, K.; Cichna-Markl, M.; Hochegger, R. Interlaboratory Validation of a DNA Metabarcoding Assay for Mammalian and Poultry Species to Detect Food Adulteration. Foods 2022, 11, 1108. [Google Scholar] [CrossRef]
- Dobrovolny, S.; Blaschitz, M.; Weinmaier, T.; Pechatschek, J.; Cichna-Markl, M.; Indra, A.; Hufnagl, P.; Hochegger, R. Development of a DNA Metabarcoding Method for the Identification of Fifteen Mammalian and Six Poultry Species in Food. Food Chem. 2019, 272, 354–361. [Google Scholar] [CrossRef] [PubMed]
- Ribani, A.; Schiavo, G.; Utzeri, V.J.; Bertolini, F.; Geraci, C.; Bovo, S.; Fontanesi, L. Application of next Generation Semiconductor Based Sequencing for Species Identification and Analysis of Within-Species Mitotypes Useful for Authentication of Meat Derived Products. Food Control 2018, 91, 58–67. [Google Scholar] [CrossRef]
- Bertolini, F.; Ghionda, M.C.; D’Alessandro, E.; Geraci, C.; Chiofalo, V.; Fontanesi, L. A Next Generation Semiconductor Based Sequencing Approach for the Identification of Meat Species in DNA Mixtures. PLoS ONE 2015, 10, e0121701. [Google Scholar] [CrossRef] [PubMed]
- Palumbo, F.; Scariolo, F.; Vannozzi, A.; Barcaccia, G. NGS-Based Barcoding with Mini-COI Gene Target Is Useful for Pet Food Market Surveys Aimed at Mislabelling Detection. Sci. Rep. 2020, 10, 17767. [Google Scholar] [CrossRef]
- Gryson, N. Effect of Food Processing on Plant DNA Degradation and PCR-Based GMO Analysis: A Review. Anal. Bioanal. Chem. 2010, 396, 2003–2022. [Google Scholar] [CrossRef]
- Piombo, E.; Abdelfattah, A.; Droby, S.; Wisniewski, M.; Spadaro, D.; Schena, L. Metagenomics Approaches for the Detection and Surveillance of Emerging and Recurrent Plant Pathogens. Microorganisms 2021, 9, 188. [Google Scholar] [CrossRef]
- Bruno, A.; Sandionigi, A.; Agostinetto, G.; Bernabovi, L.; Frigerio, J.; Casiraghi, M.; Labra, M. Food Tracking Perspective: DNA Metabarcoding to Identify Plant Composition in Complex and Processed Food Products. Genes 2019, 10, 248. [Google Scholar] [CrossRef]
- Raclariu-Manolică, A.C.; Anmarkrud, J.A.; Kierczak, M.; Rafati, N.; Thorbek, B.L.G.; Schrøder-Nielsen, A.; de Boer, H.J. DNA Metabarcoding for Quality Control of Basil, Oregano, and Paprika. Front. Plant Sci. 2021, 12, 665618. [Google Scholar] [CrossRef]
- Gense, K.; Peterseil, V.; Licina, A.; Wagner, M.; Cichna-Markl, M.; Dobrovolny, S.; Hochegger, R. Development of a DNA Metabarcoding Method for the Identification of Bivalve Species in Seafood Products. Foods 2021, 10, 2618. [Google Scholar] [CrossRef]
- Li, H. Lh3/Seqtk. Available online: https://github.com/lh3/seqtk (accessed on 24 October 2022).
- Denay, G. CVUA-RRW/BaRCoD: BaRCoD v1.1.1. Available online: https://zenodo.org/record/6976282 (accessed on 24 October 2022).
