Hierarchical Effects of Lactic Fermentation and Grain Germination on the Microbial and Metabolomic Profile of Rye Doughs
Abstract
:1. Introduction
2. Materials and Methods
2.1. Microbial Strains and Growth Conditions
2.2. Germination of Rye Flour and Micro-Malting Process
2.3. Dough Preparation and Chemical-Physical Characterization
2.4. Extraction of DNA and Full-Length 16S Metagenomics Analysis
2.5. Oligosaccharide Semi-Quantification of Rye Doughs
2.6. Untargeted Metabolomic Profiling of Rye Doughs
2.7. Statistics
3. Results
3.1. General Parameters of Rye Doughs
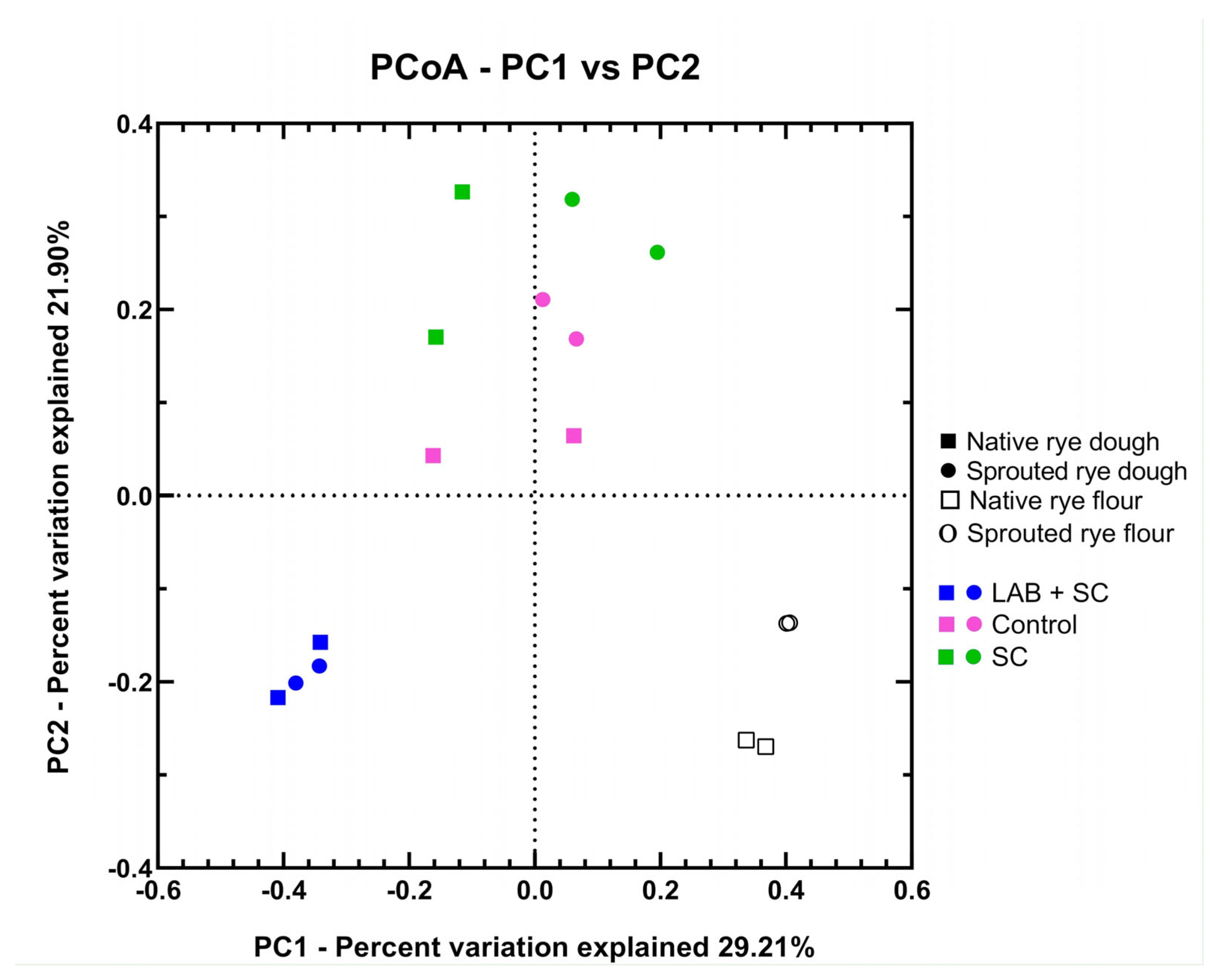
3.2. Diversity of Bacterial Communities in Rye Doughs
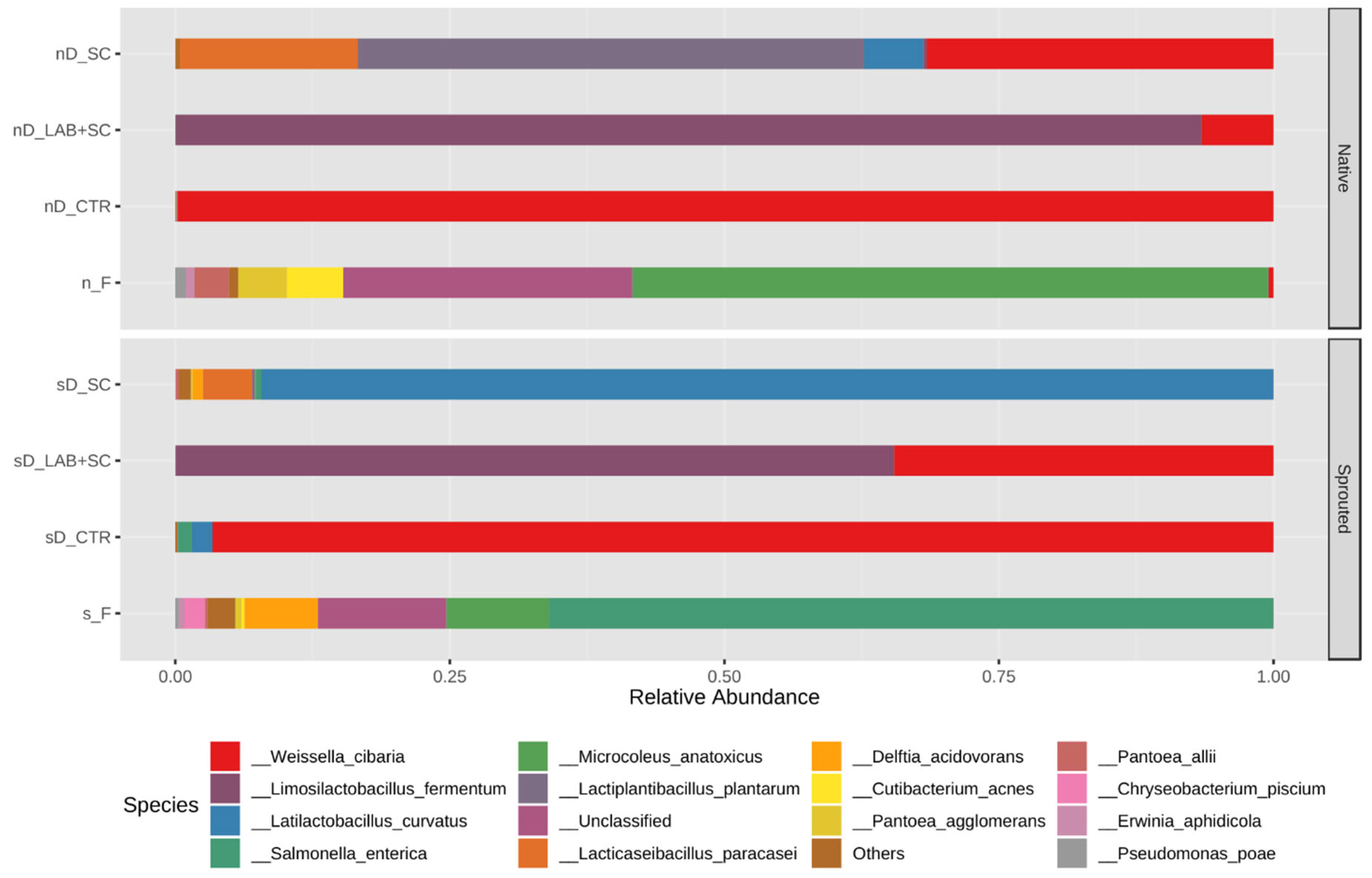
3.3. Bacterial Taxonomic Composition of Dough Samples
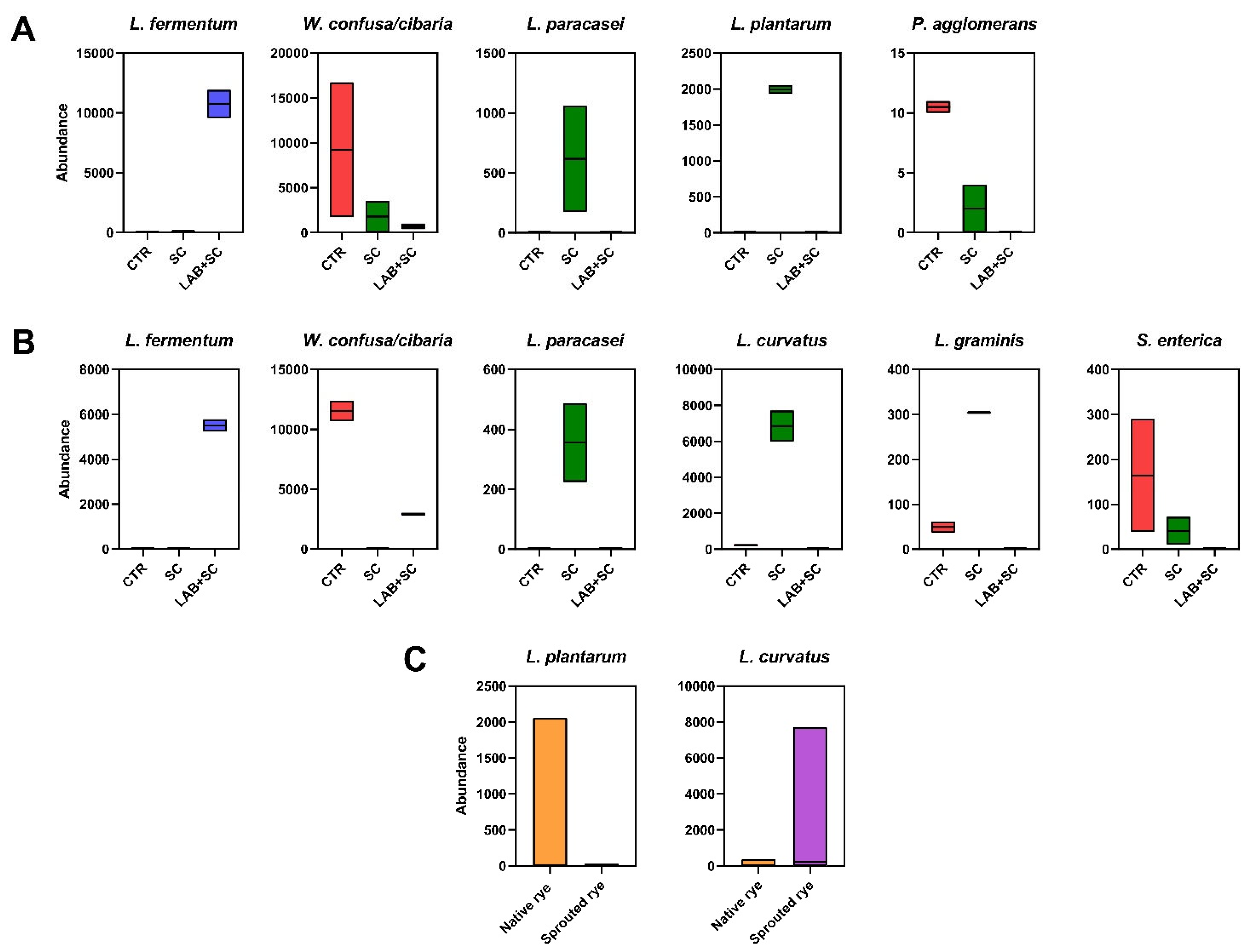
3.4. Differential Analysis of Discriminant Species between Dough Samples
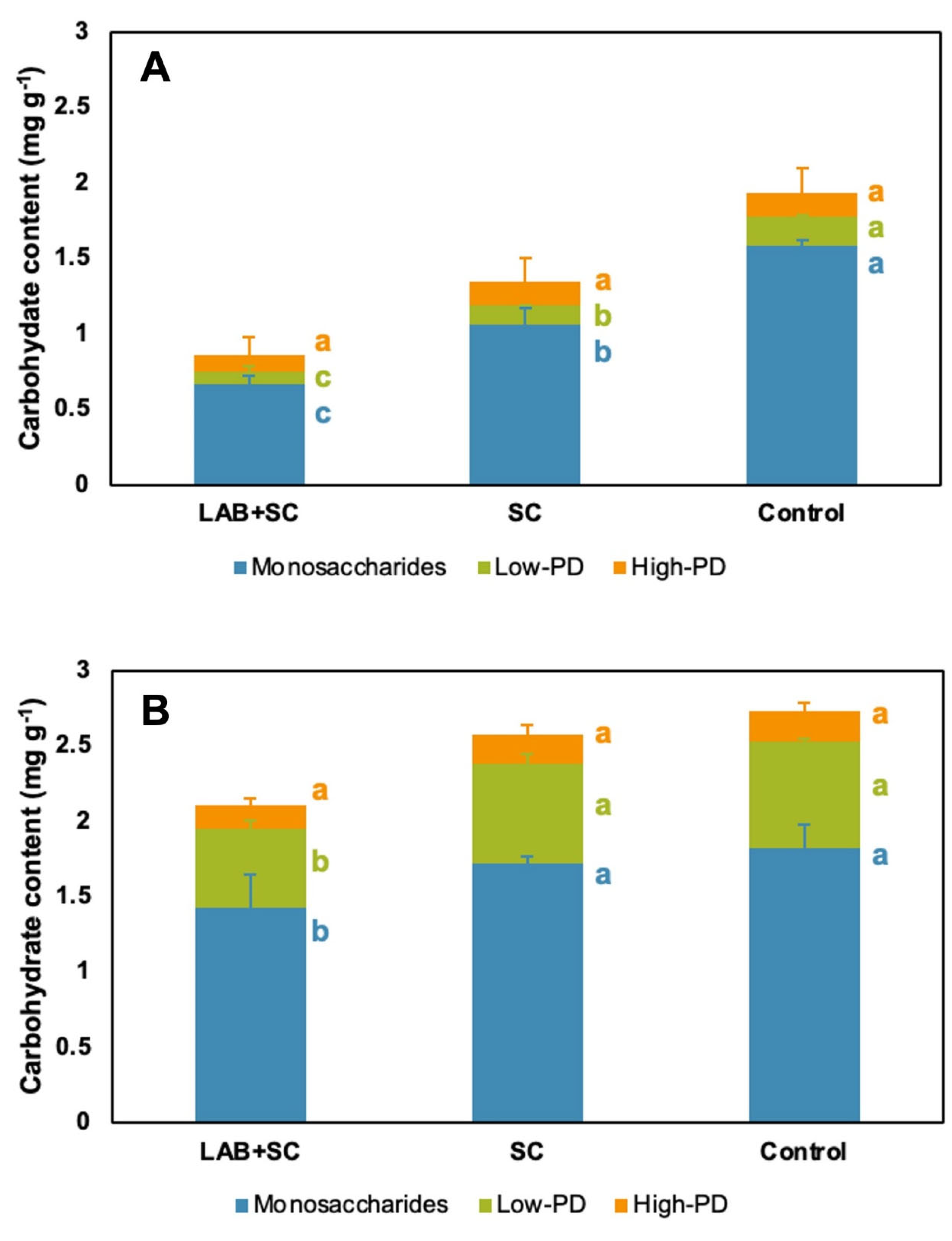
3.5. Oligosaccharide Semi-Quantification of Rye Sourdoughs
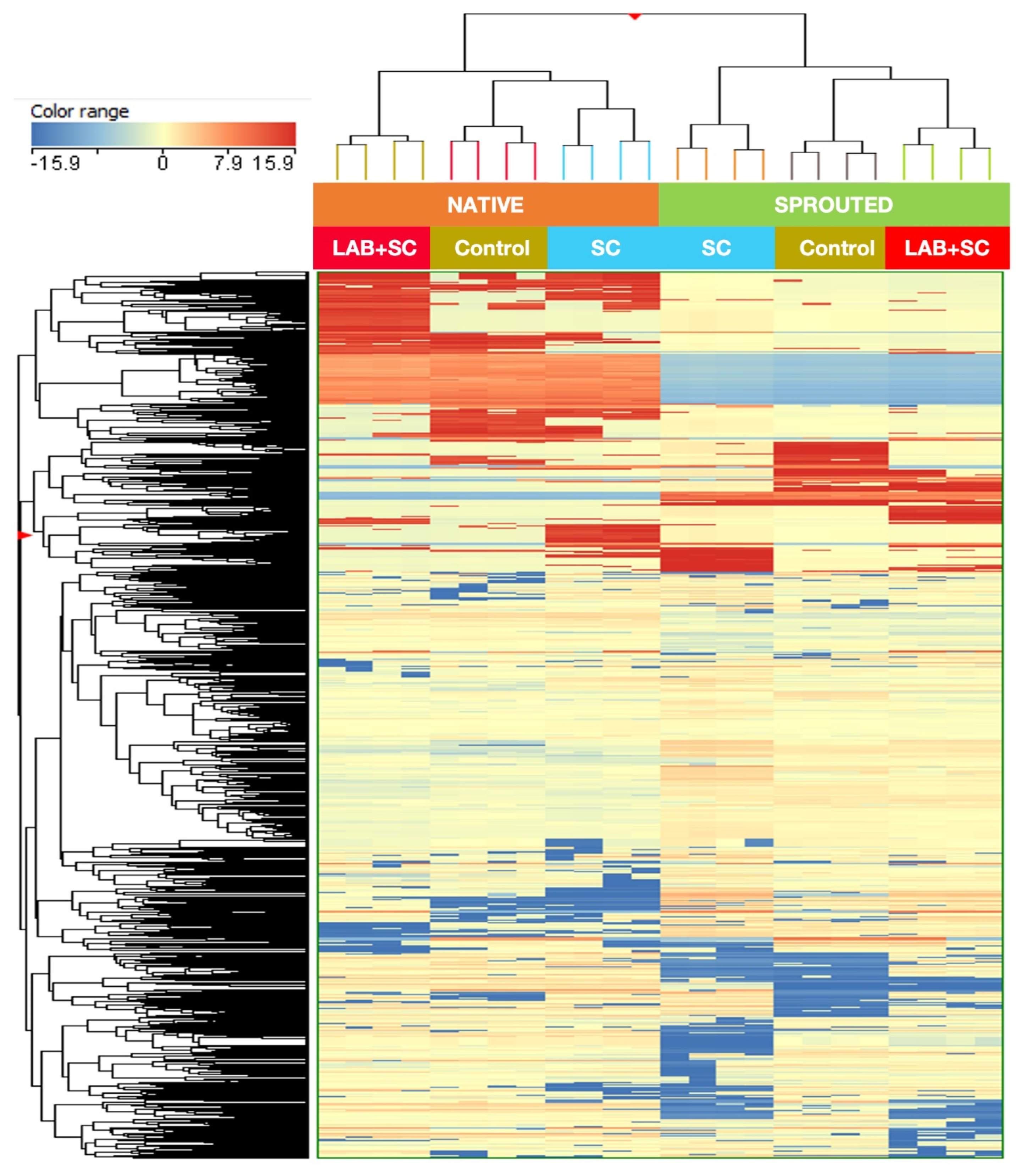
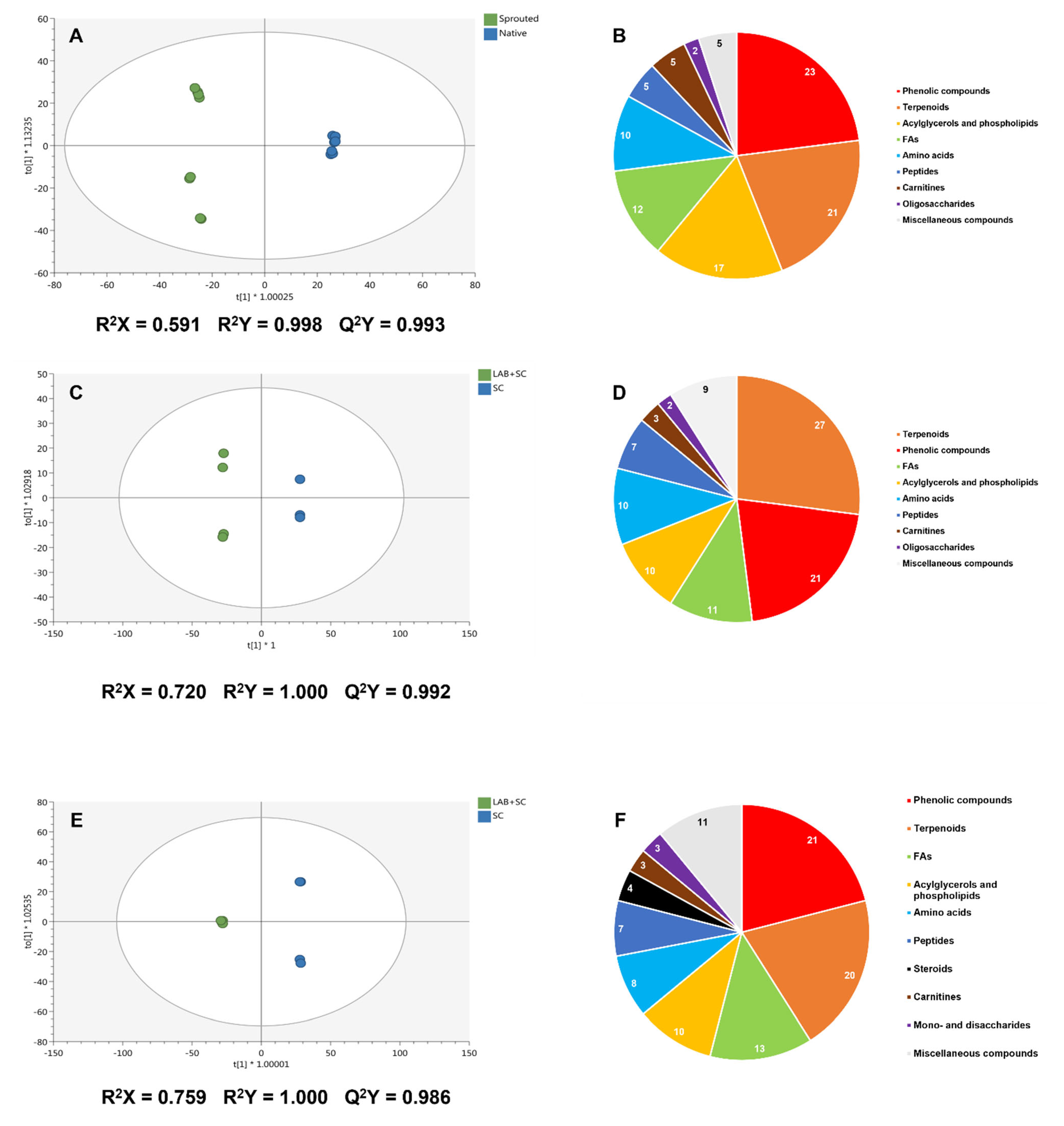
3.6. Untargeted UHPLC/QTOF-MS Metabolomic Profile of Rye Doughs
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Kaur, P.; Sandhu, K.S.; Purewal, S.S.; Kaur, M.; Singh, S.K. Rye: A wonder crop with industrially important macromolecules and health benefits. Food Res. Int. 2021, 150, 110769. [Google Scholar] [CrossRef] [PubMed]
- Jonsson, K.; Andersson, R.; Knudsen, K.E.B.; Hallmans, G.; Hanhineva, K.; Katina, K.; Kolehmainen, M.; Kyrø, C.; Langton, M.; Nordlund, E.; et al. Rye and health—Where do we stand and where do we go? Trends Food Sci. Technol. 2018, 79, 78–87. [Google Scholar] [CrossRef]
- El-Mahis, A.; Baky, M.H.; Farag, M.A. How Does Rye Compare to other Cereals? A Comprehensive Review of its Potential Nutritional Value and Better Opportunities for its Processing as a Food-Based Cereal. Food Rev. Int. 2022, 1–24. [Google Scholar] [CrossRef]
