Effect of High Hydrostatic Pressure Processing on the Microbiological Quality and Bacterial Diversity of Sous-Vide-Cooked Cod
Abstract
1. Introduction
2. Materials and Methods
2.1. Sample Preparation and Treatments
2.2. Determination of Viable Cell Concentrations
2.3. DNA Isolation and Quantification
2.4. DNA Sequencing and Analysis
2.5. Statistical Analysis
3. Results
3.1. Effect of Treatments on Microbial Loads
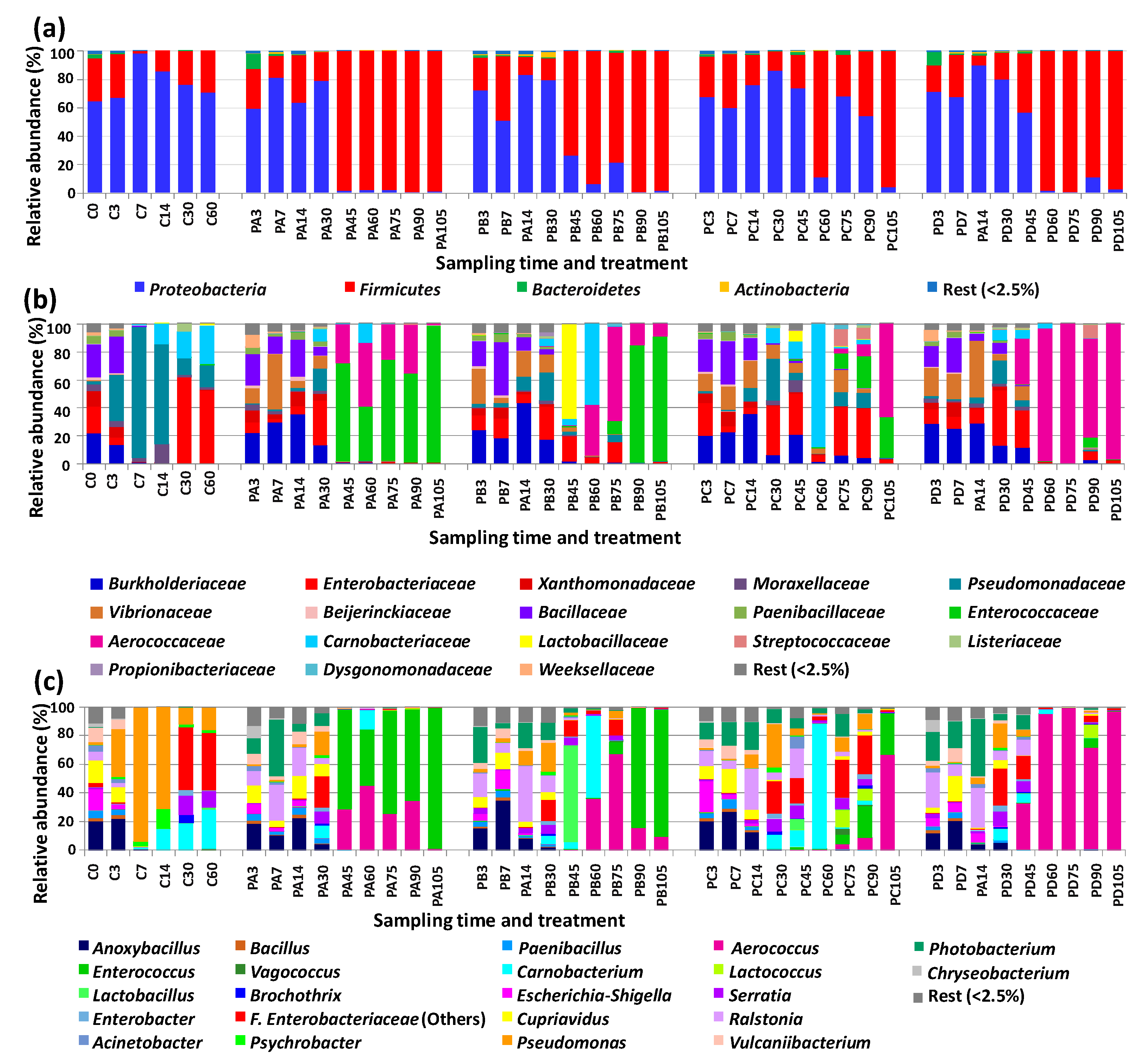
3.2. Changes in Bacterial Diversity
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Zavadlav, S.; Blažić, M.; Van de Velde, F.; Vignatti, C.; Fenoglio, C.; Piagentini, A.M.; Pirovani, M.E.; Perotti, C.M.; Bursać Kovačević, D.; Putnik, P. Sous-vide as a technique for preparing healthy and high-quality vegetable and seafood products. Foods 2020, 9, 1537. [Google Scholar] [CrossRef]
- Coşansu, S.; Mol, S.; Haskaraca, G. Sous-vide cooking: Effects on seafood quality and combination with other hurdles. Int. J. Gastron. Food Sci. 2022, 29, 100586. [Google Scholar] [CrossRef]
- Cui, Z.; Zhang, N.; Lou, W.; Manoli, T. Application of sous vide cooking to aquatic food products: A review. Food Sci. Technol. 2022, 42, e108021. [Google Scholar] [CrossRef]
- Belitz, H.-D.; Grosch, W.; Schieberle, P. Food Chemistry, 3rd ed.; Springer: Berlin/Heidelberg, Germany, 2004. [Google Scholar]
- Picouet, P.A.; Cofan-Carbo, S.; Vilaseca, H.; Ballbè, L.C.; Castells, P. Stability of sous-vide cooked salmon loins processed by high pressure. Innovat. Food Sci. Emerg. Technol. 2011, 12, 26–31. [Google Scholar] [CrossRef]
- Baldwin, D.E. Sous vide cooking: A review. Int. J. Gastron. Food Sci. 2012, 1, 15–30. [Google Scholar] [CrossRef]
- Huang, H.W.; Lung, H.M.; Yang, B.B.; Wang, C. Responses of microorganisms to high hydrostatic pressure processing. Food Control 2014, 40, 250–259. [Google Scholar] [CrossRef]
- de Oliveira, F.A.; Otavio, C.N.; Dos Santos, M.R.; dos Santos, L.M.R.; Ferreira, E.H.R.; Rosenthal, A. Effect of high pressure on fish meat quality. Trends Food Sci. Technol. 2017, 66, 1–19. [Google Scholar] [CrossRef]
- Puértolas, E.; Lavilla, L. HPP in seafood products: Impact on quality and applications. In Present and Future of High Pressure Processing; Elsevier Inc.: Amsterdam, The Netherlands, 2020; pp. 201–219. [Google Scholar]
- Abel, N.; Rotabakk, B.T.; Lerfall, J. Mild processing of seafood—A review. Compr. Rev. Food Sci. Food Saf. 2022, 21, 340–370. [Google Scholar] [CrossRef]
- Angsupanich, K.; Ledward, D.A. High pressure treatment effects on cod (Gadus morhua) muscle. Food Chemitry 1998, 63, 39–50. [Google Scholar] [CrossRef]
- Angsupanich, K.; Edde, M.; Ledward, D.A. Effects of high pressure on the myofibrillar proteins of cod and turkey muscle. J. Agric. Food Chem. 1999, 47, 92–99. [Google Scholar] [CrossRef]