- Camacho, C.; Coulouris, G.; Avagyan, V.; Ma, N.; Papadopoulos, J.; Bealer, K.; Madden, T.L. BLAST+: Architecture and Applications. BMC Bioinform. 2009, 10, 421. [Google Scholar] [CrossRef]
- Ye, J.; Coulouris, G.; Zaretskaya, I.; Cutcutache, I.; Rozen, S.; Madden, T.L. Primer-BLAST: A Tool to Design Target-Specific Primers for Polymerase Chain Reaction. BMC Bioinform. 2012, 13, 134. [Google Scholar] [CrossRef] [PubMed]
- Martin, M. Cutadapt Removes Adapter Sequences from High-Throughput Sequencing Reads. EMBnet. J. 2011, 17, 10–12. [Google Scholar] [CrossRef]
- Rognes, T.; Flouri, T.; Nichols, B.; Quince, C.; Mahé, F. VSEARCH: A Versatile Open Source Tool for Metagenomics. PeerJ 2016, 4, e2584. [Google Scholar] [CrossRef] [PubMed]
- Denay, G. CVUA-RRW/TaxidTools: 2.2.3. Available online: https://zenodo.org/record/5556006 (accessed on 26 August 2022).
- Edgar, R.C. Updating the 97% Identity Threshold for 16S Ribosomal RNA OTUs. Bioinformatics 2018, 34, 2371–2375. [Google Scholar] [CrossRef] [PubMed]
- Denay, G. CVUA-RRW/FooDMe: Foodme v1.6.3. Available online: https://zenodo.org/record/7078595 (accessed on 14 September 2022).
- Chen, S.; Zhou, Y.; Chen, Y.; Gu, J. Fastp: An Ultra-Fast All-in-One FASTQ Preprocessor. Bioinformatics 2018, 34, i884–i890. [Google Scholar] [CrossRef] [PubMed]
- Callahan, B.J.; McMurdie, P.J.; Rosen, M.J.; Han, A.W.; Johnson, A.J.A.; Holmes, S.P. DADA2: High-Resolution Sample Inference from Illumina Amplicon Data. Nat. Methods 2016, 13, 581–583. [Google Scholar] [CrossRef]
- Bazinet, A.L.; Ondov, B.D.; Sommer, D.D.; Ratnayake, S. BLAST-Based Validation of Metagenomic Sequence Assignments. PeerJ 2018, 6, e4892. [Google Scholar] [CrossRef]
- Bokulich, N.A.; Kaehler, B.D.; Rideout, J.R.; Dillon, M.; Bolyen, E.; Knight, R.; Huttley, G.A.; Gregory Caporaso, J. Optimizing Taxonomic Classification of Marker-Gene Amplicon Sequences with QIIME 2’s Q2-Feature-Classifier Plugin. Microbiome 2018, 6, 90. [Google Scholar] [CrossRef]
- Pedregosa, F.; Varoquaux, G.; Gramfort, A.; Michel, V.; Thirion, B.; Grisel, O.; Blondel, M.; Prettenhofer, P.; Weiss, R.; Dubourg, V.; et al. Scikit-Learn: Machine Learning in Python. J. Mach. Learn. Res. 2011, 12, 2825–2830. [Google Scholar]
- Wickham, H.; Averick, M.; Bryan, J.; Chang, W.; McGowan, L.D.; François, R.; Grolemund, G.; Hayes, A.; Henry, L.; Hester, J.; et al. Welcome to the Tidyverse. J. Open Source Softw. 2019, 4, 1686. [Google Scholar] [CrossRef]
- Harris, C.R.; Millman, K.J.; van der Walt, S.J.; Gommers, R.; Virtanen, P.; Cournapeau, D.; Wieser, E.; Taylor, J.; Berg, S.; Smith, N.J.; et al. Array Programming with NumPy. Nature 2020, 585, 357–362. [Google Scholar] [CrossRef] [PubMed]
- Meyer, R.; Höfelein, C.; Lüthy, J.; Candrian, U. Polymerase Chain Reaction-Restriction Fragment Length Polymorphism Analysis: A Simple Method for Species Identification in Food. J. AOAC Int. 1995, 78, 1542–1551. [Google Scholar] [CrossRef] [PubMed]
- Palumbi, S.R.; Martin, A.; Romano, S.; McMillan, W.O.; Stice, L.; Grabowski, G. The Simple Fool’s Guide to PCR, 2nd ed.; University of Hawaii at Manoa, Kewalo Marine Laboratory, Dept. of Zoology and Kewalo Marine Laboratory; University of Hawaii: Honolulu, HI, USA, 2002; pp. 25–37. [Google Scholar]
- Verband Deutscher Landwirtschaftlicher Untersuchungs-und Forschungsanstalten MB3-29.1 Molekularbiologischer Nachweis von tierischen Bestandteilen (PCR-Methode). In 125 Jahre Verband Deutscher Landwirtschaftlicher Untersuchungs-und Forschungsanstalten e.V: Eine Dokumentation; VDLUFA-Schriftenreihe; VDLUFA-Verl: Darmstadt, Germany, 2013; ISBN 978-3-941273-14-6.