- Naumenko, N.; Potoroko, I.; Kalinina, I.; Fatkullin, R.; Ivanisova, E. The Influence of the Use of Whole Grain Flour from Sprouted Wheat Grain on the Rheological and Microstructural Properties of Dough and Bread. Int. J. Food Sci. 2021, 2021, 7548759. [Google Scholar] [CrossRef]
- Gobbetti, M.; Corsetti, A.; Rossi, J. Interaction between lactic acid bacteria and yeasts in sour-dough using a rheofermentometer. World J. Microbiol. Biotechnol. 1995, 11, 625–630. [Google Scholar] [CrossRef]
- Hadaegh, H.; Ardabili, S.M.S.; Ebrahimi, M.T.; Chamani, M.; Nezhad, R.A. The Impact of Different Lactic Acid Bacteria Sourdoughs on the Quality Characteristics of Toast Bread. J. Food Qual. 2017, 2017, 7825203. [Google Scholar] [CrossRef] [Green Version]
- Fernández-Peláez, J.; Paesani, C.; Gómez, M. Sourdough Technology as a Tool for the Development of Healthier Grain-Based Products: An Update. Agronomy 2020, 10, 1962. [Google Scholar] [CrossRef]
- Muhialdin, B.J.; Saari, N.; Hussin, A.S.M. Review on the Biological Detoxification of Mycotoxins Using Lactic Acid Bacteria to Enhance the Sustainability of Foods Supply. Molecules 2020, 25, 2655. [Google Scholar] [CrossRef]
- Katina, K.; Liukkonen, K.-H.; Kaukovirta-Norja, A.; Adlercreutz, H.; Heinonen, S.-M.; Lampi, A.-M.; Pihlava, J.-M.; Poutanen, K. Fermentation-induced changes in the nutritional value of native or germinated rye. J. Cereal Sci. 2007, 46, 348–355. [Google Scholar] [CrossRef]
- Koistinen, V.M.; Mattila, O.; Katina, K.; Poutanen, K.; Aura, A.-M.; Hanhineva, K. Metabolic profiling of sourdough fermented wheat and rye bread. Sci. Rep. 2018, 8, 5684. [Google Scholar] [CrossRef] [Green Version]
- Srinivas, M.; O’Sullivan, O.; Cotter, P.D.; van Sinderen, D.; Kenny, J.G. The Application of Metagenomics to Study Microbial Communities and Develop Desirable Traits in Fermented Foods. Foods 2022, 11, 3297. [Google Scholar] [CrossRef]
- Bocchi, S.; Rocchetti, G.; Elli, M.; Lucini, L.; Lim, C.-Y.; Morelli, L. The combined effect of fermentation of lactic acid bacteria and in vitro digestion on metabolomic and oligosaccharide profile of oat beverage. Food Res. Int. 2021, 142, 110216. [Google Scholar] [CrossRef]