- Matser, A.M.; Stegeman, D.; Kals, J.; Bartels, P.V. Effects of high pressure on colour and texture of fish. High Press. Res. 2000, 19, 109–115. [Google Scholar] [CrossRef]
- Rode, T.M.; Hovda, M.B. High pressure processing extend the shelf life of fresh salmon, cod and mackerel. Food Control 2016, 70, 242–248. [Google Scholar] [CrossRef]
- Humaid, S.; Nayyar, D.; Bolton, J.; Skonberg, D.I. Physicochemical properties and consumer acceptance of high-pressure processed, sous vide-cooked lobster tails. J. Food Sci. 2019, 84, 3454–3462. [Google Scholar] [CrossRef]
- Humaid, S.; Nayyar, D.; Bolton, J.; Perkins, B.; Skonberg, D.I. Refrigerated shelf-life evaluation of high pressure processed, raw and sous vide cooked lobster. High Press. Res. 2020, 40, 444–463. [Google Scholar] [CrossRef]
- Ahmad, I.; Mark, P.; Traynor, M.P. Impact of high-pressure processing and sous vide cooking on the physicochemical, sensorial, and textural properties of fresh whiteleg shrimp (Litopenaeus setiferus). J. Aquat. Food Prod. Technol. 2022, 31, 508–524. [Google Scholar] [CrossRef]
- Espinosa, M.C.; Diaz, P.; Linares, M.B.; Teruel, M.R.; Garrido, M.D. Quality characteristics of sous vide ready to eat seabream processed by high pressure. LWT-Food Sci. Technol. 2015, 64, 657–662. [Google Scholar] [CrossRef]
- Zhou, M.; Ling, Y.; Chen, F.; Wang, C.; Qiao, Y.; Xiong, G.; Wang, L.; Wu, W.; Shi, L.; Ding, A. Effect of high hydrostatic pressure combined with sous-vide treatment on the quality of largemouth bass during storage. Foods 2022, 11, 1931. [Google Scholar] [CrossRef] [PubMed]
- Elizaquível, P.; Sánchez, G.; Aznar, R. Quantitative detection of viable foodborne E. coli O157:H7, Listeria monocytogenes and Salmonella in fresh-cut vegetables combining propidium monoazide and real-time PCR. Food Control 2012, 25, 704–708. [Google Scholar] [CrossRef]
- Nocker, A.; Sossa-Fernandez, P.; Burr, M.D.; Camper, A.K. Use of propidium monoazide for live/dead distinction in microbial ecology. Appl. Environ. Microbiol. 2007, 73, 5111–5117. [Google Scholar] [CrossRef] [PubMed]
- Klindworth, A.; Pruesse, E.; Schweer, T.; Peplies, J.; Quast, C.; Horn, M.; Glöckner, F.O. Evaluation of general 16S ribosomal RNA gene PCR primers for classical and next-generation sequencing-based diversity studies. Nucleic Acids Res. 2013, 41, e1. [Google Scholar] [CrossRef]
- Rodríguez-López, J.; Grande, M.J.; Pérez-Pulido, R.; Gálvez, A.; Lucas, R. Impact of high-hydrostatic pressure treat-ments applied singly or in combination with moderate heat on the microbial load, antimicrobial resistance, and bacterial diversity of guacamole. Microorganisms 2020, 8, 909. [Google Scholar] [CrossRef]
- FDA Circular No.2022-012. Guidelines on the Microbiological Requirements and Assessment of Certain Prepackaged Processed Food Products Repealing FDA Circular No. 2013-010 Entitled “Revised Guidelines for the Assessment of Microbiological Quality of Processed Foods”. Available online: https://www.fda.gov.ph/wp-content/uploads/2022/12/FDA-Circular-No.2022-12-2.pdf (accessed on 15 February 2023).