- Szabo, K.; Malorny, B.; Stoyke, M. Etablierung der § 64 LFGB Arbeitsgruppen “NGS—Bakteriencharakterisierung” und “NGS—Speziesidentifizierung”. J. Consum. Prot. Food Saf. 2020, 15, 85–89. [Google Scholar] [CrossRef]
- Matthes, N.; Pietsch, K.; Rullmann, A.; Näumann, G.; Pöpping, B.; Szabo, K. The Barcoding Table of Animal Species (BaTAnS): A New Tool to Select Appropriate Methods for Animal Species Identification Using DNA Barcoding. Mol. Biol. Rep. 2020, 47, 6457–6461. [Google Scholar] [CrossRef]
- BVL—Amtliche Sammlung von Untersuchungsverfahren—Barcoding-Tabelle Für Die Tierartenbestimmung (Barcoding Table of Animal Species—BaTAnS). Available online: https://www.bvl.bund.de/SharedDocs/Downloads/07_Untersuchungen/Barcoding-Tabelle%20f%C3%BCr%20die%20Tierartenbestimmung.html?nn=11009496 (accessed on 26 August 2022).
- Dobrovolny, S.; Austrian Agency for Health and Food Safety (AGES), Institute for Food Safety, Vienna, Austria. Fallow Deer Primers. Personal communication, 2022. [Google Scholar]
- Karlsson, A.O.; Holmlund, G. Identification of Mammal Species Using Species-Specific DNA Pyrosequencing. Forensic Sci. Int. 2007, 173, 16–20. [Google Scholar] [CrossRef]
- Blaxter, M.; Mann, J.; Chapman, T.; Thomas, F.; Whitton, C.; Floyd, R.; Abebe, E. Defining Operational Taxonomic Units Using DNA Barcode Data. Philos. Trans. R. Soc. B Biol. Sci. 2005, 360, 1935–1943. [Google Scholar] [CrossRef]
- Callahan, B.J.; McMurdie, P.J.; Holmes, S.P. Exact Sequence Variants Should Replace Operational Taxonomic Units in Marker-Gene Data Analysis. ISME J. 2017, 11, 2639–2643. [Google Scholar] [CrossRef]
- Westcott, S.L.; Schloss, P.D. De Novo Clustering Methods Outperform Reference-Based Methods for Assigning 16S RRNA Gene Sequences to Operational Taxonomic Units. PeerJ 2015, 3, e1487. [Google Scholar] [CrossRef]
- DLA Proficiency Tests Gmbh. Ring-Trials Reports for PtAUS3.2 and 45/2019. Private communication, 2020. [Google Scholar]
- Dully, V.; Balliet, H.; Frühe, L.; Däumer, M.; Thielen, A.; Gallie, S.; Berrill, I.; Stoeck, T. Robustness, Sensitivity and Reproducibility of EDNA Metabarcoding as an Environmental Biomonitoring Tool in Coastal Salmon Aquaculture—An Inter-Laboratory Study. Ecol. Indic. 2021, 121, 107049. [Google Scholar] [CrossRef]
- Staats, M.; Arulandhu, A.J.; Gravendeel, B.; Holst-Jensen, A.; Scholtens, I.; Peelen, T.; Prins, T.W.; Kok, E. Advances in DNA Metabarcoding for Food and Wildlife Forensic Species Identification. Anal. Bioanal. Chem. 2016, 408, 4615–4630. [Google Scholar] [CrossRef]
- García-López, R.; Cornejo-Granados, F.; Lopez-Zavala, A.A.; Cota-Huízar, A.; Sotelo-Mundo, R.R.; Gómez-Gil, B.; Ochoa-Leyva, A. OTUs and ASVs Produce Comparable Taxonomic and Diversity from Shrimp Microbiota 16S Profiles Using Tailored Abundance Filters. Genes 2021, 12, 564. [Google Scholar] [CrossRef] [PubMed]