- Falasconi, I.; Fontana, A.; Patrone, V.; Rebecchi, A.; Garrido, G.D.; Principato, L.; Callegari, M.L.; Spigno, G.; Morelli, L. Genome-Assisted Characterization of Lactobacillus fermentum, Weissella cibaria, and Weissella confusa Strains Isolated from Sorghum as Starters for Sourdough Fermentation. Microorganisms 2020, 8, 1388. [Google Scholar] [CrossRef]
- Cereals and Grains Associations. Approved Methods of Analysis; AACCI; American Association of Cereal Chemists: Saint Paul, MI, USA, 2001. [Google Scholar]
- Campbell, L.; Euston, S.R.; Ahmed, M.A. Effect of addition of thermally modified cowpea protein on sensory acceptability and textural properties of wheat bread and sponge cake. Food Chem. 2016, 194, 1230–1237. [Google Scholar] [CrossRef]
- Comasio, A.; Verce, M.; Van Kerrebroeck, S.; De Vuyst, L. Diverse microbial composition of dourdoughs from different origins. Front. Microbiol. 2020, 11, 1212. [Google Scholar] [CrossRef]
- Heuer, H.; Krsek, M.; Baker, P.; Smalla, K.; Wellington, E.M.H. Analysis of actinomycete communities by specific amplification of genes encoding 16S rRNA and gel-electrophoretic separation in denaturing gradients. Appl. Environ. Microbiol. 1997, 63, 3233–3241. [Google Scholar] [CrossRef] [Green Version]
- Schloss, P.D.; Westcott, S.L.; Ryabin, T.; Hall, J.R.; Hartmann, M.; Hollister, E.B.; Lesniewski, R.A.; Oakley, B.B.; Parks, D.H.; Robinson, C.J.; et al. Introducing mothur: Open-Source, Platform-Independent, Community-Supported Software for Describing and Comparing Microbial Communities. Appl. Environ. Microbiol. 2009, 75, 7537–7541. [Google Scholar] [CrossRef] [Green Version]
- Caporaso, J.G.; Kuczynski, J.; Stombaugh, J.; Bittinger, K.; Bushman, F.D.; Costello, E.K.; Fierer, N.; Gonzalez Peña, A.; Goodrich, J.K.; Gordon, J.I.; et al. QIIME allows analysis of high-throughput community sequencing data. Nat. Methods 2010, 7, 335–336. [Google Scholar] [CrossRef] [Green Version]