- Bondoc, I. European regulation in the veterinary sanitary and food safety area, a component of the European policies on the safety of food products and the protection of consumer Interests: A 2007 retrospective. Part one: The role of European institutions in laying down and passing laws specific to the veterinary sanitary and food safety area. Universul Jurid. 2016, 12–15. [Google Scholar]
- Bondoc, I. European Regulation in the Veterinary Sanitary and Food Safety Area, a Component of the European Policies on the Safety of Food Products and the Protection of Consumer Interests: A 2007 Retrospective. Part two: Regulations. Universul Jurid. 2016, 16–19. [Google Scholar]
- Rybka-Rodgers, S. Improvement of food safety design of cook-chill foods. Food Res. Int. 2001, 34, 449–455. [Google Scholar] [CrossRef]
- Sarker, M.R.; Akhtar, S.; Torres, J.A.; Paredes-Sabja, D. High hydrostatic pressure-induced inactivation of bacterial spores. Crit. Rev. Microbiol. 2015, 41, 18–26. [Google Scholar] [CrossRef]
- Lenz, C.A.; Reineke, K.; Knorr, D.; Vogel, R.F. High pressure thermal inactivation of Clostridium botulinum type E endospores—Kinetic modeling and mechanistic insights. Front Microbiol. 2015, 6, 652. [Google Scholar] [CrossRef] [PubMed]
- Maier, M.B.; Schweiger, T.; Lenz, C.A.; Vogel, R.F. Inactivation of non-proteolytic Clostridium botulinum type E in low-acid foods and phosphate buffer by heat and pressure. PLoS ONE 2018, 13, e0200102. [Google Scholar] [CrossRef] [PubMed]
- Narayan, V.V.; Hatha, M.A.; Morgan, H.W.; Rao, D. Isolation and characterization of aerobic thermophilic bacteria from the Savusavu hot springs in Fiji. Microbes Environ. 2008, 23, 350–352. [Google Scholar] [CrossRef] [PubMed]
- Naya-Català, F.; do Vale Pereira, G.; Piazzon, M.C.; Fernandes, A.M.; Calduch-Giner, J.A.; Sitjà-Bobadilla, A.; Conceição, L.E.C.; Pérez-Sánchez, J. Cross-talk between intestinal microbiota and host gene expression in gilthead sea bream (Sparus aurata) juveniles: Insights in fish feeds for increased circularity and resource utilization. Front Physiol. 2021, 12, 748265. [Google Scholar] [CrossRef]
- Malone, A.S.; Chung, Y.K.; Yousef, A.E. Genes of Escherichia coli O157:H7 that are involved in high-pressure resistance. Appl. Environ. Microbiol. 2006, 72, 2661–2671. [Google Scholar] [CrossRef]
- Li, H.; Mercer, R.; Behr, J.; Heinzlmeir, S.; McMullen, L.M.; Vogel, R.F.; Gänzle, M.G. Heat and pressure resistance in Escherichia coli relates to protein folding and aggregation. Front. Microbiol. 2020, 11, 111. [Google Scholar] [CrossRef]
- Gram, L.; Dalgaard, P. Fish spoilage bacteria—Problems and solutions. Curr. Opin. Biotechnol. 2002, 13, 262–266. [Google Scholar] [CrossRef]