- Chiarello, M.; McCauley, M.; Villéger, S.; Jackson, C.R. Ranking the Biases: The Choice of OTUs vs. ASVs in 16S RRNA Amplicon Data Analysis Has Stronger Effects on Diversity Measures than Rarefaction and OTU Identity Threshold. PLoS ONE 2022, 17, e0264443. [Google Scholar] [CrossRef] [PubMed]
- Altschul, S.F.; Gish, W.; Miller, W.; Myers, E.W.; Lipman, D.J. Basic Local Alignment Search Tool. J. Mol. Biol. 1990, 215, 403–410. [Google Scholar] [CrossRef] [PubMed]
- Neto, L.; Pinto, N.; Proença, A.; Amorim, A.; Conde-Sousa, E. 4SpecID: Reference DNA Libraries Auditing and Annotation System for Forensic Applications. Genes 2021, 12, 61. [Google Scholar] [CrossRef] [PubMed]
- Mölder, F.; Jablonski, K.P.; Letcher, B.; Hall, M.B.; Tomkins-Tinch, C.H.; Sochat, V.; Forster, J.; Lee, S.; Twardziok, S.O.; Kanitz, A.; et al. Sustainable Data Analysis with Snakemake. F1000Research 2021, 10, 33. [Google Scholar] [CrossRef] [PubMed]

| Method | Number of Taxids Retrieved | Median Number of Barcode per Taxid a | Median Length of Barcode [bp] b | 97% Identity | 100% Identity | ||
|---|---|---|---|---|---|---|---|
| Sequences Assigned at Max. | Sequences Assigned at Max. | ||||||
| Species Level [%] | Genus Level [%] | Species Level [%] | Genus Level [%] | ||||
| 16S_dobrovolny | 7701 | 3 | 75 | 66.77 | 85.36 | 81.98 | 93.27 |
| 16S_xing | 6383 | 3 | 204 | 78.53 | 95.07 | 93.29 | 98.21 |
| cox1_palumbi | 2293 | 5 | 522 | 90.16 | 97.82 | 97.08 | 98.63 |
| cytB_meyer | 10,909 | 25 | 333 | 91.31 | 98.47 | 95.29 | 98.89 |
| cytB_palumbi | 10,078 | 35 | 741 | 94.55 | 99.17 | 97.93 | 99.56 |
| cytB_VDLUFA | 18,136 | 16 | 220 | 89.95 | 98.52 | 94.38 | 98.85 |
| miniCOI_palumbo | 5482 | 2 | 151 | 82.55 | 95.46 | 92.86 | 97.71 |
| Organism | Taxid | Common Name | 16S Dobrovolny | 16S Xing | COX1 Palumbi | cytB Meyer | cytB Palumbi | cytB VDLUFA | MiniCOI Palumbo |
|---|---|---|---|---|---|---|---|---|---|
| Addax nasomaculatus | 59515 | Addax | + | + | + | + | + | + | + |
| Ailuropoda melanoleuca | 9646 | Giant panda | + | + | 0 | + | + | + | + |
| Alcelaphus buselaphus | 59517 | Hartebeest | + | + | 0 | + | + | + | + |
| Alcelaphus caama | 59519 | Red hartebeest | + | + | 0 | + | + | + | + |
| Alces alces | 9852 | Eurasian elk | + | + | 0 | 0 | 0 | + | + |
| Alectoris chukar | 9078 | Chukar partridge | + | + | + | + | + | + | + |
| Ammotragus lervia | 9899 | Barbary sheep | + | + | + | + | + | + | + |
| Anas platyrhynchos | 8839 | Duck | + | + | + | + | + | + | + |
| Anser anser | 8843 | Greylag goose | + | + | + | + | + | + | + |
| Anser cygnoides | 8845 | Chinese goose | + | + | 0 | + | + | + | + |
| Anser indicus | 8846 | Bar-headed goose | + | + | + | + | + | + | + |
| Anser rossii | 56281 | Ross’ goose | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| Antidorcas marsupialis | 59523 | Springbok | + | + | + | + | + | + | + |