- Rocchetti, G.; Zhang, L.; Bocchi, S.; Giuberti, G.; Ak, G.; Elbasan, F.; Yıldıztugay, E.; Ceylan, R.; Picot-Allain, M.C.N.; Mahomoodally, M.F.; et al. The functional potential of nine Allium species related to their untargeted phytochemical characterization, antioxidant capacity and enzyme inhibitory ability. Food Chem. 2022, 368, 130782. [Google Scholar] [CrossRef]
- Rocchetti, G.; Lucini, L.; Gallo, A.; Masoero, F.; Trevisan, M.; Giuberti, G. Untargeted metabolomics reveals differences in chemical fingerprints between PDO and non-PDO Grana Padano cheeses. Food Res. Int. 2018, 113, 407–413. [Google Scholar] [CrossRef]
- García-Pérez, P.; Miras-Moreno, B.; Lucini, L.; Gallego, P.P. The metabolomics reveals intraspecies variability of bioactive compounds in elicited suspension cell cultures of three Bryophyllum species. Ind. Crops Prod. 2021, 163, 113322. [Google Scholar] [CrossRef]
- Dhariwal, A.; Chong, J.; Habib, S.; King, I.L.; Agellon, L.B.; Xia, J. MicrobiomeAnalyst: A web-based tool for comprehensive statistical, visual and meta-analysis of microbiome data. Nucleic Acids Res. 2017, 45, W180–W188. [Google Scholar] [CrossRef] [PubMed]
- Love, M.I.; Huber, W.; Anders, S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014, 15, 550. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Barupal, D.K.; Fiehn, O. Chemical Similarity Enrichment Analysis (ChemRICH) as alternative to biochemical pathway mapping for metabolomic datasets. Sci. Rep. 2017, 7, 14567. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Feng, G. “Rye”, in Bioactive Factors and Processing Technology for Cereal Foods; Wang, J., Sun, B., Tsao, R., Eds.; Springer: Singapore, 2019; pp. 151–170. [Google Scholar] [CrossRef]
- Chegeni, M.; Hamaker, B. Induction of differentiation of small intestinal enterocyte cells by maltooligosaccharides. FASEB J. 2015, 29, 596.14. [Google Scholar] [CrossRef]