- Odeyemi, O.A.; Burke, C.M.; Bolch, C.C.J.; Stanley, R. Seafood spoilage microbiota and associated volatile organic compounds at different storage temperatures and packaging conditions. Int. J. Food Microbiol. 2018, 280, 87–99. [Google Scholar] [CrossRef]
- Sørensen, J.S.; Ørnfeld-Jensen, O.; Bøknæs, N.; Mejlholm, O.; Jessen, F.; Dalgaard, P. Thawed and chilled Atlantic cod (Gadus morhua L.) from Greenland -Options for improved distribution. LWT-Food Sci. Technol. 2020, 131, 109473. [Google Scholar] [CrossRef]
- Joffraud, J.J.; Leroi, F.; Roy, C.; Berdague, J.L. Characterisation of volatile compounds produced by bacteria isolated from the spoilage flora of cold-smoked salmon. Int. J. Food Microbiol. 2001, 66, 175–184. [Google Scholar] [CrossRef] [PubMed]
- Fidan, H.; Esatbeyoglu, T.; Simat, V.; Trif, M.; Tabanelli, G.; Kostka, T.; Montanari, C.; Ibrahim, S.A.; Özogul, F. Recent developments of lactic acid bacteria and their metabolites on foodborne pathogens and spoilage bacteria: Facts and gaps. Food Biosci. 2022, 47, 101741. [Google Scholar] [CrossRef]
- Franzetti, L.; Scarpellini, M.; Mora, D.; Galli, A. Carnobacterium spp. in seafood packaged in modified atmosphere. Ann. Microbiol. 2003, 53, 189–198. [Google Scholar]
- Françoise, L. Occurrence and role of lactic acid bacteria in seafood products. Food Microbiol. 2010, 27, 698–709. [Google Scholar] [CrossRef]
- Silbande, A.; Adenet, S.; Smith-Ravin, J.; Joffraud, J.J.; Rochefort, K.; Leroi, F. Quality assessment of ice-stored tropical yellowfin tuna (Thunnus albacares) and influence of vacuum and modified atmosphere packaging. Food Microbiol. 2016, 60, 62–72. [Google Scholar] [CrossRef]
- Rodrigues, M.J.; Ho, P.; Lopez-Caballero, M.E.; Vaz-Pires, P.; Nunes, M.L. Characterization and identification of microflora from soaked cod and respective salted raw materials. Food Microbiol. 2003, 20, 471–481. [Google Scholar] [CrossRef]
- Wiernasz, N.; Cornet, J.; Cardinal, M.; Pilet, M.F.; Passerini, D.; Leroi, F. Lactic acid bacteria selection for biopreservation as a part of hurdle technology approach applied on seafood. Front. Mar. Sci. 2017, 4, 119. [Google Scholar] [CrossRef]
- Lv, X.; Li, Y.; Cui, T.; Sun, M.; Bai, F.; Li, X.; Li, J.; Yi, S. Bacterial community succession and volatile compound changes during fermentation of shrimp paste from Chinese Jinzhou region. LWT-Food Sci Technol. 2020, 122, 108998. [Google Scholar] [CrossRef]
- Ke, X.; Lu, M.; Ye, X.; Gao, F.; Zhu, H.; Huang, Z. Recovery and pathogenicity analysis of Aerococcus viridans isolated from tilapia (Orecohromis niloticus) cultured in southwest of China. Aquaculture 2012, 342–343, 18–23. [Google Scholar] [CrossRef]

| Time (Days) | Control (TSA) | Control (Saline TSA) | Control (MacConkey) | HP (500 MPa, 8 min; TSA) | HP (500 MPa, 8 min; Saline TSA) | HP (500 MPa, 8 min; MacConkey) |
|---|---|---|---|---|---|---|