| Bison bison | 9901 | Bison | + | + | 0 | + | + | + | + |
| Bison bonasus | 9902 | Wisent | + | + | + | + | + | + | + |
| Bos mutus | 72004 | Yak | + | + | 0 | + | + | + | + |
| Bos taurus | 9913 | Cattle | + | + | + | + | + | + | + |
| Bubalus bubalis | 89462 | Water buffalo | + | + | + | + | + | + | + |
| Cairina moschata | 8855 | Muscovy duck | + | + | 0 | + | + | + | + |
| Canis lupus | 9612 | Grey wolf, dog | + | + | + | + | + | + | + |
| Capra aegagrus | 9923 | Wild goat | + | + | + | + | + | + | + |
| Capra hircus | 9925 | Domestic goat | + | + | + | + | + | + | + |
| Capra ibex | 72542 | Ibex | + | + | + | + | + | + | + |
| Capreolus capreolus | 9858 | Roe deer | + | + | 0 | + | + | + | + |
| Cavia porcellus | 10141 | Guinea pig | + | + | 0 | + | + | + | + |
| Cervus elaphus | 9860 | Red deer | + | + | + | + | + | + | + |
| Cervus nippon | 9863 | Sika deer | + | + | 0 | + | + | + | + |
| Columba livia | 8932 | Domestic pigeon | + | + | 0 | + | + | + | + |
| Connochaetes gnou | 59528 | Black wildebeest | + | + | 0 | + | + | + | + |
| Connochaetes taurinus | 9927 | Blue wildebeest | + | + | + | + | + | + | + |
| Coturnix coturnix | 9091 | Common quail | + | + | 0 | + | + | + | + |
| Coturnix japonica | 93934 | Japanese quail | + | + | 0 | + | + | + | + |
| Cygnus olor | 8869 | Mute swan | + | + | 0 | + | + | + | + |
| Dama dama | 30532 | Fallow deer | 0 | 0 | 0 | 0 | 0 | + | 0 |
| Damaliscus pygargus | 9931 | Bontebok | + | + | + | + | + | + | + |
| Equus asinus | 9793 | Donkey | + | + | + | + | + | + | + |
| Equus caballus | 9796 | Horse | + | + | + | + | + | + | + |
| Equus quagga | 89248 | Plain zebra | 0 | 0 | 0 | 0 | 0 | + | 0 |
| Equus zebra | 9791 | Mountain zebra | + | + | + | + | + | + | + |
| Felis catus | 9685 | Cat | + | + | + | + | + | + | + |
| Gallus gallus | 9031 | Chicken | + | + | + | + | + | + | + |
| Gazella dorcas | 37751 | Dorcas gazelle | + | + | 0 | + | + | + | + |
| Gazella subgutturosa | 59529 | Black-tailed gazelle | + | + | 0 | + | + | + | + |
| Glis glis | 41261 | Fat dormouse | + | + | 0 | + | + | + | + |
| Hippotragus niger | 37189 | Sable antelope | + | + | 0 | + | + | + | + |
| Kobus ellipsiprymnus | 9962 | Waterbuck | + | + | 0 | + | + | + | + |
| Lama glama | 9844 | Llama | + | + | + | + | + | + | + |
| Lepus europaeus | 9983 | European hare | + | + | 0 | + | + | + | + |
| Macropus fuliginosus | 9316 | Western gray kangaroo | + | + | 0 | 0 | 0 | + | 0 |
| Macropus giganteus | 9317 | Eastern gray kangaroo | + | + | 0 | + | + | + | + |
| Marmota marmota | 9993 | Alpine marmot | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| Martes martes | 29065 | European pine marten | + | + | 0 | + | + | + | + |
| Meleagris gallopavo | 9103 | Turkey | + | + | + | + | + | + | + |
| Muntiacus reevesi | 9886 | Reeves’ muntjac | + | + | 0 | + | + | + | + |
| Mus musculus | 10090 | Mouse | + | + | + | + | + | + | + |