- Teleky, B.-E.; Martău, A.G.; Ranga, F.; Chețan, F.; Vodnar, D.C. Exploitation of Lactic Acid Bacteria and Baker’s Yeast as Single or Multiple Starter Cultures of Wheat Flour Dough Enriched with Soy Flour. Biomolecules 2020, 10, 778. [Google Scholar] [CrossRef]
- Lancetti, R.; Sciarini, L.; Pérez, G.T.; Salvucci, E. Technological Performance and Selection of Lactic Acid Bacteria Isolated from Argentinian Grains as Starters for Wheat Sourdough. Curr. Microbiol. 2021, 78, 255–264. [Google Scholar] [CrossRef]
- Montemurro, M.; Pontonio, E.; Gobbetti, M.; Rizzello, C.G. Investigation of the nutritional, functional and technological effects of the sourdough fermentation of sprouted flours. Int. J. Food Microbiol. 2019, 302, 47–58. [Google Scholar] [CrossRef]
- Gobbetti, M.; Rizzello, C.G.; Di Cagno, R.; De Angelis, M. How the sourdough may affect the functional features of leavened baked goods. Food Microbiol. 2014, 37, 30–40. [Google Scholar] [CrossRef]
- Van der Meulen, R.; Scheirlinck, I.; Van Schoor, A.; Huys, G.; Vancanneyt, M.; Vandamme, P.; De Vuyst, L. Population Dynamics and Metabolite Target Analysis of Lactic Acid Bacteria during Laboratory Fermentations of Wheat and Spelt Sourdoughs. Appl. Environ. Microbiol. 2007, 73, 4741–4750. [Google Scholar] [CrossRef] [Green Version]
- Ercolini, D.; Pontonio, E.; De Filippis, F.; Minervini, F.; La Storia, A.; Gobbetti, M.; Di Cagno, R. Microbial Ecology Dynamics during Rye and Wheat Sourdough Preparation. Appl. Environ. Microbiol. 2013, 79, 7827–7836. [Google Scholar] [CrossRef] [Green Version]
- Ispirli, H.; Demirbaş, F.; Yüzer, M.O.; Dertli, E. Identification of Lactic Acid Bacteria from Spontaneous Rye Sourdough and Determination of Their Functional Characteristics. Food Biotechnol. 2018, 32, 222–235. [Google Scholar] [CrossRef]
- Boreczek, J.; Litwinek, D.; Urban, J.; Izak, D.; Buksa, K.; Gawor, J.; Gromadka, R.; Bardowski, J.K.; Kowalczyk, M. Bacterial community dynamics in spontaneous sourdoughs made from wheat, spelt, and rye wholemeal flour. Microbiologyopen 2020, 9, e1009. [Google Scholar] [CrossRef] [PubMed]