| 0 | <1.30 | <1.30 | <1.30 | <1.30 | <1.30 | <1.30 |
| 3 | 1.70 ± 1.41 a | 1.70 ± 1.41 a | <1.30 | <1.30 | <1.30 | <1.30 |
| 7 | 1.70 ± 1.41 a | 1.70 ± 1.41 a | <1.30 | <1.30 | <1.30 | <1.30 |
| 14 | 1.70 ± 1.41 a | 2.18 ± 0.21 a | <1.30 | <1.30 | <1.30 | <1.30 |
| 30 | 3.70 ± 0.03 a | 3.78 ± 0.15 a | <1.30 | <1.30 | <1.30 | <1.30 |
| 45 | 5.63 ± 0.06 a | 5.69 ± 0.02 a | <1.30 | <1.30 | <1.30 | <1.30 |
| 60 | 7.33 ± 0.03 a | 7.40 ± 0.01 a | <1.30 | 4.92 ± 0.03 b | 4.98 ± 0.01 b | <1.30 |
| Time (Days) | Control (TSA) | Control (MacConkey) | Treatment A | Treatment B | Treatment C | Treatment D |
|---|---|---|---|---|---|---|
| 0 | 2.30 ± 0.29 a | 1.90 ± 0.30 a | <1.30 | <1.30 | <1.30 | <1.30 |
| 3 | 3.13 ± 0.22 a | 2.86 ± 0.24 a | 2.13 ± 0.30 b | 1.86 ± 0.45 b | 2.02 ± 0.41 b | 1.59 ± 0.35 b,c,d |
| 7 | 5.45 ± 0.06 | 3.70 ± 0.13 b | <1.30 | <1.30 | <1.30 | <1.30 |
| 14 | 6.45 ± 0.06 | 4.70 ± 0.14 b | <1.30 | <1.30 | <1.30 | <1.30 |
| 30 | 8.73 ± 0.03 | 7.71 ± 0.05 b | <1.30 | <1.30 | <1.30 | <1.30 |
| 45 | 9.59 ± 0.15 a | 9.21 ± 0.43 a | 4.99 ± 0.13 b | 2.38 ± 0.14 b,c,d | 3.04 ± 0.09 b,c | 1.85 ± 0.39 b,c,d,e |
| 60 | 10.51 ± 0.44 | 9.68 ± 0.15 b | 6.70 ± 0.04 b | 4.79 ± 0.06 b,c | 5.17 ± 0.05 b,c | 4.26 ± 0.10 b,c,d,e |
| 75 | 11.03 ± 0.38 | 9.56 ± 0.07 b | 8.47 ± 0.10 b | 5.67 ± 0.06 b,c,d | 6.54 ± 0.05 b,c | 5.37 ± 0.07 b,c,d |
| 90 | 11.64 ± 0.09 | 10.45 ± 0.39 b | 8.77 ± 0.11 b | 5.67 ± 0.07 b,c,d | 7.07 ± 0.14 b,c | 5.10 ± 0.17 b,c,d,e |
| 105 | 9.79 ± 0.48 | 8.84 ± 0.10 b | 6.55 ± 0.05 b | 5.27 ± 0.15 b,c | 5.64 ± 0.23 b,c | 4.58 ± 0.20 b,c,d,e |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Pérez Alcalá, D.; Grande Burgos, M.J.; Rodríguez López, J.; Lucas, R.; Gálvez, A.; Pérez Pulido, R. Effect of High Hydrostatic Pressure Processing on the Microbiological Quality and Bacterial Diversity of Sous-Vide-Cooked Cod. Foods 2023, 12, 1206. https://doi.org/10.3390/foods12061206
Pérez Alcalá D, Grande Burgos MJ, Rodríguez López J, Lucas R, Gálvez A, Pérez Pulido R. Effect of High Hydrostatic Pressure Processing on the Microbiological Quality and Bacterial Diversity of Sous-Vide-Cooked Cod. Foods. 2023; 12(6):1206. https://doi.org/10.3390/foods12061206
Chicago/Turabian StylePérez Alcalá, Diego, María José Grande Burgos, Javier Rodríguez López, Rosario Lucas, Antonio Gálvez, and Rubén Pérez Pulido. 2023. "Effect of High Hydrostatic Pressure Processing on the Microbiological Quality and Bacterial Diversity of Sous-Vide-Cooked Cod" Foods 12, no. 6: 1206. https://doi.org/10.3390/foods12061206
APA StylePérez Alcalá, D., Grande Burgos, M. J., Rodríguez López, J., Lucas, R., Gálvez, A., & Pérez Pulido, R. (2023). Effect of High Hydrostatic Pressure Processing on the Microbiological Quality and Bacterial Diversity of Sous-Vide-Cooked Cod. Foods, 12(6), 1206. https://doi.org/10.3390/foods12061206