| Myodes glareolus | 447135 | Bank vole | + | + | + | + | + | + | + |
| Numida meleagris | 8996 | Helmeted guineafowl | + | + | 0 | + | 0 | + | + |
| Oryctolagus cuniculus | 9986 | Rabbit | + | + | + | + | + | + | + |
| Oryx dammah | 59534 | Scimitar-horned oryx | + | + | + | + | + | + | + |
| Oryx gazella | 9958 | Gemsbok | + | + | + | + | + | + | + |
| Osphranter robustus | 9319 | Common wallaroo | + | + | 0 | + | + | + | + |
| Osphranter rufus | 9321 | Red kangaroo | + | + | + | + | + | + | + |
| Ovibos moschatus | 37176 | Musk ox | + | + | 0 | 0 | 0 | + | + |
| Ovis aries | 9940 | Sheep | + | + | + | + | + | + | + |
| Ovis orientalis | 469796 | Asiatic mouflon | + | + | + | + | + | + | + |
| Phasianus colchicus | 9054 | Common pheasant | + | + | 0 | + | + | + | + |
| Rangifer tarandus | 9870 | Reindeer | + | + | + | + | + | + | + |
| Rattus norvegicus | 10116 | Rat | + | + | + | + | + | + | + |
| Struthio camelus | 8801 | Ostrich | + | + | 0 | + | + | + | + |
| Sus scrofa | 9823 | Pig | + | + | + | + | + | + | + |
| Syncerus caffer | 9970 | African buffalo | + | + | + | + | + | + | + |
| Tragelaphus oryx | 9945 | Eland | + | + | + | + | + | + | + |
| Tragelaphus spekii | 69298 | Sitatunga | + | + | 0 | + | + | + | + |
| Vulpes vulpes | 9627 | Red fox | + | + | + | + | + | + | + |
| Evaluation Rank | Confusion Matrix | Performance Metrics | |||
|---|---|---|---|---|---|
| True Positives | False Positives | False Negatives | Recall | Precision | |
| Species a | 490 (84%) | 56 (10%) | 39 (7%) | 93% | 90% |
| Species b | 490 (86%) | 56 (10%) | 21 (4%) | 96% | 90% |
| Genus a | 494 (95%) | 6 (1%) | 21 (4%) | 96% | 99% |
| Genus b | 494 (98%) | 6 (1%) | 3 (1%) | 99% | 99% |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Denay, G.; Preckel, L.; Petersen, H.; Pietsch, K.; Wöhlke, A.; Brünen-Nieweler, C. Benchmarking and Validation of a Bioinformatics Workflow for Meat Species Identification Using 16S rDNA Metabarcoding. Foods 2023, 12, 968. https://doi.org/10.3390/foods12050968
Denay G, Preckel L, Petersen H, Pietsch K, Wöhlke A, Brünen-Nieweler C. Benchmarking and Validation of a Bioinformatics Workflow for Meat Species Identification Using 16S rDNA Metabarcoding. Foods. 2023; 12(5):968. https://doi.org/10.3390/foods12050968
Chicago/Turabian StyleDenay, Grégoire, Laura Preckel, Henning Petersen, Klaus Pietsch, Anne Wöhlke, and Claudia Brünen-Nieweler. 2023. "Benchmarking and Validation of a Bioinformatics Workflow for Meat Species Identification Using 16S rDNA Metabarcoding" Foods 12, no. 5: 968. https://doi.org/10.3390/foods12050968
APA StyleDenay, G., Preckel, L., Petersen, H., Pietsch, K., Wöhlke, A., & Brünen-Nieweler, C. (2023). Benchmarking and Validation of a Bioinformatics Workflow for Meat Species Identification Using 16S rDNA Metabarcoding. Foods, 12(5), 968. https://doi.org/10.3390/foods12050968