- De Vero, L.; Iosca, G.; Gullo, M.; Pulvirenti, A. Functional and Healthy Features of Conventional and Non-Conventional Sourdoughs. Appl. Sci. 2021, 11, 3694. [Google Scholar] [CrossRef]
- Di Cagno, R.; Pontonio, E.; Buchin, S.; De Angelis, M.; Lattanzi, A.; Valerio, F.; Gobbetti, M.; Calasso, M. Diversity of the Lactic Acid Bacterium and Yeast Microbiota in the Switch from Firm- to Liquid-Sourdough Fermentation. Appl. Environ. Microbiol. 2014, 80, 3161–3172. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zheng, J.; Wittouck, S.; Salvetti, E.; Franz, C.M.A.P.; Harris, H.M.B.; Mattarelli, P.; O’Toole, P.W.; Pot, B.; Vandamme, P.; Walter, J.; et al. A taxonomic note on the genus Lactobacillus: Description of 23 novel genera, emended description of the genus Lactobacillus Beijerinck 1901, and union of Lactobacillaceae and Leuconostocaceae. Int. J. Syst. Evol. Microbiol. 2020, 70, 2782–2858. [Google Scholar] [CrossRef]
- Vrancken, G.; Rimaux, T.; Weckx, S.; Leroy, F.; De Vuyst, L. Influence of Temperature and Backslopping Time on the Microbiota of a Type I Propagated Laboratory Wheat Sourdough Fermentation. Appl. Environ. Microbiol. 2011, 77, 2716–2726. [Google Scholar] [CrossRef] [Green Version]
- Gänzle, M.G.; Follador, R. Metabolism of Oligosaccharides and Starch in Lactobacilli: A Review. Front. Microbiol. 2012, 3, 340. [Google Scholar] [CrossRef] [Green Version]
- Dziki, D. Rye Flour and Rye Bran: New Perspectives for Use. Processes 2022, 10, 293. [Google Scholar] [CrossRef]
- García-Pérez, P.; Gallego, P.P. Plant Phenolics as Dietary Antioxidants: Insights on Their Biosynthesis, Sources, Health-Promoting Effects, Sustainable Production, and Effects on Lipid Oxidation. In Lipid Oxidation in Food and Biological Systems; Bravo-Diaz, C., Ed.; Springer: Cham, Switzerland, 2022. [Google Scholar] [CrossRef]
- Corso, M.; Perreau, F.; Rajjou, L.; Ben Malek, R.; Lepiniec, L.; Mouille, G. Specialized metabolites in seeds. Adv. Bot. Res. 2021, 98, 35–70. [Google Scholar] [CrossRef]
- Litwinek, D.; Boreczek, J.; Gambuś, H.; Buksa, K.; Berski, W.; Kowalczyk, M. Developing lactic acid bacteria starter cultures for wholemeal rye flour bread with improved functionality, nutritional value, taste, appearance and safety. PLoS ONE 2022, 17, e0261677. [Google Scholar] [CrossRef]
- Gänzle, M.G. Enzymatic and bacterial conversions during sourdough fermentation. Food Microbiol. 2014, 37, 2–10. [Google Scholar] [CrossRef]
- De Angelis, M.; Mariotti, L.; Rossi, J.; Servili, M.; Fox, P.F.; Rollán, G.; Gobbetti, M. Arginine Catabolism by Sourdough Lactic Acid Bacteria: Purification and Characterization of the Arginine Deiminase Pathway Enzymes from Lactobacillus sanfranciscensis CB1. Appl. Environ. Microbiol. 2002, 68, 6193–6201. [Google Scholar] [CrossRef] [Green Version]
- Zhao, C.J.; Gänzle, M.G. Synthesis of Taste-Active γ-Glutamyl Dipeptides during Sourdough Fermentation by Lactobacillus reuteri. J. Agric. Food Chem. 2016, 64, 7561–7568. [Google Scholar] [CrossRef]
| Parameter | Rye flour | Starter | p-Value | ||||
|---|---|---|---|---|---|---|---|
| Control | SC | LAB + SC | Starter | Germination | Starter × Germination | ||
| pH | Native | 4.19 ± 0.11 a | 4.57 ± 0.36 b | 3.97 ± 0.10 a | 0.0005 | 0.005 | 0.06 |
| Sprouted | 4.57 ± 0.10 a | 5.33 ± 0.01 b | 4.03 ± 0.01 a | ||||
| TTA (%) | Native | 0.39 ± 0.01 a | 0.42 ± 0.03 a | 0.82 ± 0.06 bA | <0.0001 | <0.0001 | <0.0001 |
| Sprouted | 0.50 ± 0.02 a | 0.56 ± 0.08 a | 2.08 ± 0.05 bB | ||||
| aw | Native | 1.02 ± 0.002 a | 0.94 ± 0.02 b | 0.95 ± 0.01 b | 0.0001 | 0.044 | 0.026 |
| Sprouted | 0.99 ± 0.01 a | 0.94 ± 0.01 b | 0.96 ± 0.01 ab | ||||
| Dough rise (%) | Native | 70 ± 4.24 a | 143 ± 4.95 b | 220 ± 18.38 c | <0.0001 | 0.22 | 0.06 |
| Sprouted | 90 ± 4.24 a | 130 ± 4.24 ab | 180 ± 28.30 b | ||||
| Total LAB (log CFU/g) | Native | 9.16 ± 0.04 a | 6.70 ± 0.09 bA | 10.32 ± 0.38 c | <0.0001 | 0.008 | 0.09 |
| Sprouted | 8.94 ± 0.01 a | 5.29 ± 0.01 bB | 9.53 ± 0.12 c | ||||
| Total yeast (log CFU/g) | Native | 6.27 ± 0.01 a | 8.33 ± 0.10 b | 8.24 ± 0.17 b | <0.0001 | 0.56 | 0.05 |
| Sprouted | 5.47 ± 0.66 a | 8.73 ± 0.03 b | 8.34 ± 0.01 b |
| Samples * | OTUs | Chao1 | Shannon | Gini-Simpson | Good’s Coverage |
|---|---|---|---|---|---|
| nDLAB + SC1 | 217 | 386.5 | 0.733 | 0.132 | 0.991 |
| nDLAB + SC2 | 348 | 485.8 | 1.486 | 0.296 | 0.987 |
| nDSC1 | 194 | 230.1 | 2.414 | 0.610 | 0.993 |
| nDSC2 | 236 | 271.3 | 3.108 | 0.696 | 0.984 |
| nControl1 | 58 | 100.0 | 0.093 | 0.013 | 0.998 |
| nControl2 | 415 | 449.0 | 3.995 | 0.751 | 0.993 |
| sDLAB + SC1 | 466 | 602.5 | 2.579 | 0.595 | 0.981 |
| sDLAB + SC2 | 454 | 565.8 | 2.624 | 0.600 | 0.981 |
| sDSC1 | 327 | 438.0 | 1.712 | 0.278 | 0.988 |
| sDSC2 | 417 | 533.4 | 2.587 | 0.444 | 0.982 |
| sControl1 | 227 | 312.1 | 0.989 | 0.173 | 0.993 |
| sControl2 | 305 | 430.4 | 1.199 | 0.205 | 0.989 |
| nF1 | 284 | 300.8 | 3.211 | 0.740 | 0.998 |
| nF2 | 504 | 541.1 | 4.947 | 0.884 | 0.996 |
| sF1 | 596 | 737.5 | 5.014 | 0.841 | 0.974 |
| sF2 | 609 | 799.0 | 5.028 | 0.794 | 0.967 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Mancino, W.; Carnevali, P.; Terzi, V.; Pérez, P.G.; Zhang, L.; Giuberti, G.; Morelli, L.; Patrone, V.; Lucini, L. Hierarchical Effects of Lactic Fermentation and Grain Germination on the Microbial and Metabolomic Profile of Rye Doughs. Foods 2023, 12, 998. https://doi.org/10.3390/foods12050998
Mancino W, Carnevali P, Terzi V, Pérez PG, Zhang L, Giuberti G, Morelli L, Patrone V, Lucini L. Hierarchical Effects of Lactic Fermentation and Grain Germination on the Microbial and Metabolomic Profile of Rye Doughs. Foods. 2023; 12(5):998. https://doi.org/10.3390/foods12050998
Chicago/Turabian StyleMancino, Walter, Paola Carnevali, Valeria Terzi, Pascual García Pérez, Leilei Zhang, Gianluca Giuberti, Lorenzo Morelli, Vania Patrone, and Luigi Lucini. 2023. "Hierarchical Effects of Lactic Fermentation and Grain Germination on the Microbial and Metabolomic Profile of Rye Doughs" Foods 12, no. 5: 998. https://doi.org/10.3390/foods12050998










